Non-Coding RNAs of Mitochondrial Origin: Roles in Cell Division and Implications in Cancer
Abstract
1. Introduction
2. The Mitochondrion
2.1. Role of Mitochondria in Human Health
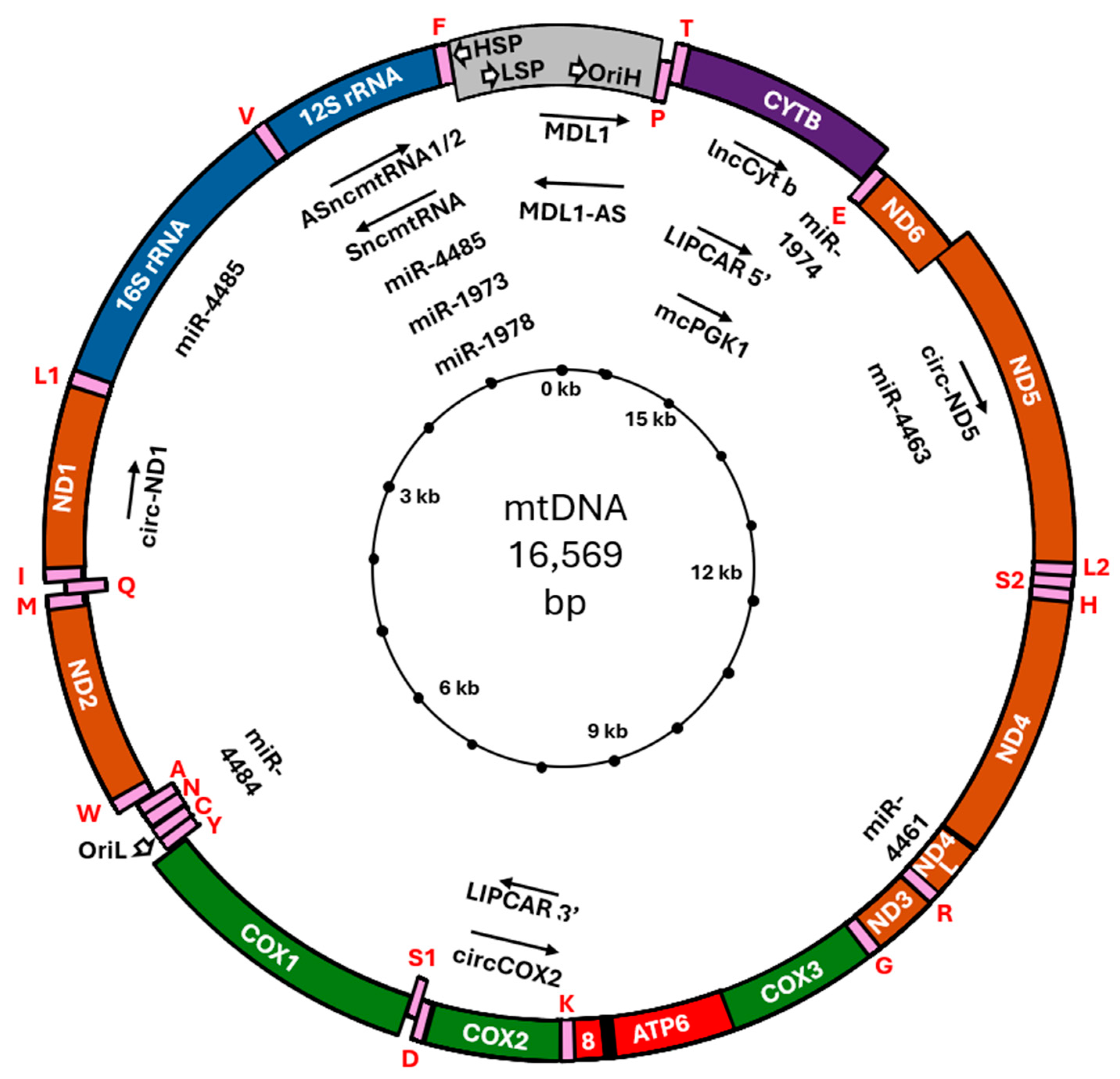
2.2. Discovery and Characterization of Mitochondrial DNA
3. Mitochondria and Cancer
4. Non-Coding RNA and Cancer
5. Mitochondrial lncRNA
5.1. SncmtRNA, ASncmtRNA-1, ASncmtRNA-2, miR-4485 and miR-1973
5.2. LIPCAR
5.3. lncCyt b
5.4. MDL1 and MDL1AS
5.5. Circ-COX2
5.6. mcPGK1
5.7. Circ-ND1 and Circ-ND5
6. Mitochondrial sncRNA
6.1. A Brief Overview of Validated vs. Non-Validated mtDNA-Encoded miRNAs
6.2. miR-1974
6.3. miR-1978
6.4. miR-4461
6.5. miR-4463
6.6. miR-4484
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mayevsky, A. Introduction. In Mitochondrial Function In Vivo Evaluated by NADH Fluorescence; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–6. [Google Scholar] [CrossRef]
- Ernster, L.; Schatz, G. Mitochondria: A Historical Review. J. Cell Biol. 1981, 91, 227s–255s. [Google Scholar] [CrossRef]
- Michaelis, L. Die Vitale Färbung, Eine Darstellungsmethode Der Zellgranula. Arch. Für Mikrosk. Anat. 1899, 55, 558–575. [Google Scholar] [CrossRef]
- Lazarow, A.; Cooperstein, S.J. Studies on the Mechanism of Janus Green B Staining of Mitochondria. I. Review of the Literature. Exp. Cell Res. 1953, 5, 56–69. [Google Scholar] [CrossRef]
- Palade, G.E. The Fine Structure of Mitochondria. Anat. Rec. 1952, 114, 427–451. [Google Scholar] [CrossRef]
- Warburg, O. Über Die Empfindlichkeit Der Sauerstoffatmung Gegenüber Indifferenten Narkotika—Nebst Einer Bemerkung Über Die Sauerstoffatmenden Leberzellengranula. Pflug. Arch. Gesamte Physiol. Menschen Tiere 1914, 158, 19–28. [Google Scholar] [CrossRef]
- Siekevitz, P. Powerhouse of the Cell. Sci. Am. 1957, 197, 131–144. [Google Scholar] [CrossRef]
- Yan, C.; Duanmu, X.; Zeng, L.; Liu, B.; Song, Z. Mitochondrial DNA: Distribution, Mutations, and Elimination. Cells 2019, 8, 379. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Metabolic Regulation of Mitochondrial Dynamics. J. Cell Biol. 2016, 212, 379. [Google Scholar] [CrossRef]
- Vakifahmetoglu-Norberg, H.; Ouchida, A.T.; Norberg, E. The Role of Mitochondria in Metabolism and Cell Death. Biochem. Biophys. Res. Commun. 2017, 482, 426–431. [Google Scholar] [CrossRef]
- Abate, M.; Festa, A.; Falco, M.; Lombardi, A.; Luce, A.; Grimaldi, A.; Zappavigna, S.; Sperlongano, P.; Irace, C.; Caraglia, M.; et al. Mitochondria as Playmakers of Apoptosis, Autophagy and Senescence. Semin. Cell Dev. Biol. 2020, 98, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Holt, I.J.; Harding, A.E.; Morgan-Hughes, J.A. Deletions of Muscle Mitochondrial DNA in Patients with Mitochondrial Myopathies. Nature 1988, 331, 717–719. [Google Scholar] [CrossRef]
- Zeviani, M.; Moraes, C.T.; DiMauro, S.; Nakase, H.; Bonilla, E.; Schon, E.A.; Rowland, L.P. Deletions of Mitochondrial DNA in Kearns-Sayre Syndrome. Neurology 1988, 38, 1339–1346. [Google Scholar] [CrossRef]
- Wallace, D.C.; Singh, G.; Lott, M.T.; Hodge, J.A.; Schurr, T.G.; Lezza, A.M.S.; Elsas, L.J.; Nikoskelainen, E.K. Mitochondrial DNA Mutation Associated with Leber’s Hereditary Optic Neuropathy. Science 1988, 242, 1427–1430. [Google Scholar] [CrossRef]
- Kadowaki, T.; Kadowaki, H.; Mori, Y.; Tobe, K.; Sakuta, R.; Suzuki, Y.; Tanabe, Y.; Sakura, H.; Awata, T.; Goto, Y.; et al. A Subtype of Diabetes Mellitus Associated with a Mutation of Mitochondrial DNA. N. Engl. J. Med. 1994, 330, 962–968. [Google Scholar] [CrossRef]
- Ballinger, S.W.; Shoffner, J.M.; Hedaya, E.V.; Trounce, I.; Polak, M.A.; Koontz, D.A.; Wallace, D.C. Maternally Transmitted Diabetes and Deafness Associated with a 10.4 Kb Mitochondrial DNA Deletion. Nat. Genet. 1992, 1, 11–15. [Google Scholar] [CrossRef]
- Zambrano, K.; Barba, D.; Castillo, K.; Noboa, L.; Argueta-Zamora, D.; Robayo, P.; Arizaga, E.; Caicedo, A.; Gavilanes, A.W.D. Fighting Parkinson’s Disease: The Return of the Mitochondria. Mitochondrion 2022, 64, 34–44. [Google Scholar] [CrossRef]
- Bhatia, S.; Rawal, R.; Sharma, P.; Singh, T.; Singh, M.; Singh, V. Mitochondrial Dysfunction in Alzheimer’s Disease: Opportunities for Drug Development. Curr. Neuropharmacol. 2022, 20, 675. [Google Scholar] [CrossRef]
- Kim, J.E.; Park, H.; Choi, S.H.; Kong, M.J.; Kang, T.C. TRPC6-Mediated ERK1/2 Activation Increases Dentate Granule Cell Resistance to Status Epilepticus Via Regulating Lon Protease-1 Expression and Mitochondrial Dynamics. Cells 2019, 8, 1376. [Google Scholar] [CrossRef]
- Kho, A.R.; Choi, B.Y.; Lee, S.H.; Hong, D.K.; Jeong, J.H.; Kang, B.S.; Kang, D.H.; Park, K.H.; Park, J.B.; Suh, S.W. The Effects of Sodium Dichloroacetate on Mitochondrial Dysfunction and Neuronal Death Following Hypoglycemia-Induced Injury. Cells 2019, 8, 405. [Google Scholar] [CrossRef]
- Tan, Y.; Jin, Y.; Wang, Q.; Huang, J.; Wu, X.; Ren, Z. Perilipin 5 Protects against Cellular Oxidative Stress by Enhancing Mitochondrial Function in HepG2 Cells. Cells 2019, 8, 1241. [Google Scholar] [CrossRef]
- Migliaccio, V.; Di Gregorio, I.; Putti, R.; Lionetti, L. Mitochondrial Involvement in the Adaptive Response to Chronic Exposure to Environmental Pollutants and High-Fat Feeding in a Rat Liver and Testis. Cells 2019, 8, 834. [Google Scholar] [CrossRef]
- Graf, A.; Trofimova, L.; Ksenofontov, A.; Baratova, L.; Bunik, V. Hypoxic Adaptation of Mitochondrial Metabolism in Rat Cerebellum Decreases in Pregnancy. Cells 2020, 9, 139. [Google Scholar] [CrossRef]
- Picca, A.; Mankowski, R.T.; Kamenov, G.; Anton, S.D.; Manini, T.M.; Buford, T.W.; Saini, S.K.; Calvani, R.; Landi, F.; Bernabei, R.; et al. Advanced Age Is Associated with Iron Dyshomeostasis and Mitochondrial DNA Damage in Human Skeletal Muscle. Cells 2019, 8, 1525. [Google Scholar] [CrossRef]
- Wacquier, B.; Combettes, L.; Van Nhieu, G.T.; Dupont, G. Interplay Between Intracellular Ca2+ Oscillations and Ca2+-Stimulated Mitochondrial Metabolism. Sci. Rep. 2016, 6, 19316. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Guo, Y.; Shi, X.; Chen, X.; Feng, W.; Wu, L.L.; Zhang, J.; Yu, S.; Wang, Y.; et al. An Overview: The Diversified Role of Mitochondria in Cancer Metabolism. Int. J. Biol. Sci. 2023, 19, 897. [Google Scholar] [CrossRef]
- Miwa, S.; Kashyap, S.; Chini, E.; von Zglinicki, T. Mitochondrial Dysfunction in Cell Senescence and Aging. J. Clin. Investig. 2022, 132, e158447. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Miwa, S.; Carroll, B.; von Zglinicki, T. Mitochondria in Cell Senescence: Is Mitophagy the Weakest Link? EBioMedicine 2017, 21, 7–13. [Google Scholar] [CrossRef]
- Nacarelli, T.; Lau, L.; Fukumoto, T.; Zundell, J.; Fatkhutdinov, N.; Wu, S.; Aird, K.M.; Iwasaki, O.; Kossenkov, A.V.; Schultz, D.; et al. NAD+ Metabolism Governs the Proinflammatory Senescence-Associated Secretome. Nat. Cell Biol. 2019, 21, 397–407. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA Damage Response in Cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Brillo, V.; Chieregato, L.; Leanza, L.; Muccioli, S.; Costa, R. Mitochondrial Dynamics, ROS, and Cell Signaling: A Blended Overview. Life 2021, 11, 332. [Google Scholar] [CrossRef]
- Dencher, N.A.; Frenzel, M.; Reifschneider, N.H.; Sugawa, M.; Krause, F. Proteome Alterations in Rat Mitochondria Caused by Aging. Ann. N. Y. Acad. Sci. 2007, 1100, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Camacho, I.; Flores-Herrera, O.; Zazueta, C. The Relevance of the Supramolecular Arrangements of the Respiratory Chain Complexes in Human Diseases and Aging. Mitochondrion 2019, 47, 266–272. [Google Scholar] [CrossRef]
- Boengler, K.; Kosiol, M.; Mayr, M.; Schulz, R.; Rohrbach, S. Mitochondria and Ageing: Role in Heart, Skeletal Muscle and Adipose Tissue. J. Cachexia Sarcopenia Muscle 2017, 8, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Meves, F. Die Chondriosomen Als Träger Erblicher Anlagen. Cytologische Studien Am Hühnerembryo. Arch. Für Mikrosk. Anat. 1908, 72, 816–867. [Google Scholar] [CrossRef]
- Regaud, C. Participation Du Chondriome à La Formationdes Grains de Ségrégation Dans Les Cellules Des Tubes Contournés Du Rein. C. R. Soc. Biol. 1909, 66, 1034–1036. [Google Scholar]
- Nass, M.M.; Nass, S. Intramitochondrial fibers with dna characteristics. i. fixation and electron staining reactions. J. Cell Biol. 1963, 19, 593–611. [Google Scholar] [CrossRef]
- Schatz, G.; Haslbrunner, E.; Tuppy, H. Deoxyribonucleic acid associated with yeast mitochondria. Biochem. Biophys. Res. Commun. 1964, 15, 127–132. [Google Scholar] [CrossRef]
- Luck, D.J.; Reich, E. DNA in mitochondria of neurospora crassa. Proc. Natl. Acad. Sci. USA 1964, 52, 931–938. [Google Scholar] [CrossRef]
- Rossmann, M.P.; Dubois, S.M.; Agarwal, S.; Zon, L.I. Mitochondrial Function in Development and Disease. Dis. Model. Mech. 2021, 14, dmm048912. [Google Scholar] [CrossRef]
- Nicholls, T.J.; Gustafsson, C.M. Separating and Segregating the Human Mitochondrial Genome. Trends Biochem. Sci. 2018, 43, 869–881. [Google Scholar] [CrossRef]
- Gustafsson, C.M.; Falkenberg, M.; Larsson, N.G. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem. 2016, 85, 133–160. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Niikura, T.; Tajima, H.; Yasukawa, T.; Sudo, H.; Ito, Y.; Kita, Y.; Kawasumi, M.; Kouyama, K.; Doyu, M.; et al. A Rescue Factor Abolishing Neuronal Cell Death by a Wide Spectrum of Familial Alzheimer’s Disease Genes and Abeta. Proc. Natl. Acad. Sci. USA 2001, 98, 6336–6341. [Google Scholar] [CrossRef] [PubMed]
- Coradduzza, D.; Congiargiu, A.; Chen, Z.; Cruciani, S.; Zinellu, A.; Carru, C.; Medici, S. Humanin and Its Pathophysiological Roles in Aging: A Systematic Review. Biology 2023, 12, 558. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Son, J.M.; Benayoun, B.A.; Lee, C. The Mitochondrial-Encoded Peptide MOTS-c Translocates to the Nucleus to Regulate Nuclear Gene Expression in Response to Metabolic Stress. Cell Metab. 2018, 28, 516–524.e7. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Zeng, J.; Drew, B.G.; Sallam, T.; Martin-Montalvo, A.; Wan, J.; Kim, S.J.; Mehta, H.; Hevener, A.L.; De Cabo, R.; et al. The Mitochondrial-Derived Peptide MOTS-c Promotes Metabolic Homeostasis and Reduces Obesity and Insulin Resistance. Cell Metab. 2015, 21, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Majidpoor, J.; Mortezaee, K. Angiogenesis as a Hallmark of Solid Tumors—Clinical Perspectives. Cell Oncol. 2021, 44, 715–737. [Google Scholar] [CrossRef] [PubMed]
- Martins Pinto, M.; Paumard, P.; Bouchez, C.; Ransac, S.; Duvezin-Caubet, S.; Mazat, J.P.; Rigoulet, M.; Devin, A. The Warburg Effect and Mitochondrial Oxidative Phosphorylation: Friends or Foes? Biochim. Biophys. Acta (BBA) Bioenerg. 2023, 1864, 148931. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. THE METABOLISM OF TUMORS IN THE BODY. J. Gen. Physiol. 1927, 8, 519. [Google Scholar] [CrossRef]
- Chen, Z.; Han, F.; Du, Y.; Shi, H.; Zhou, W. Hypoxic Microenvironment in Cancer: Molecular Mechanisms and Therapeutic Interventions. Signal Transduct. Target. Ther. 2023, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Eales, K.L.; Hollinshead, K.E.R.; Tennant, D.A. Hypoxia and Metabolic Adaptation of Cancer Cells. Oncogenesis 2016, 5, e190. [Google Scholar] [CrossRef] [PubMed]
- Kopinski, P.K.; Singh, L.N.; Zhang, S.; Lott, M.T.; Wallace, D.C. Mitochondrial DNA Variation and Cancer. Nat. Rev. Cancer 2021, 21, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Petros, J.A.; Baumann, A.K.; Ruiz-Pesini, E.; Amin, M.B.; Sun, C.Q.; Hall, J.; Lim, S.D.; Issa, M.M.; Flanders, W.D.; Hosseini, S.H.; et al. MtDNA Mutations Increase Tumorigenicity in Prostate Cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Kalsbeek, A.M.F.; Chan, E.F.K.; Grogan, J.; Petersen, D.C.; Jaratlerdsiri, W.; Gupta, R.; Lyons, R.J.; Haynes, A.M.; Horvath, L.G.; Kench, J.G.; et al. Mutational Load of the Mitochondrial Genome Predicts Pathological Features and Biochemical Recurrence in Prostate Cancer. Aging 2016, 8, 2702–2712. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.F.; Sabelnykova, V.Y.; Weischenfeldt, J.; Simon, R.; Aguiar, J.A.; Alkallas, R.; Heisler, L.E.; Zhang, J.; Watson, J.D.; Chua, M.L.K.; et al. Mitochondrial Mutations Drive Prostate Cancer Aggression. Nat. Commun. 2017, 8, 656. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.F.; Denroche, R.E.; Aguiar, J.A.; Notta, F.; Connor, A.A.; Wilson, J.M.; Stein, L.D.; Gallinger, S.; Boutros, P.C. Mutations in Mitochondrial DNA From Pancreatic Ductal Adenocarcinomas Associate With Survival Times of Patients and Accumulate as Tumors Progress. Gastroenterology 2018, 154, 1620–1624.e5. [Google Scholar] [CrossRef]
- Ju, Y.S.; Alexandrov, L.B.; Gerstung, M.; Martincorena, I.; Nik-Zainal, S.; Ramakrishna, M.; Davies, H.R.; Papaemmanuil, E.; Gundem, G.; Shlien, A.; et al. Origins and Functional Consequences of Somatic Mitochondrial DNA in Human Cancer. Elife 2014, 3, 17. [Google Scholar] [CrossRef]
- Grandhi, S.; Bosworth, C.; Maddox, W.; Sensiba, C.; Akhavanfard, S.; Ni, Y.; LaFramboise, T. Heteroplasmic Shifts in Tumor Mitochondrial Genomes Reveal Tissue-Specific Signals of Relaxed and Positive Selection. Hum. Mol. Genet. 2017, 26, 2912. [Google Scholar] [CrossRef]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive Molecular Characterization of Mitochondrial Genomes in Human Cancers. Nat. Genet. 2020, 52, 342. [Google Scholar] [CrossRef]
- Reznik, E.; Miller, M.L.; Şenbabaoğlu, Y.; Riaz, N.; Sarungbam, J.; Tickoo, S.K.; Al-Ahmadie, H.A.; Lee, W.; Seshan, V.E.; Hakimi, A.A.; et al. Mitochondrial DNA Copy Number Variation across Human Cancers. Elife 2016, 5, e10769. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.M.; Whitehall, J.C.; Greaves, L.C. Mitochondrial DNA Mutations in Ageing and Cancer. Mol. Oncol. 2022, 16, 3276. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. Mitochondria and Cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Mambo, E.; Sidransky, D. Mitochondrial DNA Mutations in Human Cancer. Oncogene 2006, 25, 4663–4674. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Choudhury, A.R.; Tiwari, H.K. Numtogenesis as a Mechanism for Development of Cancer. Semin. Cancer Biol. 2017, 47, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Harutyunyan, T. The Known Unknowns of Mitochondrial Carcinogenesis: De Novo NUMTs and Intercellular Mitochondrial Transfer. Mutagenesis 2024, 39, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Baba, T.; Zhan, Q.; Kamimura, N.; Cuthbert, J.A. HeLaTG Cells Have Mitochondrial DNA Inserted into the C-Myc Oncogene. Oncogene 1991, 6, 1869–1874. [Google Scholar] [PubMed]
- Lutz-Bonengel, S.; Niederstätter, H.; Naue, J.; Koziel, R.; Yang, F.; Sänger, T.; Huber, G.; Berger, C.; Pflugradt, R.; Strobl, C.; et al. Evidence for Multi-Copy Mega-NUMTs in the Human Genome. Nucleic Acids Res. 2021, 49, 1517–1531. [Google Scholar] [CrossRef] [PubMed]
- Balciuniene, J.; Balciunas, D. A Nuclear MtDNA Concatemer (Mega-NUMT) Could Mimic Paternal Inheritance of Mitochondrial Genome. Front. Genet. 2019, 10, 518. [Google Scholar] [CrossRef]
- Liehr, T. Repetitive Elements in Humans. Int. J. Mol. Sci. 2021, 22, 2072. [Google Scholar] [CrossRef]
- Chen, D.; Xue, W.; Xiang, J. The Intra-Nucleus Integration of Mitochondrial DNA (MtDNA)in Cervical Mucosa Cells and Its Relation with c-Myc Expression. J. Exp. Clin. Cancer Res. 2008, 27, 36. [Google Scholar] [CrossRef] [PubMed]
- Srinivasainagendra, V.; Sandel, M.W.; Singh, B.; Sundaresan, A.; Mooga, V.P.; Bajpai, P.; Tiwari, H.K.; Singh, K.K. Migration of Mitochondrial DNA in the Nuclear Genome of Colorectal Adenocarcinoma. Genome Med. 2017, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.S.; Tubio, J.M.C.; Mifsud, W.; Fu, B.; Davies, H.R.; Ramakrishna, M.; Li, Y.; Yates, L.; Gundem, G.; Tarpey, P.S.; et al. Frequent Somatic Transfer of Mitochondrial DNA into the Nuclear Genome of Human Cancer Cells. Genome Res. 2015, 25, 814. [Google Scholar] [CrossRef]
- Brischigliaro, M.; Sierra-Magro, A.; Ahn, A.; Barrientos, A. Mitochondrial Ribosome Biogenesis and Redox Sensing. FEBS Open Bio 2024. [Google Scholar] [CrossRef] [PubMed]
- Khawaja, A.; Cipullo, M.; Krüger, A.; Rorbach, J. Insights into Mitoribosomal Biogenesis from Recent Structural Studies. Trends Biochem. Sci. 2023, 48, 629–641. [Google Scholar] [CrossRef]
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis—Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Russo, G. Ribosomal Proteins Control or Bypass P53 during Nucleolar Stress. Int. J. Mol. Sci. 2017, 18, 140. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Rice, W.G.; Grummt, I. The RNA Polymerase i Transcription Machinery: An Emerging Target for the Treatment of Cancer. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 131–156. [Google Scholar] [CrossRef]
- Oran, A.R.; Adams, C.M.; Zhang, X.Y.; Gennaro, V.J.; Pfeiffer, H.K.; Mellert, H.S.; Seidel, H.E.; Mascioli, K.; Kaplan, J.; Gaballa, M.R.; et al. Multi-Focal Control of Mitochondrial Gene Expression by Oncogenic MYC Provides Potential Therapeutic Targets in Cancer. Oncotarget 2016, 7, 72395–72414. [Google Scholar] [CrossRef]
- Molavi, G.; Samadi, N.; Hosseingholi, E.Z. The Roles of Moonlight Ribosomal Proteins in the Development of Human Cancers. J. Cell Physiol. 2019, 234, 8327–8341. [Google Scholar] [CrossRef]
- Zong, W.X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667. [Google Scholar] [CrossRef]
- Alberghina, L. The Warburg Effect Explained: Integration of Enhanced Glycolysis with Heterogeneous Mitochondria to Promote Cancer Cell Proliferation. Int. J. Mol. Sci. 2023, 24, 15787. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Antonaros, F.; Vitale, L.; Strippoli, P.; Pelleri, M.C.; Caracausi, M. Human Protein-Coding Genes and Gene Feature Statistics in 2019. BMC Res. Notes 2019, 12, 315. [Google Scholar] [CrossRef]
- Hatje, K.; Mühlhausen, S.; Simm, D.; Kollmar, M. The Protein-Coding Human Genome: Annotating High-Hanging Fruits. Bioessays 2019, 41, e1900066. [Google Scholar] [CrossRef]
- Rao, M. Long Non Coding RNA Biology; Rao, M.R.S., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; Volume 1008, ISBN 978-981-10-5202-6. [Google Scholar]
- Cipriano, A.; Ballarino, M. The Ever-Evolving Concept of the Gene: The Use of RNA/Protein Experimental Techniques to Understand Genome Functions. Front. Mol. Biosci. 2018, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Piergentili, R.; Basile, G.; Nocella, C.; Carnevale, R.; Marinelli, E.; Patrone, R.; Zaami, S. Using NcRNAs as Tools in Cancer Diagnosis and Treatment—The Way towards Personalized Medicine to Improve Patients’ Health. Int. J. Mol. Sci. 2022, 23, 9353. [Google Scholar] [CrossRef]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef]
- Zhou, Q.; Shu, X.; Chai, Y.; Liu, W.; Li, Z.; Xi, Y. The Non-Coding Competing Endogenous RNAs in Acute Myeloid Leukemia: Biological and Clinical Implications. Biomed. Pharmacother. 2023, 163, 114807. [Google Scholar] [CrossRef]
- Cen, L.; Liu, R.; Liu, W.; Li, Q.; Cui, H. Competing Endogenous RNA Networks in Glioma. Front. Genet. 2021, 12, 675498. [Google Scholar] [CrossRef]
- Abdollahzadeh, R.; Daraei, A.; Mansoori, Y.; Sepahvand, M.; Amoli, M.M.; Tavakkoly-Bazzaz, J. Competing Endogenous RNA (CeRNA) Cross Talk and Language in CeRNA Regulatory Networks: A New Look at Hallmarks of Breast Cancer. J. Cell Physiol. 2019, 234, 10080–10100. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Kan, C.H.; Liu, H.; Liu, Y.H.; Wu, C.C.; Kuo, Y.P.; Chang, I.Y.F.; Chang, K.P.; Yu, J.S.; Tan, B.C.M. Modular Scaffolding by LncRNA HOXA10-AS Promotes Oral Cancer Progression. Cell Death Dis. 2022, 13, 629. [Google Scholar] [CrossRef] [PubMed]
- Andric, V.; Nevers, A.; Hazra, D.; Auxilien, S.; Menant, A.; Graille, M.; Palancade, B.; Rougemaille, M. A Scaffold LncRNA Shapes the Mitosis to Meiosis Switch. Nat. Commun. 2021, 12, 770. [Google Scholar] [CrossRef]
- Liu, B.; Xiang, W.; Liu, J.; Tang, J.; Wang, J.; Liu, B.; Long, Z.; Wang, L.; Yin, G.; Liu, J. The Regulatory Role of Antisense LncRNAs in Cancer. Cancer Cell Int. 2021, 21, 459. [Google Scholar] [CrossRef]
- Jiang, B.; Yuan, Y.; Yi, T.; Dang, W. The Roles of Antisense Long Noncoding RNAs in Tumorigenesis and Development through Cis-Regulation of Neighbouring Genes. Biomolecules 2023, 13, 684. [Google Scholar] [CrossRef]
- Yagi, M.; Uchiumi, T.; Takazaki, S.; Okuno, B.; Nomura, M.; Yoshida, S.I.; Kanki, T.; Kang, D. P32/GC1qR Is Indispensable for Fetal Development and Mitochondrial Translation: Importance of Its RNA-Binding Ability. Nucleic Acids Res. 2012, 40, 9717–9737. [Google Scholar] [CrossRef]
- Wang, M.; Pestov, D.G. 5′-End Surveillance by Xrn2 Acts as a Shared Mechanism for Mammalian Pre-RRNA Maturation and Decay. Nucleic Acids Res. 2011, 39, 1811–1822. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, R.; Verheyden, Y.; Ishikawa, H.; Goedert, L.; Nicolas, E.; Saraf, K.; Armaos, A.; Delli Ponti, R.; Izumikawa, K.; Mestdagh, P.; et al. SAMMSON Fosters Cancer Cell Fitness by Concertedly Enhancing Mitochondrial and Cytosolic Translation. Nat. Struct. Mol. Biol. 2018, 25, 1035–1046. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma Addiction to the Long Non-Coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Naeli, P.; Winter, T.; Hackett, A.P.; Alboushi, L.; Jafarnejad, S.M. The Intricate Balance between MicroRNA-Induced MRNA Decay and Translational Repression. FEBS J. 2023, 290, 2508–2524. [Google Scholar] [CrossRef]
- Riolo, G.; Cantara, S.; Marzocchi, C.; Ricci, C. MiRNA Targets: From Prediction Tools to Experimental Validation. Methods Protoc. 2020, 4, 1. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Silva, B.V.; Gao, T.; Xu, Z.; Cui, J. Dynamic and Modularized MicroRNA Regulation and Its Implication in Human Cancers. Sci. Rep. 2017, 7, 13356. [Google Scholar] [CrossRef]
- Kumar, S.; Gonzalez, E.A.; Rameshwar, P.; Etchegaray, J.P. Non-Coding RNAs as Mediators of Epigenetic Changes in Malignancies. Cancers 2020, 12, 3657. [Google Scholar] [CrossRef]
- Borralho, P.M.; Rodrigues, C.M.P.; Steer, C.J. MicroRNAs in Mitochondria: An Unexplored Niche. Adv. Exp. Med. Biol. 2015, 887, 31–51. [Google Scholar] [CrossRef]
- Duarte, F.V.; Palmeira, C.M.; Rolo, A.P. The Role of MicroRNAs in Mitochondria: Small Players Acting Wide. Genes 2014, 5, 865. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Thankachan, S.; Abu Fawaz, P.P.; Venkatesh, T.; Prasada Kabekkodu, S.; Suresh, P.S. Deciphering the Role of MitomiRs in Cancer: A Comprehensive Review. Mitochondrion 2023, 70, 118–130. [Google Scholar] [CrossRef]
- Duarte, F.V.; Palmeira, C.M.; Rolo, A.P. The Emerging Role of MitomiRs in the Pathophysiology of Human Disease. Adv. Exp. Med. Biol. 2015, 888, 123–154. [Google Scholar] [CrossRef]
- Purohit, P.K.; Saini, N. Mitochondrial MicroRNA (MitomiRs) in Cancer and Complex Mitochondrial Diseases: Current Status and Future Perspectives. Cell Mol. Life Sci. 2021, 78, 1405–1421. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.; Bhadra, U. A Complex Genome-MicroRNA Interplay in Human Mitochondria. Biomed. Res. Int. 2015, 2015, 206382. [Google Scholar] [CrossRef]
- Sripada, L.; Tomar, D.; Prajapati, P.; Singh, R.; Singh, A.K.; Singh, R. Systematic Analysis of Small RNAs Associated with Human Mitochondria by Deep Sequencing: Detailed Analysis of Mitochondrial Associated MiRNA. PLoS ONE 2012, 7, e44873. [Google Scholar] [CrossRef] [PubMed]
- Barrey, E.; Saint-Auret, G.; Bonnamy, B.; Damas, D.; Boyer, O.; Gidrol, X. Pre-MicroRNA and Mature MicroRNA in Human Mitochondria. PLoS ONE 2011, 6, e20220. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xiang, H.; Ning, C.; Liu, H.; Liu, J.F.; Zhao, X. Mitochondrial DNA Enrichment Reduced NUMT Contamination in Porcine NGS Analyses. Brief. Bioinform. 2020, 21, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Biró, B.; Gál, Z.; Fekete, Z.; Klecska, E.; Hoffmann, O.I. Mitochondrial Genome Plasticity of Mammalian Species. BMC Genom. 2024, 25, 278. [Google Scholar] [CrossRef]
- Maude, H.; Davidson, M.; Charitakis, N.; Diaz, L.; Bowers, W.H.T.; Gradovich, E.; Andrew, T.; Huntley, D. NUMT Confounding Biases Mitochondrial Heteroplasmy Calls in Favor of the Reference Allele. Front. Cell Dev. Biol. 2019, 7, 201. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Visavadiya, N.P.; Pandya, J.D.; Nelson, P.T.; Sullivan, P.G.; Springer, J.E. Mitochondria-Associated MicroRNAs in Rat Hippocampus Following Traumatic Brain Injury. Exp. Neurol. 2015, 265, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.N.S.; Kar, A.N.; Kowalak, J.A.; Gale, J.R.; Aschrafi, A.; Chen, C.Y.; Gioio, A.E.; Kaplan, B.B. Axonal Localization and Mitochondrial Association of Precursor MicroRNA 338. Cell Mol. Life Sci. 2016, 73, 4327–4340. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Tian, T.; Chen, W.; Lv, X.; Lei, X.; Zhang, H.; Sun, S.; Cai, L.; Pan, G.; He, L.; et al. Mitochondrial MiRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Res. 2019, 79, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear MiRNA Regulates the Mitochondrial Genome in the Heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef]
- Bian, Z.; Li, L.M.; Tang, R.; Hou, D.X.; Chen, X.; Zhang, C.Y.; Zen, K. Identification of Mouse Liver Mitochondria-Associated MiRNAs and Their Potential Biological Functions. Cell Res. 2010, 20, 1076–1078. [Google Scholar] [CrossRef]
- Bandiera, S.; Rüberg, S.; Girard, M.; Cagnard, N.; Hanein, S.; Chrétien, D.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. Nuclear Outsourcing of RNA Interference Components to Human Mitochondria. PLoS ONE 2011, 6, e20746. [Google Scholar] [CrossRef] [PubMed]
- Macgregor-Das, A.M.; Das, S. A MicroRNA’s Journey to the Center of the Mitochondria. Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H206–H215. [Google Scholar] [CrossRef] [PubMed]
- Salama, R.M.; Eissa, N.; Doghish, A.S.; Abulsoud, A.I.; Abdelmaksoud, N.M.; Mohammed, O.A.; Abdel Mageed, S.S.; Darwish, S.F. Decoding the Secrets of Longevity: Unraveling Nutraceutical and MiRNA-Mediated Aging Pathways and Therapeutic Strategies. Front. Aging 2024, 5, 1373741. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef]
- Havens, M.A.; Reich, A.A.; Duelli, D.M.; Hastings, M.L. Biogenesis of Mammalian MicroRNAs by a Non-Canonical Processing Pathway. Nucleic Acids Res. 2012, 40, 4626–4640. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Kummer, E.; Ban, N. Mechanisms and Regulation of Protein Synthesis in Mitochondria. Nat. Rev. Mol. Cell Biol. 2021, 22, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.; Ma, H.Y.; Park, C.; Ortogero, N.; Song, R.; Hennig, G.W.; Zheng, H.; Lin, Y.M.; Moro, L.; Hsieh, J.T.; et al. The Mitochondrial Genome Encodes Abundant Small Noncoding RNAs. Cell Res. 2013, 23, 759–774. [Google Scholar] [CrossRef]
- Villegas, J.; Müller, I.; Arredondo, J.; Pinto, R.; Burzio, L.O. A Putative RNA Editing from U to C in a Mouse Mitochondrial Transcript. Nucleic Acids Res. 2002, 30, 1895–1901. [Google Scholar] [CrossRef][Green Version]
- Villegas, J.; Zarraga, A.M.; Muller, I.; Montecinos, L.; Werner, E.; Brito, M.; Meneses, A.M.; Burzio, L.O. A Novel Chimeric Mitochondrial RNA Localized in the Nucleus of Mouse Sperm. DNA Cell Biol. 2000, 19, 579–588. [Google Scholar] [CrossRef]
- Villegas, J.; Araya, P.; Bustos-Obregon, E.; Burzio, L.O. Localization of the 16S Mitochondrial RRNA in the Nucleus of Mammalian Spermatogenic Cells. Mol. Hum. Reprod. 2002, 8, 977–983. [Google Scholar] [CrossRef]
- Villegas, J.; Burzio, V.; Villota, C.; Landerer, E.; Martinez, R.; Santander, M.; Martinez, R.; Pinto, R.; Vera, M.I.; Boccardo, E.; et al. Expression of a Novel Non-Coding Mitochondrial RNA in Human Proliferating Cells. Nucleic Acids Res. 2007, 35, 7336–7347. [Google Scholar] [CrossRef]
- Burzio, V.A.; Villota, C.; Villegas, J.; Landerer, E.; Boccardo, E.; Villa, L.L.; Martínez, R.; Lopez, C.; Gaete, F.; Toro, V.; et al. Expression of a Family of Noncoding Mitochondrial RNAs Distinguishes Normal from Cancer Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 9430–9434. [Google Scholar] [CrossRef]
- Landerer, E.; Villegas, J.; Burzio, V.A.; Oliveira, L.; Villota, C.; Lopez, C.; Restovic, F.; Martinez, R.; Castillo, O.; Burzio, L.O. Nuclear Localization of the Mitochondrial NcRNAs in Normal and Cancer Cells. Cell Oncol. 2011, 34, 297–305. [Google Scholar] [CrossRef]
- Rivas, A.; Burzio, V.; Landerer, E.; Borgna, V.; Gatica, S.; Ávila, R.; López, C.; Villota, C.; De La Fuente, R.; Echenique, J.; et al. Determination of the Differential Expression of Mitochondrial Long Non-Coding RNAs as a Noninvasive Diagnosis of Bladder Cancer. BMC Urol. 2012, 12, 37. [Google Scholar] [CrossRef]
- Varas-Godoy, M.; Lladser, A.; Farfan, N.; Villota, C.; Villegas, J.; Tapia, J.C.; Burzio, L.O.; Burzio, V.A.; Valenzuela, P.D.T. In Vivo Knockdown of Antisense Non-Coding Mitochondrial RNAs by a Lentiviral-Encoded ShRNA Inhibits Melanoma Tumor Growth and Lung Colonization. Pigment. Cell Melanoma Res. 2018, 31, 64–72. [Google Scholar] [CrossRef]
- Araya, M.; Sepúlveda, F.; Villegas, J.; Alarcón, L.; Burzio, L.O.; Burzio, V.A.; Borgna, V. Knockdown of Antisense Noncoding Mitochondrial RNA Reduces Tumorigenicity of Patient-Derived Clear Cell Renal Carcinoma Cells in an Orthotopic Xenograft Mouse Model. Cancers 2024, 16, 830. [Google Scholar] [CrossRef]
- Borgna, V.; Villegas, J.; Burzio, V.A.; Belmar, S.; Araya, M.; Jeldes, E.; Lobos-González, L.; Silva, V.; Villota, C.; Oliveira-Cruz, L.; et al. Mitochondrial ASncmtRNA-1 and ASncmtRNA-2 as Potent Targets to Inhibit Tumor Growth and Metastasis in the RenCa Murine Renal Adenocarcinoma Model. Oncotarget 2017, 8, 43692–43708. [Google Scholar] [CrossRef]
- Lobos-González, L.; Silva, V.; Araya, M.; Restovic, F.; Echenique, J.; Oliveira-Cruz, L.; Fitzpatrick, C.; Briones, M.; Villegas, J.; Villota, C.; et al. Targeting Antisense Mitochondrial NcRNAs Inhibits Murine Melanoma Tumor Growth and Metastasis through Reduction in Survival and Invasion Factors. Oncotarget 2016, 7, 58331–58350. [Google Scholar] [CrossRef]
- Vidaurre, S.; Fitzpatrick, C.; Burzio, V.A.; Briones, M.; Villota, C.; Villegas, J.; Echenique, J.; Oliveira-Cruz, L.; Araya, M.; Borgna, V.; et al. Down-Regulation of the Antisense Mitochondrial Non-Coding RNAs (NcRNAs) Is a Unique Vulnerability of Cancer Cells and a Potential Target for Cancer Therapy. J. Biol. Chem. 2014, 289, 27182–27198. [Google Scholar] [CrossRef]
- Tu, H.; Costa, M. XIAP’s Profile in Human Cancer. Biomolecules 2020, 10, 1493. [Google Scholar] [CrossRef]
- Warrier, N.M.; Agarwal, P.; Kumar, P. Emerging Importance of Survivin in Stem Cells and Cancer: The Development of New Cancer Therapeutics. Stem Cell Rev. Rep. 2020, 16, 828–852. [Google Scholar] [CrossRef]
- Fitzpatrick, C.; Bendek, M.F.; Briones, M.; Farfán, N.; Silva, V.A.; Nardocci, G.; Montecino, M.; Boland, A.; Deleuze, J.F.; Villegas, J.; et al. Mitochondrial NcRNA Targeting Induces Cell Cycle Arrest and Tumor Growth Inhibition of MDA-MB-231 Breast Cancer Cells through Reduction of Key Cell Cycle Progression Factors. Cell Death Dis. 2019, 10, 423. [Google Scholar] [CrossRef]
- Bendek, M.F.; Fitzpatrick, C.; Jeldes, E.; Boland, A.; Deleuze, J.F.; Farfán, N.; Villegas, J.; Nardocci, G.; Montecino, M.; Burzio, L.O.; et al. Inverse Modulation of Aurora Kinase A and Topoisomerase IIα in Normal and Tumor Breast Cells upon Knockdown of Mitochondrial ASncmtRNA. Noncoding RNA 2023, 9, 59. [Google Scholar] [CrossRef]
- Bergerot, P.; Burns, K.; Prajapati, D.; Fox, R.; Salgia, M.; Pal, S.K. Advances in the Treatment of Metastatic Renal Cell Carcinoma. Cancer Treat. Res. 2018, 175, 127–137. [Google Scholar] [CrossRef]
- Jin, J.; Xie, Y.; Zhang, J.S.; Wang, J.Q.; Dai, S.J.; He, W.F.; Li, S.Y.; Ashby, C.R.; Chen, Z.S.; He, Q. Sunitinib Resistance in Renal Cell Carcinoma: From Molecular Mechanisms to Predictive Biomarkers. Drug Resist. Updat. 2023, 67, 100929. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef]
- Villota, C.; Varas-Godoy, M.; Jeldes, E.; Campos, A.; Villegas, J.; Borgna, V.; Burzio, L.O.; Burzio, V.A. HPV-18 E2 Protein Downregulates Antisense Noncoding Mitochondrial RNA-2, Delaying Replicative Senescence of Human Keratinocytes. Aging 2018, 11, 33–47. [Google Scholar] [CrossRef]
- Villota, C.; Campos, A.; Vidaurre, S.; Oliveira-Cruz, L.; Boccardo, E.; Burzio, V.A.; Varas, M.; Villegas, J.; Villa, L.L.; Valenzuela, P.D.T.; et al. Expression of Mitochondrial Non-Coding RNAs (NcRNAs) Is Modulated by High Risk Human Papillomavirus (HPV) Oncogenes. J. Biol. Chem. 2012, 287, 21303–21315. [Google Scholar] [CrossRef]
- Mukherjee, A.G.; Ramesh Wanjari, U.; Valsala Gopalakrishnan, A.; Jayaraj, R.; Katturajan, R.; Kannampuzha, S.; Murali, R.; Namachivayam, A.; Evan Prince, S.; Vellingiri, B.; et al. HPV-Associated Cancers: Insights into the Mechanistic Scenario and Latest Updates. Med. Oncol. 2023, 40, 212. [Google Scholar] [CrossRef]
- Bianchessi, V.; Badi, I.; Bertolotti, M.; Nigro, P.; D’Alessandra, Y.; Capogrossi, M.C.; Zanobini, M.; Pompilio, G.; Raucci, A.; Lauri, A. The Mitochondrial LncRNA ASncmtRNA-2 Is Induced in Aging and Replicative Senescence in Endothelial Cells. J. Mol. Cell Cardiol. 2015, 81, 62–70. [Google Scholar] [CrossRef]
- Canovas, P.M.; Guadagno, T.M. Functional Analysis of Survivin in Spindle Assembly in Xenopus Egg Extracts. J. Cell Biochem. 2007, 100, 217–229. [Google Scholar] [CrossRef]
- Babkoff, A.; Cohen-Kfir, E.; Aharon, H.; Ronen, D.; Rosenberg, M.; Wiener, R.; Ravid, S. A Direct Interaction between Survivin and Myosin II Is Required for Cytokinesis. J. Cell Sci. 2019, 132, jcs233130. [Google Scholar] [CrossRef]
- Vivek, R.; Kannan, S.; Achiraman, S.; Thirumurugan, R.; Ganesh, D.S.; Krishnan, M. Survivin Deficiency Leads to Imparalization of Cytokinesis in Cancer Cells. Asian Pac. J. Cancer Prev. 2011, 12, 1675–1679. [Google Scholar]
- Dhawan, M.S.; Aggarwal, R.R.; Boyd, E.; Comerford, K.; Zhang, J.; Méndez, B.; Valenzuela, P.; Grabowsky, J.; Thomas, S.; Munster, P.N. Phase 1 Study of ANDES-1537: A Novel Antisense Oligonucleotide against Non-Coding Mitochondrial DNA in Advanced Solid Tumors. J. Clin. Oncol. 2018, 36, 2557. [Google Scholar] [CrossRef]
- Du, R.; Huang, C.; Liu, K.; Li, X.; Dong, Z. Targeting AURKA in Cancer: Molecular Mechanisms and Opportunities for Cancer Therapy. Mol. Cancer 2021, 20, 15. [Google Scholar] [CrossRef]
- Sasai, K.; Treekitkarnmongkol, W.; Kai, K.; Katayama, H.; Sen, S. Functional Significance of Aurora Kinases-P53 Protein Family Interactions in Cancer. Front. Oncol. 2016, 6, 247. [Google Scholar] [CrossRef]
- Naso, F.D.; Boi, D.; Ascanelli, C.; Pamfil, G.; Lindon, C.; Paiardini, A.; Guarguaglini, G. Nuclear Localisation of Aurora-A: Its Regulation and Significance for Aurora-A Functions in Cancer. Oncogene 2021, 40, 3917–3928. [Google Scholar] [CrossRef]
- Mou, P.K.; Yang, E.J.; Shi, C.; Ren, G.; Tao, S.; Shim, J.S. Aurora Kinase A, a Synthetic Lethal Target for Precision Cancer Medicine. Exp. Mol. Med. 2021, 53, 835–847. [Google Scholar] [CrossRef]
- Zhao, F.; Chang, J.; Zhao, P.; Wang, W.; Sun, X.; Ma, X.; Yin, M.; Wang, Y.; Yang, Y. Oncogenetic Function and Prognostic Value of DNA Topoisomerase II Alpha in Human Malignances: A Pan-Cancer Analysis. Front. Genet. 2022, 13, 856692. [Google Scholar] [CrossRef]
- Pommier, Y.; Nussenzweig, A.; Takeda, S.; Austin, C. Human Topoisomerases and Their Roles in Genome Stability and Organization. Nat. Rev. Mol. Cell Biol. 2022, 23, 407–427. [Google Scholar] [CrossRef]
- Uusküla-Reimand, L.; Wilson, M.D. Untangling the Roles of TOP2A and TOP2B in Transcription and Cancer. Sci. Adv. 2022, 8, eadd4920. [Google Scholar] [CrossRef]
- Delgado, J.L.; Hsieh, C.M.; Chan, N.L.; Hiasa, H. Topoisomerases as Anticancer Targets. Biochem. J. 2018, 475, 373–398. [Google Scholar] [CrossRef]
- Farfán, N.; Sanhueza, N.; Briones, M.; Burzio, L.O.; Burzio, V.A. Antisense Noncoding Mitochondrial RNA-2 Gives Rise to MiR-4485-3p by Dicer Processing in Vitro. Biol. Res. 2021, 54, 33. [Google Scholar] [CrossRef]
- Dorn, G.W. LIPCAR: A Mitochondrial Lnc in the Noncoding RNA Chain? Circ. Res. 2014, 114, 1548–1550. [Google Scholar] [CrossRef]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; De Groote, P.; Pinet, F.; Thum, T. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients with Heart Failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef]
- Frismantiene, A.; Philippova, M.; Erne, P.; Resink, T.J. Smooth Muscle Cell-Driven Vascular Diseases and Molecular Mechanisms of VSMC Plasticity. Cell. Signal. 2018, 52, 48–64. [Google Scholar] [CrossRef]
- Lu, Q.B.; Wang, H.P.; Tang, Z.H.; Cheng, H.; Du, Q.; Wang, Y.B.; Feng, W.B.; Li, K.X.; Cai, W.W.; Qiu, L.Y.; et al. Nesfatin-1 Functions as a Switch for Phenotype Transformation and Proliferation of VSMCs in Hypertensive Vascular Remodeling. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2154–2168. [Google Scholar] [CrossRef]
- Owens, G.K.; Kumar, M.S.; Wamhoff, B.R. Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease. Physiol. Rev. 2004, 84, 767–801. [Google Scholar] [CrossRef]
- Kong, P.; Wang, X.; Gao, Y.K.; Zhang, D.D.; Huang, X.F.; Song, Y.; Zhang, W.D.; Guo, R.J.; Li, H.; Han, M. RGS5 Maintaining Vascular Homeostasis Is Altered by the Tumor Microenvironment. Biol. Direct 2023, 18, 78. [Google Scholar] [CrossRef]
- Murgai, M.; Ju, W.; Eason, M.; Kline, J.; Beury, D.W.; Kaczanowska, S.; Miettinen, M.M.; Kruhlak, M.; Lei, H.; Shern, J.F.; et al. KLF4-Dependent Perivascular Cell Plasticity Mediates Pre-Metastatic Niche Formation and Metastasis. Nat. Med. 2017, 23, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yan, X.; Cao, J.; Yang, Z.; Cao, X.; Zhang, Y.; Liang, L.; Zheng, M.; Liu, X.; Zhang, J.; et al. SM22α+ Vascular Mural Cells Are Essential for Vessel Stability in Tumors and Undergo Phenotype Transition Regulated by Notch Signaling. J. Exp. Clin. Cancer Res. 2020, 39, 124. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, D.; Chen, H.; Wei, X.; Xu, X. Expression of Long Noncoding RNA LIPCAR Promotes Cell Proliferation, Cell Migration, and Change in Phenotype of Vascular Smooth Muscle Cells. Med. Sci. Monit. 2019, 25, 7645–7651. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Hu, H.; Fang, Z.; Cui, J.; Liu, F. CTRP6 Inhibits PDGF-BB-Induced Vascular Smooth Muscle Cell Proliferation and Migration. Biomed. Pharmacother. 2018, 103, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shao, L.; Yu, J.; Huang, J.; Feng, Q. PDGF-BB Promotes Vascular Smooth Muscle Cell Migration by Enhancing Pim-1 Expression via Inhibiting MiR-214. Ann. Transl. Med. 2021, 9, 1728. [Google Scholar] [CrossRef] [PubMed]
- Poznyak, A.V.; Nikiforov, N.G.; Markin, A.M.; Kashirskikh, D.A.; Myasoedova, V.A.; Gerasimova, E.V.; Orekhov, A.N. Overview of OxLDL and Its Impact on Cardiovascular Health: Focus on Atherosclerosis. Front. Pharmacol. 2021, 11, 613780. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Liu, C.P.; Shiao, W.C.; Jayakumar, T.; Li, Y.S.; Chang, N.C.; Huang, S.Y.; Hsieh, C.Y. Inhibitory Effect of PDGF-BB and Serum-Stimulated Responses in Vascular Smooth Muscle Cell Proliferation by Hinokitiol via up-Regulation of P21 and P53. Arch. Med. Sci. 2018, 14, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Bongolo, C.C.; Thokerunga, E.; Fidele, N.B.; Souraka, T.D.M.; Kisembo, P.; Rugera, S.P.; Worley, P.F.; Tu, J.C. Upregulation of the Long Non-Coding RNA, LIPCAR Promotes Proliferation, Migration, and Metastasis of Hepatocellular Carcinoma. Cancer Biomark. 2022, 35, 245–256. [Google Scholar] [CrossRef]
- Wang, H.; Song, T.; Zhao, Y.; Zhao, J.; Wang, X.; Fu, X. Long Non-Coding RNA LICPAR Regulates Atrial Fibrosis via TGF-β/Smad Pathway in Atrial Fibrillation. Tissue Cell 2020, 67, 101440. [Google Scholar] [CrossRef]
- Cao, F.; Li, Z.; Ding, W.M.; Yan, L.; Zhao, Q.Y. LncRNA PVT1 Regulates Atrial Fibrosis via MiR-128-3p-SP1-TGF-Β1-Smad Axis in Atrial Fibrillation. Mol. Med. 2019, 25, 7. [Google Scholar] [CrossRef]
- Hu, H.H.; Chen, D.Q.; Wang, Y.N.; Feng, Y.L.; Cao, G.; Vaziri, N.D.; Zhao, Y.Y. New Insights into TGF-β/Smad Signaling in Tissue Fibrosis. Chem. Biol. Interact. 2018, 292, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Li, N.; Xu, J.; Li, W.; Fang, X. Quantitative Single-Molecule Study of TGF-β/Smad Signaling. Acta Biochim. Biophys. Sin. 2018, 50, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yin, Y.; Li, Y.; Shi, N.; Xie, W.; Luo, W.; Wang, L.; Zhu, B.; Liu, W.; Jiang, X.; et al. FLOT2 Promotes Nasopharyngeal Carcinoma Progression through Suppression of TGF-β Pathway via Facilitating CD109 Expression. iScience 2023, 27, 108580. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, W.; Miao, X.; Wang, X. KIFC1 Aggravates Non-Small-Cell Lung Cancer Cell Proliferation and Metastasis via Provoking TGF-β/SMAD Signal. Cell Mol. Biol. 2023, 69, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, Y.; Wang, X.; Guo, H.; Chen, L.; Hu, G.; Cui, Y.; Liang, S.; Zuo, J.; Luo, Z.; et al. OLFM2 Promotes Epithelial-Mesenchymal Transition, Migration, and Invasion in Colorectal Cancer through the TGF-β/Smad Signaling Pathway. BMC Cancer 2024, 24, 204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.Y.; Chen, Y.; Wang, J.; Wang, Q.; Lu, H. TGF-β Signaling and Resistance to Cancer Therapy. Front. Cell Dev. Biol. 2021, 9, 786728. [Google Scholar] [CrossRef]
- Lian, G.Y.; Wang, Q.M.; Mak, T.S.K.; Huang, X.R.; Yu, X.Q.; Lan, H.Y. Inhibition of Tumor Invasion and Metastasis by Targeting TGF-β-Smad-MMP2 Pathway with Asiatic Acid and Naringenin. Mol. Ther. Oncolytics 2021, 20, 277–289. [Google Scholar] [CrossRef]
- Xin, X.; Cheng, X.; Zeng, F.; Xu, Q.; Hou, L. The Role of TGF-β/SMAD Signaling in Hepatocellular Carcinoma: From Mechanism to Therapy and Prognosis. Int. J. Biol. Sci. 2024, 20, 1436–1451. [Google Scholar] [CrossRef]
- Lu, Y.; Jiang, F.; Zheng, X.; Katakowski, M.; Buller, B.; To, S.S.T.; Chopp, M. TGF-Β1 Promotes Motility and Invasiveness of Glioma Cells through Activation of ADAM17. Oncol. Rep. 2011, 25, 1329–1335. [Google Scholar] [CrossRef]
- Eser, P.; Jänne, P.A. TGFβ Pathway Inhibition in the Treatment of Non-Small Cell Lung Cancer. Pharmacol. Ther. 2018, 184, 112–130. [Google Scholar] [CrossRef]
- Rackham, O.; Shearwood, A.M.J.; Mercer, T.R.; Davies, S.M.K.; Mattick, J.S.; Filipovska, A. Long Noncoding RNAs Are Generated from the Mitochondrial Genome and Regulated by Nuclear-Encoded Proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, S.; Zhou, L.; Li, X.; Meng, Y.; Li, Y.; Li, L.; Jiao, B.; Bai, L.; Yu, Y.; et al. Aberrant Shuttling of Long Noncoding RNAs during the Mitochondria-Nuclear Crosstalk in Hepatocellular Carcinoma Cells. Am. J. Cancer Res. 2019, 9, 1008. [Google Scholar]
- Zhang, X.; Yuan, S.; Liu, J.; Tang, Y.; Wang, Y.; Zhan, J.; Fan, J.; Nie, X.; Zhao, Y.; Wen, Z.; et al. Overexpression of Cytosolic Long Noncoding RNA Cytb Protects against Pressure-Overload-Induced Heart Failure via Sponging MicroRNA-103-3p. Mol. Ther. Nucleic Acids 2022, 27, 1127–1145. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Miao, J.; Zhang, M.; Wang, X.; Wang, Z.; Han, J.; Tong, D.; Huang, C. MiRNA-103a-3p Promotes Human Gastric Cancer Cell Proliferation by Targeting and Suppressing ATF7 in Vitro. Mol. Cells 2018, 41, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Chen, C.W.; Yang, L.Q.; Yang, W.W.; Du, Z.H.; Li, Y.R.; Li, S.L.; Ge, X.Y. MicroRNA-103a-3p Promotes Metastasis by Targeting TPD52 in Salivary Adenoid Cystic Carcinoma. Int. J. Oncol. 2020, 57, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, Z.; Zhang, Y.; Li, T.; Bao, Y.; Zhang, S. Inhibition of MiR-103a-3p Suppresses the Proliferation in Oral Squamous Cell Carcinoma Cells via Targeting RCAN1. Neoplasma 2020, 67, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Sun, W.H.; Wu, H.Q.; Liu, Z.D.; Wang, P. Knockdown of MicroRNA-103a-3p Inhibits the Malignancy of Thyroid Cancer Cells through Hippo Signaling Pathway by Upregulating LATS1. Neoplasma 2020, 67, 1266–1278. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Mao, L.; Xu, W.; Fang, W.; Wang, N.; Ye, D.; Dong, Z.; Guan, H.; Guan, C. MiR-103a-3p Suppresses Cell Proliferation and Invasion by Targeting Tumor Protein D52 in Prostate Cancer. J. Investig. Surg. 2021, 34, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Chen, J.; Wei, T.; Wang, J.; Chen, W. Inhibiting Roles of FOXA2 in Liver Cancer Cell Migration and Invasion by Transcriptionally Suppressing MicroRNA-103a-3p and Activating the GREM2/LATS2/YAP Axis. Cytotechnology 2021, 73, 523–537. [Google Scholar] [CrossRef]
- Xu, Q.; Liao, Z.; Gong, Z.; Liu, X.; Yang, Y.; Wang, Z.; Yang, W.; Hou, L.; Yang, J.; Song, J.; et al. Down-Regulation of EVA1A by MiR-103a-3p Promotes Hepatocellular Carcinoma Cells Proliferation and Migration. Cell Mol. Biol. Lett. 2022, 27, 93. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, X. MiR-103a-3p Contributes to the Progression of Colorectal Cancer by Regulating GREM2 Expression. Yonsei Med. J. 2022, 63, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huhe, M.; Lou, J. MicroRNA-103a-3p Promotes Cell Proliferation and Invasion in Non-Small-Cell Lung Cancer Cells through Akt Pathway by Targeting PTEN. Biomed. Res. Int. 2021, 2021, 7590976. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Lv, M.; Chen, J. Screening Differential Circular RNA Expression Profiles Reveals the Regulatory Role of CircTCF25-MiR-103a-3p/MiR-107-CDK6 Pathway in Bladder Carcinoma. Sci. Rep. 2016, 6, 30919. [Google Scholar] [CrossRef] [PubMed]
- She, Y.; Han, Y.; Zhou, G.; Jia, F.; Yang, T.; Shen, Z. Hsa_circ_0062389 Promotes the Progression of Non-Small Cell Lung Cancer by Sponging MiR-103a-3p to Mediate CCNE1 Expression. Cancer Genet. 2020, 241, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qiu, S.; Zheng, X.; Qiu, Y.; Yao, S.; Ge, Y.; Zhou, C. LINC00662 Modulates Cervical Cancer Cell Proliferation, Invasion, and Apoptosis via Sponging MiR-103a-3p and Upregulating PDK4. Mol. Carcinog. 2021, 60, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lin, F.; Xu, C.; Xu, Y. LINC00662 Facilitates Osteosarcoma Progression via Sponging MiR-103a-3p and Regulating SIK2 Expression. J. Tissue Eng. Regen. Med. 2021, 15, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, D.; Deng, G.; Wang, Q.; Li, L.; Zhang, J.; Wu, H. LncRNA Prostate Cancer-Associated Transcript 18 Upregulates Activating Transcription Factor 7 to Prevent Metastasis of Triple-Negative Breast Cancer via Sponging MiR-103a-3p. Bioengineered 2021, 12, 12070–12086. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qiao, K.; Zhu, K.; Li, X.; Zhao, C.; Li, J.; Feng, D.; Fang, Y.; Wang, P.; Qian, C.; et al. Long Noncoding RNA HCG18 Promotes Malignant Phenotypes of Breast Cancer Cells via the HCG18/MiR-103a-3p/UBE2O/MTORC1/HIF-1α-Positive Feedback Loop. Front. Cell Dev. Biol. 2021, 9, 675082. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Jiang, Q.; Liu, S.; Gu, J.; Hu, K.; Wang, Z. Circ_0002099 Is a Novel Molecular Therapeutic Target for Bladder Cancer. Drug Dev. Res. 2022, 83, 1890–1905. [Google Scholar] [CrossRef]
- Gao, S.; Tian, X.; Chang, H.; Sun, Y.; Wu, Z.; Cheng, Z.; Dong, P.; Zhao, Q.; Ruan, J.; Bu, W. Two Novel LncRNAs Discovered in Human Mitochondrial DNA Using PacBio Full-Length Transcriptome Data. Mitochondrion 2018, 38, 41–47. [Google Scholar] [CrossRef]
- Li, J.; Bai, R.; Yang, W.; Miao, H.; Li, Y.; Dai, H.; Li, L.; Zhao, Y.; Song, X.; Ling Li, C. The Mitochondrial-Derived LncRNA MDL1 Mediates a Mitochondria-to-Nucleus Retrograde Regulation by Inhibiting the Nuclear Translocation of P53. MedComm—Oncol. 2022, 1, e15. [Google Scholar] [CrossRef]
- Ahn, B.Y.; Trinh, D.L.N.; Zajchowski, L.D.; Lee, B.; Elwi, A.N.; Kim, S.W. Tid1 Is a New Regulator of P53 Mitochondrial Translocation and Apoptosis in Cancer. Oncogene 2010, 29, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M. Census and Evaluation of P53 Target Genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef]
- Garrido, P.; Casas-Benito, A.; Larrayoz, I.M.; Narro-Íñiguez, J.; Rubio-Mediavilla, S.; Zozaya, E.; Martín-Carnicero, A.; Martínez, A. Expression of Mitochondrial Long Non-Coding RNAs, MDL1 and MDL1AS, Are Good Prognostic and/or Diagnostic Biomarkers for Several Cancers, Including Colorectal Cancer. Cancers 2024, 16, 960. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, H.; Liu, W.; Zhu, H.; Fu, J.; Yang, C.; Fan, L.; Wang, L.; Liu, Y.; Xu, W.; et al. Circ-RPL15: A Plasma Circular RNA as Novel Oncogenic Driver to Promote Progression of Chronic Lymphocytic Leukemia. Leukemia 2020, 34, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wu, Z.; Xia, Y.; Qin, S.; Li, Y.; Wu, J.; Liang, J.; Wang, L.; Zhu, H.; Fan, L.; et al. Downregulation of Circ_0132266 in Chronic Lymphocytic Leukemia Promoted Cell Viability through MiR-337-3p/PML Axis. Aging 2019, 11, 3561–3573. [Google Scholar] [CrossRef]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular RNA Circ-CBFB Promotes Proliferation and Inhibits Apoptosis in Chronic Lymphocytic Leukemia through Regulating MiR-607/FZD3/Wnt/β-Catenin Pathway. Biochem. Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, H.; Wang, C.; Liu, W.; Liu, M.; Zhu, Y.; Xu, W.; Jin, H.; Li, J. Mitochondrial Genome-Derived CircRNA Mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol. Ther. Nucleic Acids 2020, 20, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The Exosome Journey: From Biogenesis to Uptake and Intracellular Signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Bahrami, A.; Moradi Binabaj, M.; Ferns, G.A. Exosomes: Emerging Modulators of Signal Transduction in Colorectal Cancer from Molecular Understanding to Clinical Application. Biomed. Pharmacother. 2021, 141, 111882. [Google Scholar] [CrossRef]
- Hussen, B.M.; Mohamadtahr, S.; Abdullah, S.R.; Hidayat, H.J.; Rasul, M.F.; Hama Faraj, G.S.; Ghafouri-Fard, S.; Taheri, M.; Khayamzadeh, M.; Jamali, E. Exosomal Circular RNAs: New Player in Breast Cancer Progression and Therapeutic Targets. Front. Genet. 2023, 14, 1126944. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zou, R.; Li, D.; Gao, X.; Lu, X. Exosomal CircSTRBP from Cancer Cells Facilitates Gastric Cancer Progression via Regulating MiR-1294/MiR-593-3p/E2F2 Axis. J. Cell Mol. Med. 2024, 28, e18217. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Han, J.; Zhang, X.; Zhang, X.; Song, J.; Gao, Z.; Qian, H.; Jin, J.; Liang, Z. Exosomal CircRNAs in Gastrointestinal Cancer: Role in Occurrence, Development, Diagnosis and Clinical Application (Review). Oncol. Rep. 2024, 51, 19. [Google Scholar] [CrossRef]
- Bomben, R.; Rossi, F.M.; Vit, F.; Bittolo, T.; Zucchetto, A.; Papotti, R.; Tissino, E.; Pozzo, F.; Degan, M.; Polesel, J.; et al. Clinical Impact of TP53 Disruption in Chronic Lymphocytic Leukemia Patients Treated with Ibrutinib: A Campus CLL Study. Leukemia 2023, 37, 914–918. [Google Scholar] [CrossRef]
- Wang, S.; Peng, Z.; Wang, S.; Yang, L.; Chen, Y.; Kong, X.; Song, S.; Pei, P.; Tian, C.; Yan, H.; et al. KRAB-Type Zinc-Finger Proteins PITA and PISA Specifically Regulate P53-Dependent Glycolysis and Mitochondrial Respiration. Cell Res. 2018, 28, 572–592. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, M.A.; Liu, J.; Pelicano, H.; Hammoudi, N.; Croce, C.M.; Keating, M.J.; Huang, P. Alterations of Mitochondrial Biogenesis in Chronic Lymphocytic Leukemia Cells with Loss of P53. Mitochondrion 2016, 31, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Jitschin, R.; Hofmann, A.D.; Bruns, H.; Gießl, A.; Bricks, J.; Berger, J.; Saul, D.; Eckart, M.J.; Mackensen, A.; Mougiakakos, D. Mitochondrial Metabolism Contributes to Oxidative Stress and Reveals Therapeutic Targets in Chronic Lymphocytic Leukemia. Blood 2014, 123, 2663–2672. [Google Scholar] [CrossRef]
- Yao, J. Carbonyl Cyanide 3-Chlorophenylhydrazone Promotes of Mitophagy in Gastric Cancer Cells MKN1 and MKN45 via PINK1/Parkin Pathway. 2024. [Google Scholar] [CrossRef]
- Fernández-Mosquera, L.; DIogo, C.V.; Yambire, K.F.; Santos, G.L.; Luna Sánchez, M.; Bénit, P.; Rustin, P.; Lopez, L.C.; Milosevic, I.; Raimundo, N. Acute and Chronic Mitochondrial Respiratory Chain Deficiency Differentially Regulate Lysosomal Biogenesis. Sci. Rep. 2017, 7, 45076. [Google Scholar] [CrossRef]
- Dijk, S.N.; Protasoni, M.; Elpidorou, M.; Kroon, A.M.; Taanman, J.W. Mitochondria as Target to Inhibit Proliferation and Induce Apoptosis of Cancer Cells: The Effects of Doxycycline and Gemcitabine. Sci. Rep. 2020, 10, 4363. [Google Scholar] [CrossRef]
- Scatena, C.; Roncella, M.; Di Paolo, A.; Aretini, P.; Menicagli, M.; Fanelli, G.; Marini, C.; Mazzanti, C.M.; Ghilli, M.; Sotgia, F.; et al. Doxycycline, an Inhibitor of Mitochondrial Biogenesis, Effectively Reduces Cancer Stem Cells (CSCs) in Early Breast Cancer Patients: A Clinical Pilot Study. Front. Oncol. 2018, 8, 452. [Google Scholar] [CrossRef] [PubMed]
- LaMoia, T.E.; Butrico, G.M.; Kalpage, H.A.; Goedeke, L.; Hubbard, B.T.; Vatner, D.F.; Gaspar, R.C.; Zhang, X.M.; Cline, G.W.; Nakahara, K.; et al. Metformin, Phenformin, and Galegine Inhibit Complex IV Activity and Reduce Glycerol-Derived Gluconeogenesis. Proc. Natl. Acad. Sci. USA 2022, 119, e212228711. [Google Scholar] [CrossRef] [PubMed]
- Madiraju, A.K.; Erion, D.M.; Rahimi, Y.; Zhang, X.M.; Braddock, D.T.; Albright, R.A.; Prigaro, B.J.; Wood, J.L.; Bhanot, S.; MacDonald, M.J.; et al. Metformin Suppresses Gluconeogenesis by Inhibiting Mitochondrial Glycerophosphate Dehydrogenase. Nature 2014, 510, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Turbov, J.; Rosales, R.; Thaete, L.G.; Rodriguez, G.C. Combination Simvastatin and Metformin Synergistically Inhibits Endometrial Cancer Cell Growth. Gynecol. Oncol. 2019, 154, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Tian, X.; Zhang, B.; Li, M.; Wang, Y.; Yang, C.; Wu, J.; Wei, X.; Qu, Q.; Yu, Y.; et al. Low-Dose Metformin Targets the Lysosomal AMPK Pathway through PEN2. Nature 2022, 603, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; An, H.; Liu, T.; Qin, C.; Sesaki, H.; Guo, S.; Radovick, S.; Hussain, M.; Maheshwari, A.; Wondisford, F.E.; et al. Metformin Improves Mitochondrial Respiratory Activity through Activation of AMPK. Cell Rep. 2019, 29, 1511–1523.e5. [Google Scholar] [CrossRef] [PubMed]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a Potent and Selective BCL-2 Inhibitor, Achieves Antitumor Activity While Sparing Platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Niedermeier, M.; Hennessy, B.T.; Knight, Z.A.; Henneberg, M.; Hu, J.; Kurtova, A.V.; Wierda, W.G.; Keating, M.J.; Shokat, K.M.; Burger, J.A. Isoform-Selective Phosphoinositide 3′-Kinase Inhibitors Inhibit CXCR4 Signaling and Overcome Stromal Cell-Mediated Drug Resistance in Chronic Lymphocytic Leukemia: A Novel Therapeutic Approach. Blood 2009, 113, 5549–5557. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, I.; Bouchard, E.D.J.; Beiggi, S.; Poeppl, A.G.; Johnston, J.B.; Gibson, S.B.; Banerji, V. On-Target Effect of FK866, a Nicotinamide Phosphoribosyl Transferase Inhibitor, by Apoptosis-Mediated Death in Chronic Lymphocytic Leukemia Cells. Clin. Cancer Res. 2014, 20, 4861–4872. [Google Scholar] [CrossRef]
- Bhagwat, S.V.; Gokhale, P.C.; Crew, A.P.; Cooke, A.; Yao, Y.; Mantis, C.; Kahler, J.; Workman, J.; Bittner, M.; Dudkin, L.; et al. Preclinical Characterization of OSI-027, a Potent and Selective Inhibitor of MTORC1 and MTORC2: Distinct from Rapamycin. Mol. Cancer Ther. 2011, 10, 1394–1406. [Google Scholar] [CrossRef]
- Chen, Z.; He, Q.; Lu, T.; Wu, J.; Shi, G.; He, L.; Zong, H.; Liu, B.; Zhu, P. McPGK1-Dependent Mitochondrial Import of PGK1 Promotes Metabolic Reprogramming and Self-Renewal of Liver TICs. Nat. Commun. 2023, 14, 1121. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Fang, Y.; Lyu, Z.; Zhu, Y.; Yang, L. Exploring the Dynamic Interplay between Cancer Stem Cells and the Tumor Microenvironment: Implications for Novel Therapeutic Strategies. J. Transl. Med. 2023, 21, 686. [Google Scholar] [CrossRef] [PubMed]
- Alnasser, S.M. Advances and Challenges in Cancer Stem Cells for Onco-Therapeutics. Stem Cells Int. 2023, 2023, 8722803. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Li, J.; Hu, S.; Deng, Y.; Yin, H.; Bao, X.; Zhang, Q.C.; Wang, G.; Wang, B.; et al. Identification of MecciRNAs and Their Roles in the Mitochondrial Entry of Proteins. Sci. China Life Sci. 2020, 63, 1429–1449. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; An, X.; Xiao, Y.; Sun, X.; Li, S.; Wang, Y.; Sun, W.; Yu, D. Mitochondrial-Related MicroRNAs and Their Roles in Cellular Senescence. Front. Physiol. 2024, 14, 1279548. [Google Scholar] [CrossRef] [PubMed]
- Özata, D.M.; Caramuta, S.; Velázquez-Fernández, D.; Akçakaya, P.; Xie, H.; Höög, A.; Zedenius, J.; Bäckdahl, M.; Larsson, C.; Lui, W.O. The Role of MicroRNA Deregulation in the Pathogenesis of Adrenocortical Carcinoma. Endocr. Relat. Cancer 2011, 18, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Duregon, E.; Rapa, I.; Votta, A.; Giorcelli, J.; Daffara, F.; Terzolo, M.; Scagliotti, G.V.; Volante, M.; Papotti, M. MicroRNA Expression Patterns in Adrenocortical Carcinoma Variants and Clinical Pathologic Correlations. Hum. Pathol. 2014, 45, 1555–1562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bai, G.; Zhu, W.; Bai, D.; Bi, G. Identification of MiRNA-MRNA Network Associated with Acute Myeloid Leukemia Survival. Med. Sci. Monit. 2017, 23, 4705–4714. [Google Scholar] [CrossRef]
- Zhao, Y.; Ye, G.; Wang, Y.; Luo, D. MiR-4461 Inhibits Tumorigenesis of Renal Cell Carcinoma by Targeting PPP1R3C. Cancer Biother. Radiopharm. 2022, 37, 503–514. [Google Scholar] [CrossRef]
- Nie, H.; Hu, X.; Xiong, H.; Zeng, L.; Chen, W.; Su, T. Change and Pathological Significance of Glycogen Content in Oral Squamous Cell Carcinoma and Oral Submucous Fibrosis. Tissue Cell 2024, 87, 102337. [Google Scholar] [CrossRef]
- Shen, G.M.; Zhang, F.L.; Liu, X.L.; Zhang, J.W. Hypoxia-Inducible Factor 1-Mediated Regulation of PPP1R3C Promotes Glycogen Accumulation in Human MCF-7 Cells under Hypoxia. FEBS Lett. 2010, 584, 4366–4372. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The Hallmarks of Cancer Metabolism: Still Emerging. Cell Metab. 2022, 34, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Ardizzoia, A.; Jemma, A.; Redaelli, S.; Silva, M.; Bentivegna, A.; Lavitrano, M.; Conconi, D. AhRR and PPP1R3C: Potential Prognostic Biomarkers for Serous Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 11455. [Google Scholar] [CrossRef]
- Lee, S.K.; Moon, J.W.; Lee, Y.W.; Lee, J.O.; Kim, S.J.; Kim, N.; Kim, J.; Kim, H.S.; Park, S.H. The Effect of High Glucose Levels on the Hypermethylation of Protein Phosphatase 1 Regulatory Subunit 3C (PPP1R3C) Gene in Colorectal Cancer. J. Genet. 2015, 94, 75–85. [Google Scholar] [CrossRef]
- Lee, H.S.; Yun, J.H.; Jung, J.; Yang, Y.; Kim, B.J.; Lee, S.J.; Yoon, J.H.; Moon, Y.; Kim, J.M.; Kwon, Y. Il Identification of Differentially-Expressed Genes by DNA Methylation in Cervical Cancer. Oncol. Lett. 2015, 9, 1691–1698. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Li, J.J.; Jiang, F.; Shi, W.J.; Chang, G.Y. MicroRNA-4461 Derived from Bone Marrow Mesenchymal Stem Cell Exosomes Inhibits Tumorigenesis by Downregulating COPB2 Expression in Colorectal Cancer. Biosci. Biotechnol. Biochem. 2020, 84, 338–346. [Google Scholar] [CrossRef]
- Yan, X.; Yang, P.; Liu, H.; Zhao, Y.; Wu, Z.; Zhang, B. MiR-4461 Inhibits the Progression of Gallbladder Carcinoma via Regulating EGFR/AKT Signaling. Cell Cycle 2022, 21, 1166–1177. [Google Scholar] [CrossRef]
- Dou, L.; Zhang, Y. MiR-4461 Regulates the Proliferation and Metastasis of Ovarian Cancer Cells and Cisplatin Resistance. Front. Oncol. 2021, 11, 614035. [Google Scholar] [CrossRef] [PubMed]
- Modesto, A.A.; Moraes, M.R.; Valente, C.M.; Costa, M.S.; Leal, D.F.; Pereira, E.E.; Fernandes, M.R.; Pinheiro, J.A.; Pantoja, K.B.; Moreira, F.C.; et al. Association between INDELs in MicroRNAs and Susceptibility to Gastric Cancer in Amazonian Population. Genes 2022, 14, 60. [Google Scholar] [CrossRef]
- Tan, J.; Lu, T.; Xu, J.; Hou, Y.; Chen, Z.; Zhou, K.; Ding, Y.; Jiang, B.; Zhu, Y. MicroRNA-4463 Facilitates the Development of Colon Cancer by Suppression of the Expression of PPP1R12B. Clin. Transl. Oncol. 2022, 24, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Tang, W.; Ye, W.; Song, L.; Chen, Z. ADAMTS9-AS2 Regulates PPP1R12B by Adsorbing MiR-196b-5p and Affects Cell Cycle-Related Signaling Pathways Inhibiting the Malignant Process of Esophageal Cancer. Cell Cycle 2022, 21, 1710–1725. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Tang, W.; Wu, H.; Fan, X.; Luo, J.; Feng, J.; Wen, K.; Wu, G. The PEAK1-PPP1R12B Axis Inhibits Tumor Growth and Metastasis by Regulating Grb2/PI3K/Akt Signalling in Colorectal Cancer. Cancer Lett. 2019, 442, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Fokkelman, M.; BalcloGlu, H.E.; Klip, J.E.; Yan, K.; Verbeek, F.J.; Danen, E.H.J.; Van De Water, B. Cellular Adhesome Screen Identifies Critical Modulators of Focal Adhesion Dynamics, Cellular Traction Forces and Cell Migration Behaviour. Sci. Rep. 2016, 6, 31707. [Google Scholar] [CrossRef] [PubMed]
- Kas, S.M.; De Ruiter, J.R.; Schipper, K.; Annunziato, S.; Schut, E.; Klarenbeek, S.; Drenth, A.P.; Van Der Burg, E.; Klijn, C.; Hoeve, J.J.T.; et al. Insertional Mutagenesis Identifies Drivers of a Novel Oncogenic Pathway in Invasive Lobular Breast Carcinoma. Nat. Genet. 2017, 49, 1219–1230. [Google Scholar] [CrossRef]
- Zhang, K.; Zhao, S.; Wang, Q.; Yang, H.S.; Zhu, J.; Ma, R. Identification of MicroRNAs in Nipple Discharge as Potential Diagnostic Biomarkers for Breast Cancer. Ann. Surg. Oncol. 2015, 22 (Suppl. S3), 536–544. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Zhang, W.; Chen, X.; Tian, Y.; Zhao, S.; Zhang, K.; Zhu, J.; Ma, R.; Wang, J. Breast Cancer Nipple Discharge Exosomal MicroRNAs Are Stable under Degradative Conditions. Chin. J. Physiol. 2023, 66, 181–187. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, X.; Han, X.; Pandey, V.; Lobie, P.E.; Zhu, T. The Potential of Long Noncoding RNAs for Precision Medicine in Human Cancer. Cancer Lett. 2021, 501, 12–19. [Google Scholar] [CrossRef]
| Silencing Strategy | Cell Line/Animal Model System 1 | Cellular Effects | Molecular Target and Regulatory Effect | Ref. |
|---|---|---|---|---|
| ASO-1537 | HeLa | Apoptosis induction. Cell proliferation and anchorage inhibition. | n/a | [142] |
| SiHa | Cell death induction. Anchorage inhibition | Survivin downregulation | ||
| H292 | Cell death induction | Survivin downregulation | ||
| SKMEL-2 | Cell death induction. Anchorage inhibition | Survivin and XIAP downregulation | ||
| PC3 | Cell death induction | Survivin and XIAP downregulation | ||
| OVCAR-3 | Cell death induction. Anchorage inhibition | n/a | ||
| MCF7 | Cell death induction | n/a | ||
| MDA-MB-231 | Cell death induction | n/a | ||
| HepG2 | Cell death induction | n/a | ||
| DU145 | Cell death induction | n/a | ||
| Caco-2 | Cell death induction | n/a | ||
| A498 | Cell death induction | n/a | ||
| U87 | Cell death induction | n/a | ||
| ASO-1537 | Primary and metastatic ccRCC cultures | Apoptosis induction. Cell proliferation and metastasis inhibition | n/a | [139] |
| NOD/SCID mice inoculated with primary and metastatic ccRCC cancer cells | Tumor growth inhibition | n/a | ||
| ASO-1537 | MDA-MB-231 | Apoptosis induction. Cell cycle arrest. Invasive capacity and stemness inhibition | CCNB1, CCND1, CDK1 CDK4 and Survivin downregulation hsa-miR-4485-3p, hsa-miR-4485-5p, hsa-miR-1973, hsa-miR-5096 and hsa-miR-3609 upregulation | [145] |
| MCF7 | Cell death induction. Invasive capacity and stemness inhibition | n/a | ||
| ZR-75-1 | Cell death induction. Invasive capacity and stemness inhibition | n/a | ||
| Balb/c mice inoculated with MDA-MB-231 cells | Tumor growth inhibition | n/a | ||
| ASO-1537 | MDA-MB-231 | AURKA, TOP2A downregulation | [146] | |
| MCF7 | AURKA, TOP2A downregulation | |||
| HMEC | AURKA, TOP2A Upregulation | |||
| ASO-1560 | B16F10 | Cell proliferation, stemness, invasiveness, anchorage and metastasis inhibition. Apoptosis induction | Survivin downregulation | [141] |
| C57BL/6 mice inoculated with B16F10 melanoma cells | Tumor growth and metastasis inhibition | Survivin downregulation | ||
| ASO-1232AS | RenCa | Cell proliferation, metastasis, anchorage, invasiveness inhibition. Apoptosis induction | Bcl-xL, Bcl-2, Survivin, N-cadherin and P-cadherin downregulation | [140] |
| Balb/c mice inoculated with RenCa cells | Tumor growth and metastasis inhibition. Survival increased | Survivin and MMP-9 downregulation | ||
| shRNA-Lv-sh-912 and shRNA-Lv-sh-1560 | B16F10 | Apoptosis induction | n/a | [138] |
| shRNA-Lv-sh-912 | A375 | Apoptosis induction | n/a | |
| shRNA-Lv-sh-1560 | C57BL6/J mice inoculated with B16F10 cells | Tumor growth, number and size metastasis reduction | n/a |
| Cell Line | AURKA | TOPO2A | |
|---|---|---|---|
| MDA-MB231 | T | ↓ | ↓ |
| P | ↓ | ↓ | |
| MCF-7 | T | ↓ | ↓ |
| P | ↓ | ↓ | |
| HMEC | T | ↓ | NC |
| P | ↑ | ↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piergentili, R.; Sechi, S. Non-Coding RNAs of Mitochondrial Origin: Roles in Cell Division and Implications in Cancer. Int. J. Mol. Sci. 2024, 25, 7498. https://doi.org/10.3390/ijms25137498
Piergentili R, Sechi S. Non-Coding RNAs of Mitochondrial Origin: Roles in Cell Division and Implications in Cancer. International Journal of Molecular Sciences. 2024; 25(13):7498. https://doi.org/10.3390/ijms25137498
Chicago/Turabian StylePiergentili, Roberto, and Stefano Sechi. 2024. "Non-Coding RNAs of Mitochondrial Origin: Roles in Cell Division and Implications in Cancer" International Journal of Molecular Sciences 25, no. 13: 7498. https://doi.org/10.3390/ijms25137498
APA StylePiergentili, R., & Sechi, S. (2024). Non-Coding RNAs of Mitochondrial Origin: Roles in Cell Division and Implications in Cancer. International Journal of Molecular Sciences, 25(13), 7498. https://doi.org/10.3390/ijms25137498







