Deciphering Drug Resistance: Investigating the Emerging Role of Hyaluronan Metabolism and Signaling and Tumor Extracellular Matrix in Cancer Chemotherapy
Abstract
1. Introduction
2. ECM and HA
3. HA and Its Interaction with the ECM in Cancer
4. Exploring New Roles of HA Metabolism in Cancer Treatment
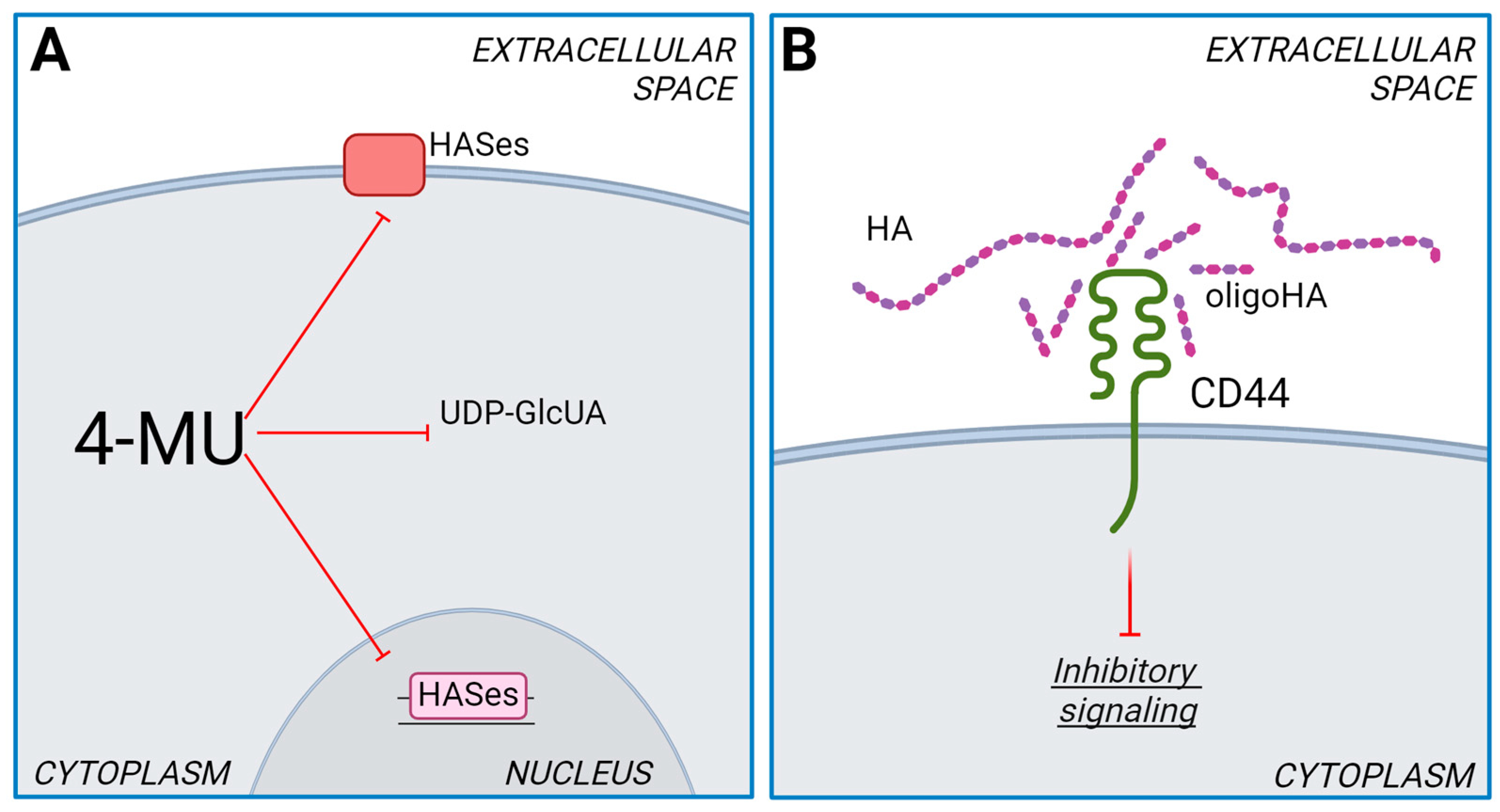
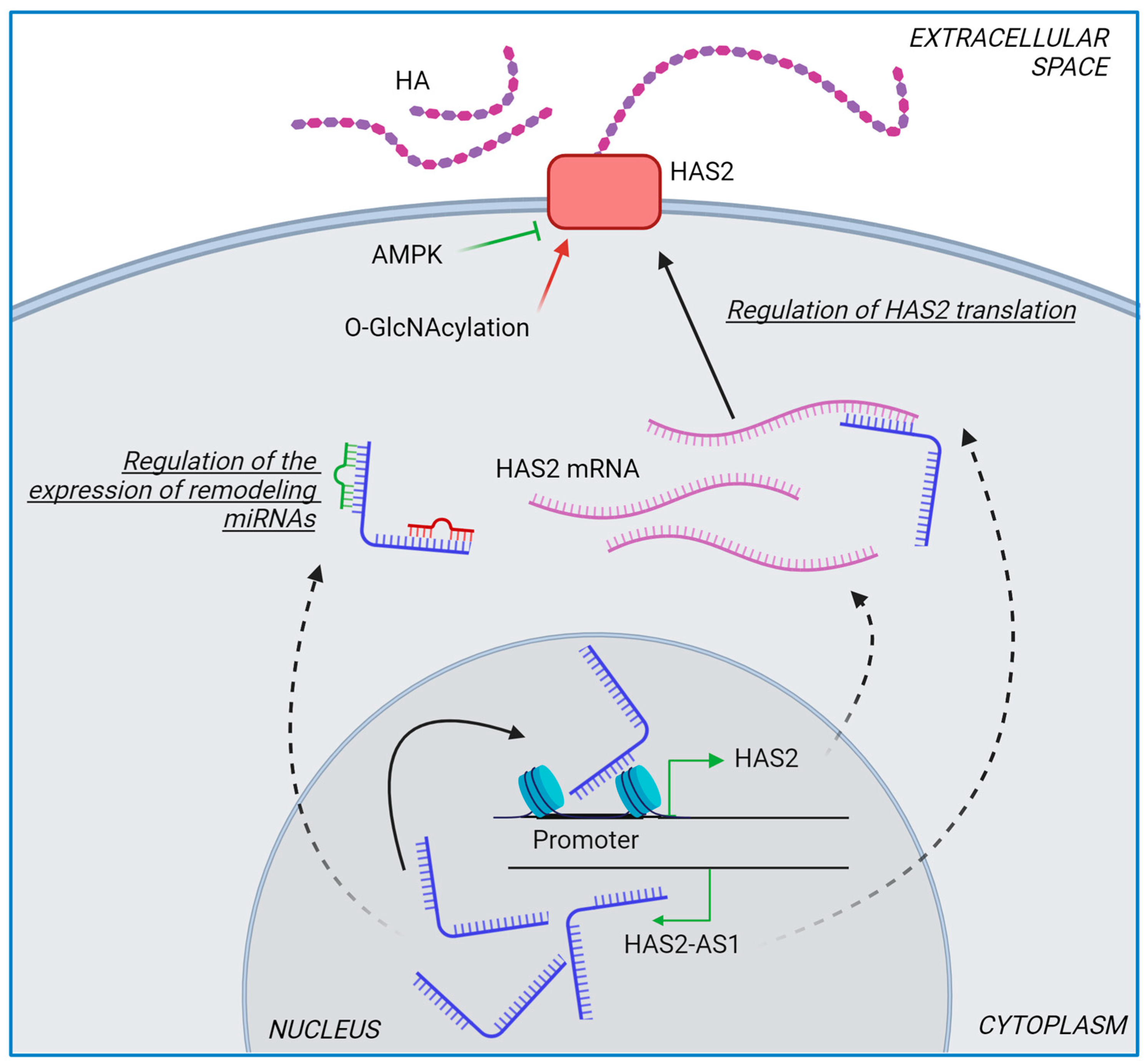
4.1. Role of HASes in Chemotherapy
4.2. Role of HYALs in Chemotherapy
4.3. Role of UGDH in Chemotherapy
4.4. Role of CD44-HA Interaction in Drug Resistance
5. Future Perspectives and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Zhou, X.; Yang, J.; Shi, H.; Li, H.; Zhao, X.; Ma, X. The Role of Tumor-Stroma Interactions in Drug Resistance Within Tumor Microenvironment. Front. Cell Dev. Biol. 2021, 9, 637675. [Google Scholar] [CrossRef] [PubMed]
- Eble, J.A.; Niland, S. The Extracellular Matrix in Tumor Progression and Metastasis. Clin. Exp. Metastasis 2019, 36, 171–198. [Google Scholar] [CrossRef] [PubMed]
- Caon, I.; Bartolini, B.; Parnigoni, A.; Caravà, E.; Moretto, P.; Viola, M.; Karousou, E.; Vigetti, D.; Passi, A. Revisiting the Hallmarks of Cancer: The Role of Hyaluronan. Semin. Cancer Biol. 2020, 62, 9–19. [Google Scholar] [CrossRef]
- Karousou, E.; Parnigoni, A.; Moretto, P.; Passi, A.; Viola, M.; Vigetti, D. Hyaluronan in the Cancer Cells Microenvironment. Cancers 2023, 15, 798. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, L.; Wang, L.; Lu, L.; Li, Y.; Huang, G.; Song, J. ‘Two-Faces’ of Hyaluronan, a Dynamic Barometer of Disease Progression in Tumor Microenvironment. Discov. Oncol. 2023, 14, 11. [Google Scholar] [CrossRef]
- Nagy, N.; Kuipers, H.F.; Marshall, P.L.; Wang, E.; Kabser, G.; Bollyky, P.L. Hyaluronan in Immune Dysregulation and Autoimmune Diseases. Matrix Biol. 2019, 78–79, 292–313. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Liang, J.; Noble, P.W. Hyaluronan in Tissue Injury and Repair. Annu. Rev. Cell Dev. Biol. 2007, 23, 435–461. [Google Scholar] [CrossRef]
- Cox, T.R. The Matrix in Cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef]
- Karamanos, N.K.; Piperigkou, Z.; Theocharis, A.D.; Watanabe, H.; Franchi, M.; Baud, S.; Brézillon, S.; Götte, M.; Passi, A.; Vigetti, D.; et al. Proteoglycan Chemical Diversity Drives Multifunctional Cell Regulation and Therapeutics. Chem. Rev. 2018, 118, 9152–9232. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular Matrix Structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, N.K.; Theocharis, A.D.; Piperigkou, Z.; Manou, D.; Passi, A.; Skandalis, S.S.; Vynios, D.H.; Orian-Rousseau, V.; Ricard-Blum, S.; Schmelzer, C.E.H.; et al. A Guide to the Composition and Functions of the Extracellular Matrix. FEBS J. 2021, 288, 6850–6912. [Google Scholar] [CrossRef] [PubMed]
- Franchi, M.; Piperigkou, Z.; Mastronikolis, N.S.; Karamanos, N. Extracellular Matrix Biomechanical Roles and Adaptation in Health and Disease. FEBS J. 2024, 291, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Parnigoni, A.; Viola, M.; Karousou, E.; Rovera, S.; Giaroni, C.; Passi, A.; Vigetti, D. Hyaluronan in Pathophysiology of Vascular Diseases: Specific Roles in Smooth Muscle Cells, Endothelial Cells, and Macrophages. Am. J. Physiol. Cell Physiol. 2022, 323, C505–C519. [Google Scholar] [CrossRef] [PubMed]
- Vigetti, D.; Karousou, E.; Viola, M.; Deleonibus, S.; De Luca, G.; Passi, A. Hyaluronan: Biosynthesis and Signaling. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 2452–2459. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Yamamoto, H.; Tobisawa, Y.; Irie, F. TMEM2: A Missing Link in Hyaluronan Catabolism Identified? Matrix Biol. 2019, 78–79, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Tavianatou, A.G.; Caon, I.; Franchi, M.; Piperigkou, Z.; Galesso, D.; Karamanos, N.K. Hyaluronan: Molecular Size-Dependent Signaling and Biological Functions in Inflammation and Cancer. FEBS J. 2019, 286, 2883–2908. [Google Scholar] [CrossRef] [PubMed]
- Cyphert, J.M.; Trempus, C.S.; Garantziotis, S. Size Matters: Molecular Weight Specificity of Hyaluronan Effects in Cell Biology. Int. J. Cell Biol. 2015, 2015, 563818. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Chanmee, T.; Itano, N. Hyaluronan: Metabolism and Function. Biomolecules 2020, 10, 1525. [Google Scholar] [CrossRef]
- Caon, I.; D’angelo, M.L.; Bartolini, B.; Caravà, E.; Parnigoni, A.; Contino, F.; Cancemi, P.; Moretto, P.; Karamanos, N.K.; Passi, A.; et al. The Secreted Protein C10orf118 Is a New Regulator of Hyaluronan Synthesis Involved in Tumour-Stroma Cross-Talk. Cancers 2021, 13, 1105. [Google Scholar] [CrossRef]
- Zhang, G.; Guo, L.; Yang, C.; Liu, Y.; He, Y.; Du, Y.; Wang, W.; Gao, F. A Novel Role of Breast Cancer-Derived Hyaluronan on Inducement of M2-like Tumor-Associated Macrophages Formation. Oncoimmunology 2016, 5, e1172154. [Google Scholar] [CrossRef] [PubMed]
- Kokoretsis, D.; Maniaki, E.K.; Kyriakopoulou, K.; Koutsakis, C.; Piperigkou, Z.; Karamanos, N.K. Hyaluronan as “Agent Smith” in Cancer Extracellular Matrix Pathobiology: Regulatory Roles in Immune Response, Cancer Progression and Targeting. IUBMB Life 2022, 74, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Hingorani, S.R. Hyaluronan, Fluid Pressure, and Stromal Resistance in Pancreas Cancer. Br. J. Cancer 2013, 108, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Kim, Y.K.; Kim, G.B.; Nam, G.H.; Kim, S.A.; Park, Y.; Yang, Y.; Kim, I.S. Degradation of Tumour Stromal Hyaluronan by Small Extracellular Vesicle-PH20 Stimulates CD103+ Dendritic Cells and in Combination with PD-L1 Blockade Boosts Anti-Tumour Immunity. J. Extracell. Vesicles 2019, 8, 1670893. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Jiang, D.; Noble, P.W. Hyaluronan as a Therapeutic Target in Human Diseases. Adv. Drug Deliv. Rev. 2016, 97, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Bin Emran, T.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.R.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug Resistance in Cancer: Role of ATP-Dependent Transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Ricciardelli, C.; Ween, M.P.; Lokman, N.A.; Tan, I.A.; Pyragius, C.E.; Oehler, M.K. Chemotherapy-Induced Hyaluronan Production: A Novel Chemoresistance Mechanism in Ovarian Cancer. BMC Cancer 2013, 13, 476. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.W.; Peyrollier, K.; Xia, W.; Gilad, E. Hyaluronan-CD44 Interaction Activates Stem Cell Marker Nanog, Stat-3-Mediated MDR1 Gene Expression, and Ankyrin-Regulated Multidrug Efflux in Breast and Ovarian Tumor Cells. J. Biol. Chem. 2008, 283, 17635–17651. [Google Scholar] [CrossRef]
- Slomiany, M.G.; Dai, L.; Bomar, P.A.; Knackstedt, T.J.; Kranc, D.A.; Tolliver, L.; Maria, B.L.; Toole, B.P. Abrogating Drug Resistance in Malignant Peripheral Nerve Sheath Tumors by Disrupting Hyaluronan-CD44 Interactions with Small Hyaluronan Oligosaccharides. Cancer Res. 2009, 69, 4992–4998. [Google Scholar] [CrossRef]
- Schulz, T.; Schumacher, U.; Prehm, P. Hyaluronan Export by the ABC Transporter MRP5 and Its Modulation by Intracellular CGMP. J. Biol. Chem. 2007, 282, 20999–21004. [Google Scholar] [CrossRef] [PubMed]
- Prehm, P.; Schumacher, U. Inhibition of Hyaluronan Export from Human Fibroblasts by Inhibitors of Multidrug Resistance Transporters. Biochem. Pharmacol. 2004, 68, 1401–1410. [Google Scholar] [CrossRef]
- Vitale, D.L.; Spinelli, F.M.; Del Dago, D.; Icardi, A.; Demarchi, G.; Caon, I.; García, M.; Bolontrade, M.F.; Passi, A.; Cristina, C.; et al. Co-Treatment of Tumor Cells with Hyaluronan plus Doxorubicin Affects Endothelial Cell Behavior Independently of VEGF Expression. Oncotarget 2018, 9, 36585–36602. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rajabi, M.; Mousa, S.A. The Role of Angiogenesis in Cancer Treatment. Biomedicines 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Shackleton, M.; Quintana, E.; Fearon, E.R.; Morrison, S.J. Heterogeneity in Cancer: Cancer Stem Cells versus Clonal Evolution. Cell 2009, 138, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.; Wilson, G.D. The Importance of CD44 as a Stem Cell Biomarker and Therapeutic Target in Cancer. Stem Cells Int. 2016, 2016, 2087204. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Izumikawa, T.; Higashide, M.; Mochizuki, N.; Chokchaitaweesuk, C.; Khansai, M.; Nakajima, K.; Kakizaki, I.; Kongtawelert, P.; et al. Hyaluronan Production Regulates Metabolic and Cancer Stem-like Properties of Breast Cancer Cells via Hexosamine Biosynthetic Pathway-Coupled HIF-1 Signaling. J. Biol. Chem. 2016, 291, 24105–24120. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Cuevas, C.; Chang, A.E.; Goel, V.K.; Von Hoff, D.D.; Hingorani, S.R. Enzymatic Targeting of the Stroma Ablates Physical Barriers to Treatment of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2012, 21, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hou, P.; Fang, J.; Zhu, J.; Zha, J.; Liu, R.; Ding, Y.; Zuo, M.; Li, P.; Cao, L.; et al. Mesenchymal Stromal Cells Confer Breast Cancer Doxorubicin Resistance by Producing Hyaluronan. Oncogene 2023, 42, 3221–3235. [Google Scholar] [CrossRef]
- Kohi, S.; Sato, N.; Cheng, X.B.; Koga, A.; Higure, A.; Hirata, K. A Novel Epigenetic Mechanism Regulating Hyaluronan Production in Pancreatic Cancer Cells. Clin. Exp. Metastasis 2016, 33, 225–230. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Earle, C.; Shiina, M. Activation of Matrix Hyaluronan-Mediated CD44 Signaling, Epigenetic Regulation and Chemoresistance in Head and Neck Cancer Stem Cells. Int. J. Mol. Sci. 2017, 18, 1849. [Google Scholar] [CrossRef]
- Mijnes, J.; Veeck, J.; Gaisa, N.T.; Burghardt, E.; de Ruijter, T.C.; Gostek, S.; Dahl, E.; Pfister, D.; Schmid, S.C.; Knüchel, R.; et al. Promoter Methylation of DNA Damage Repair (DDR) Genes in Human Tumor Entities: RBBP8/CtIP Is Almost Exclusively Methylated in Bladder Cancer. Clin. Epigenetics 2018, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, O.A.; Esteller, M. Epigenetic Modifications in Breast Cancer and Their Role in Personalized Medicine. Am. J. Pathol. 2013, 183, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. We Need to Talk about the Warburg Effect. Nat. Metab. 2020, 2, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Vigetti, D.; Deleonibus, S.; Moretto, P.; Bowen, T.; Fischer, J.W.; Grandoch, M.; Oberhuber, A.; Love, D.C.; Hanover, J.A.; Cinquetti, R.; et al. Natural Antisense Transcript for Hyaluronan Synthase 2 (HAS2-AS1) Induces Transcription of HAS2 via Protein O-GlcNAcylation. J. Biol. Chem. 2014, 289, 28816–28826. [Google Scholar] [CrossRef] [PubMed]
- Vigetti, D.; Deleonibus, S.; Moretto, P.; Karousou, E.; Viola, M.; Bartolini, B.; Hascall, V.C.; Tammi, M.; De Luca, G.; Passi, A. Role of UDP-N-Acetylglucosamine (GlcNAc) and O-GlcNacylation of Hyaluronan Synthase 2 in the Control of Chondroitin Sulfate and Hyaluronan Synthesis. J. Biol. Chem. 2012, 287, 35544–35555. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; Sakamoto, K.; Bayascas, J.R. LKB1-Dependent Signaling Pathways. Annu. Rev. Biochem. 2006, 75, 137–163. [Google Scholar] [CrossRef] [PubMed]
- Fnu, G.; Weber, G.F. Osteopontin Induces Mitochondrial Biogenesis in Deadherent Cancer Cells. Oncotarget 2023, 14, 957–969. [Google Scholar] [CrossRef]
- Thanee, M.; Dokduang, H.; Kittirat, Y.; Phetcharaburanin, J.; Klanrit, P.; Titapun, A.; Namwat, N.; Khuntikeo, N.; Wangwiwatsin, A.; Saya, H.; et al. CD44 Modulates Metabolic Pathways and Altered ROS-Mediated Akt Signal Promoting Cholangiocarcinoma Progression. PLoS ONE 2021, 16, e0245871. [Google Scholar] [CrossRef]
- Nam, K.S.; Sunhwa, O.H.; Shin, I. Ablation of CD44 Induces Glycolysis-To-Oxidative Phosphorylation Transition via Modulation of the c-Src-Akt-LKB1-AMPKα Pathway. Biochem. J. 2016, 473, 3013–3030. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordat, J.P.; Maire, P.; Giallongo, A. Hypoxia Response Elements in the Aldolase A, Enolase 1, and Lactate Dehydrogenase a Gene Promoters Contain Essential Binding Sites for Hypoxia-Inducible Factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, F.M.; Vitale, D.L.; Sevic, I.; Alaniz, L. Hyaluronan in the Tumor Microenvironment. In Tumor Microenvironment: Extracellular Matrix Components–Part A; Springer: Cham, Switzerland, 2020; Volume 1245. [Google Scholar]
- Parnigoni, A.; Moretto, P.; Viola, M.; Karousou, E.; Passi, A.; Vigetti, D. Effects of Hyaluronan on Breast Cancer Aggressiveness. Cancers 2023, 15, 3813. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.; Kuipers, H.F.; Frymoyer, A.R.; Ishak, H.D.; Bollyky, J.B.; Wight, T.N.; Bollyky, P.L. 4-Methylumbelliferone Treatment and Hyaluronan Inhibition as a Therapeutic Strategy in Inflammation, Autoimmunity, and Cancer. Front. Immunol. 2015, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.; Gurevich, I.; Kuipers, H.F.; Ruppert, S.M.; Marshall, P.L.; Xie, B.J.; Sun, W.; Malkovskiy, A.V.; Rajadas, J.; Grandoch, M.; et al. 4-Methylumbelliferyl Glucuronide Contributes to Hyaluronan Synthesis Inhibition. J. Biol. Chem. 2019, 294, 7864–7877. [Google Scholar] [CrossRef]
- Kakizaki, I.; Kojima, K.; Takagaki, K.; Endo, M.; Kannagi, R.; Ito, M.; Maruo, Y.; Sato, H.; Yasuda, T.; Mita, S.; et al. A Novel Mechanism for the Inhibition of Hyaluronan Biosynthesis by 4-Methylumbelliferone. J. Biol. Chem. 2004, 279, 33281–33289. [Google Scholar] [CrossRef] [PubMed]
- Vigetti, D.; Rizzi, M.; Viola, M.; Karousou, E.; Genasetti, A.; Clerici, M.; Bartolini, B.; Hascall, V.C.; De Luca, G.; Passi, A. The Effects of 4-Methylumbelliferone on Hyaluronan Synthesis, MMP2 Activity, Proliferation, and Motility of Human Aortic Smooth Muscle Cells. Glycobiology 2009, 19, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Caon, I.; Parnigoni, A.; Viola, M.; Karousou, E.; Passi, A.; Vigetti, D. Cell Energy Metabolism and Hyaluronan Synthesis. J. Histochem. Cytochem. 2021, 69, 35–47. [Google Scholar] [CrossRef]
- Caon, I.; Bartolini, B.; Moretto, P.; Parnigoni, A.; Caravà, E.; Vitale, D.L.; Alaniz, L.; Viola, M.; Karousou, E.; de Luca, G.; et al. Sirtuin 1 Reduces Hyaluronan Synthase 2 Expression by Inhibiting Nuclear Translocation of NF-ΚB and Expression of the Long-Noncoding RNA HAS2–AS1. J. Biol. Chem. 2020, 295, 3485–3496. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Matsumoto, S.; Fujita, Y.; Kuroda, A.; Menju, T.; Sonobe, M.; Kondo, N.; Torii, I.; Nakano, T.; Lara, P.N.; et al. Trametinib plus 4-Methylumbelliferone Exhibits Antitumor Effects by ERK Blockade and CD44 Downregulation and Affects PD-1 and PD-L1 in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2017, 12, 477–490. [Google Scholar] [CrossRef]
- Ricciardelli, C.; Lokman, N.A.; Sabit, I.; Gunasegaran, K.; Bonner, W.M.; Pyragius, C.E.; Macpherson, A.M.; Oehler, M.K. Novel Ex Vivo Ovarian Cancer Tissue Explant Assay for Prediction of Chemosensitivity and Response to Novel Therapeutics. Cancer Lett. 2018, 421, 51–58. [Google Scholar] [CrossRef]
- Malvicini, M.; Fiore, E.; Ghiaccio, V.; Piccioni, F.; Rizzo, M.; Olmedo Bonadeo, L.; García, M.; Rodríguez, M.; Bayo, J.; Peixoto, E.; et al. Tumor Microenvironment Remodeling by 4-Methylumbelliferone Boosts the Antitumor Effect of Combined Immunotherapy in Murine Colorectal Carcinoma. Mol. Ther. 2015, 23, 1444–1455. [Google Scholar] [CrossRef] [PubMed]
- Vitale, D.L.; Icardi, A.; Rosales, P.; Spinelli, F.M.; Sevic, I.; Alaniz, L.D. Targeting the Tumor Extracellular Matrix by the Natural Molecule 4-Methylumbelliferone: A Complementary and Alternative Cancer Therapeutic Strategy. Front. Oncol. 2021, 11, 710061. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Cao, M.; Liu, H.; He, Y.; Xu, J.; Du, Y.; Liu, Y.; Wang, W.; Cui, L.; Hu, J.; et al. The High and Low Molecular Weight Forms of Hyaluronan Have Distinct Effects on CD44 Clustering. J. Biol. Chem. 2012, 287, 43094–43107. [Google Scholar] [CrossRef] [PubMed]
- Rosser, J.I.; Nagy, N.; Goel, R.; Kaber, G.; Demirdjian, S.; Saxena, J.; Bollyky, J.B.; Frymoyer, A.R.; Pacheco-Navarro, A.E.; Burgener, E.B.; et al. Oral Hymecromone Decreases Hyaluronan in Human Study Participants. J. Clin. Investig. 2022, 132, e157983. [Google Scholar] [CrossRef] [PubMed]
- Itano, N.; Sawai, T.; Yoshida, M.; Lenas, P.; Yamada, Y.; Imagawa, M.; Shinomura, T.; Hamaguchi, M.; Yoshida, Y.; Ohnuki, Y.; et al. Three Isoforms of Mammalian Hyaluronan Synthases Have Distinct Enzymatic Properties. J. Biol. Chem. 1999, 274, 25085–25092. [Google Scholar] [CrossRef] [PubMed]
- Parnigoni, A.; Caon, I.; Teo, W.X.; Hua, S.H.; Moretto, P.; Bartolini, B.; Viola, M.; Karousou, E.; Yip, G.W.; Götte, M.; et al. The Natural Antisense Transcript HAS2-AS1 Regulates Breast Cancer Cells Aggressiveness Independently from Hyaluronan Metabolism. Matrix Biol. 2022, 109, 140–161. [Google Scholar] [CrossRef] [PubMed]
- Passi, A.; Vigetti, D.; Buraschi, S.; Iozzo, R.V. Dissecting the Role of Hyaluronan Synthases in the Tumor Microenvironment. FEBS J. 2019, 286, 2937–2949. [Google Scholar] [CrossRef] [PubMed]
- Parnigoni, A.; Caon, I.; Moretto, P.; Viola, M.; Karousou, E.; Passi, A.; Vigetti, D. The Role of the Multifaceted Long Non-Coding RNAs: A Nuclear-Cytosolic Interplay to Regulate Hyaluronan Metabolism. Matrix Biol. Plus 2021, 11, 100060. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Sun, L.; Cui, J.; Liu, L.; He, Q. Long Noncoding RNA HAS2-AS1 Accelerates Non-Small Cell Lung Cancer Chemotherapy Resistance by Targeting LSD1/EphB3 Pathway. Am. J. Transl. Res. 2020, 12, 950–958. [Google Scholar]
- Locke, K.W. ENHANZE® Drug Delivery Technology: A Novel Approach to Subcutaneous Administration Using Recombinant Human Hyaluronidase PH20. Drug Deliv. 2019, 26, 98–106, Erratum in Drug Deliv. 2019, 26. [Google Scholar] [CrossRef]
- European Medicines Agency. Herceptin Summary of Product Characteristics; European Medicines Agency: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Roche Canada. Pr HERCEPTINVR SC Product Monograph; Roche Canada: Laval, QC, Canada, 2018. [Google Scholar]
- Halozyme Therapeutics Inc. FDA Accepts Biologics License Application for Subcutaneous Formulation of Herceptin; Halozyme Therapeutics Inc.: San Diego, CA, USA, 2018. [Google Scholar]
- Zhao, M.; Liu, J.; Tang, Y.; Zhang, L.; Ge, X.; Chen, M.; Wen, Q.; Zhu, L.; Ma, Q. Hyaluronidase Responsive Second Near-Infrared Fluorescent Nanocomplex for Combined HER2 Blockade and Chemotherapy of HER2+ Breast Cancer. Biomater. Adv. 2022, 141, 213115. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. MabThera Summary of Product Characteristics; European Medicines Agency: Amsterdam, The Netherlands, 2018. [Google Scholar]
- U.S. Food and Drug Administration. Rituxan Hycela Fda Approval; U.S. Food and Drug Administration: Rockville, MD, USA, 2017. [Google Scholar]
- Roche Canada. Pr Rituxanvr SC Product Monograph; Roche Canada: Laval, QC, Canada, 2018. [Google Scholar]
- Veronese, F.M.; Mero, A. The Impact of PEGylation on Biological Therapies. BioDrugs 2008, 22, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Morosi, L.; Meroni, M.; Ubezio, P.; Fuso Nerini, I.; Minoli, L.; Porcu, L.; Panini, N.; Colombo, M.; Blouw, B.; Kang, D.W.; et al. PEGylated Recombinant Human Hyaluronidase (PEGPH20) Pre-Treatment Improves Intra-Tumour Distribution and Efficacy of Paclitaxel in Preclinical Models. J. Exp. Clin. Cancer Res. 2021, 40, 286. [Google Scholar] [CrossRef] [PubMed]
- Ko, A.H.; Kim, K.P.; Siveke, J.T.; Lopez, C.D.; Lacy, J.; O’Reilly, E.M.; Macarulla, T.; Manji, G.A.; Lee, J.; Ajani, J.; et al. Atezolizumab Plus PEGPH20 Versus Chemotherapy in Advanced Pancreatic Ductal Adenocarcinoma and Gastric Cancer: MORPHEUS Phase Ib/II Umbrella Randomized Study Platform. Oncologist 2023, 28, 553-e472. [Google Scholar] [CrossRef] [PubMed]
- Burotto, M.; Zvirbule, Z.; Mochalova, A.; Runglodvatana, Y.; Herraez-Baranda, L.; Liu, S.N.; Chan, P.; Shearer-Kang, E.; Liu, X.; Tosti, N.; et al. IMscin001 Part 2: A Randomised Phase III, Open-Label, Multicentre Study Examining the Pharmacokinetics, Efficacy, Immunogenicity, and Safety of Atezolizumab Subcutaneous versus Intravenous Administration in Previously Treated Locally Advanced or Metastatic Non-Small-Cell Lung Cancer and Pharmacokinetics Comparison with Other Approved Indications. Ann. Oncol. 2023, 34, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Clinical Trials. Gov—NIH—National Library of Medicine, National Center for Biotechnology Information. Available online: https://clinicaltrials.gov/search?cond=cancer&intr=hyaluronidase&viewType=Table&aggFilters=status:act%20rec%20not&term=chemotherapy (accessed on 16 May 2024).
- Bazan-Peregrino, M.; Garcia-Carbonero, R.; Laquente, B.; Álvarez, R.; Mato-Berciano, A.; Gimenez-Alejandre, M.; Morgado, S.; Rodríguez-García, A.; Maliandi, M.V.; Riesco, M.C.; et al. VCN-01 Disrupts Pancreatic Cancer Stroma and Exerts Antitumor Effects. J. Immunother. Cancer 2021, 9, e003254. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; Xu, C.; Dong, J.; Wei, J. An Oncolytic Vaccinia Virus Encoding Hyaluronidase Reshapes the Extracellular Matrix to Enhance Cancer Chemotherapy and Immunotherapy. J. Immunother. Cancer 2024, 12, e008431. [Google Scholar] [CrossRef] [PubMed]
- Price, M.J.; Nguyen, A.D.; Byemerwa, J.K.; Flowers, J.; Baëta, C.D.; Goodwin, C.R. UDP-Glucose Dehydrogenase (UGDH) in Clinical Oncology and Cancer Biology. Oncotarget 2023, 14, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Sharma, A.K.; Dellinger, R.W.; Blevins-Primeau, A.S.; Balliet, R.M.; Chen, G.; Boyiri, T.; Amin, S.; Lazarus, P. Glucuronidation of Active Tamoxifen Metabolites by the Human UDP Glucuronosyltransferases. Drug Metab. Dispos. 2007, 35, 2006–2014. [Google Scholar] [CrossRef]
- Sten, T.; Bichlmaier, I.; Kuuranne, T.; Leinonen, A.; Yli-Kauhaluoma, J.; Finel, M. UDP-Glucuronosyltransferases (UGTs) 2B7 and UGT2B17 Display Converse Specificity in Testosterone and Epitestosterone Glucuronidation, Whereas UGT2A1 Conjugates Both Androgens Similarly. Drug Metab. Dispos. 2009, 37, 417–423. [Google Scholar] [CrossRef]
- Sawyer, M.B.; Pituskin, E.; Damaraju, S.; Bies, R.R.; Vos, L.J.; Prado, C.M.M.; Kuzma, M.; Scarfe, A.G.; Clemons, M.; Tonkin, K.; et al. A Uridine Glucuronosyltransferase 2B7 Polymorphism Predicts Epirubicin Clearance and Outcomes in Early-Stage Breast Cancer. Clin. Breast Cancer 2016, 16, 139–144.e3. [Google Scholar] [CrossRef]
- Zhang, W.; Gou, P.; Dupret, J.M.; Chomienne, C.; Rodrigues-Lima, F. Etoposide, an Anticancer Drug Involved in Therapy-Related Secondary Leukemia: Enzymes at Play. Transl. Oncol. 2021, 14, 101169. [Google Scholar] [CrossRef] [PubMed]
- Dean, L. Irinotecan Therapy and UGT1A1 Genotype. In Medical Genetics Summaries; National Center for Biotechnology Information: Bethesda, MD, USA, 2012. [Google Scholar]
- Vigetti, D.; Ori, M.; Viola, M.; Genasetti, A.; Karousou, E.; Rizzi, M.; Pallotti, F.; Nardi, I.; Hascall, V.C.; De Luca, G.; et al. Molecular Cloning and Characterization of UDP-Glucose Dehydrogenase from the Amphibian Xenopus Laevis and Its Involvement in Hyaluronan Synthesis. J. Biol. Chem. 2006, 281, 8254–8263. [Google Scholar] [CrossRef] [PubMed]
- Egger, S.; Chaikuad, A.; Kavanagh, K.L.; Oppermann, U.; Nidetzky, B. UDP-Glucose Dehydrogenase: Structure and Function of a Potential Drug Target. Biochem. Soc. Trans. 2010, 38, 1378–1385. [Google Scholar] [CrossRef] [PubMed]
- Tukey, R.H.; Strassburg, C.P. Human UDP-Glucuronosyltransferases: Metabolism, Expression, and Disease. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 581–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, C.; Bou-Tayeh, B.; Lisle, R.; Kriaucionis, S.; Shi, Y. UXS1 Regulates UDP-GlcA Levels to Support Growth of UGDH-High Cancer Cells. bioRxiv, 2024; bioRxiv:08.579288. [Google Scholar]
- Wei, Q.; Galbenus, R.; Raza, A.; Cerny, R.L.; Simpson, M.A. Androgen-Stimulated UDP-Glucose Dehydrogenase Expression Limits Prostate Androgen Availability without Impacting Hyaluronan Levels. Cancer Res. 2009, 69, 2332–2339. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, B.M.; Howell, M.E.; Ma, L.; Enders, J.R.; Lehman, D.; Corey, E.; Barycki, J.J.; Simpson, M.A. Altered Glucuronidation Deregulates Androgen Dependent Response Profiles and Signifies Castration Resistance in Prostate Cancer. Oncotarget 2021, 12, 1886–1902. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Casale, G.P.; Tian, J.; Lele, S.M.; Pisarev, V.M.; Simpson, M.A.; Hemstreet, G.P. UDP-Glucose Dehydrogenase as a Novel Field-Specific Candidate Biomarker of Prostate Cancer. Int. J. Cancer 2010, 126, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Khasraw, M.; Bell, R.; Dang, C. Epirubicin: Is It like Doxorubicin in Breast Cancer? A Clinical Review. Breast 2012, 21, 142–149. [Google Scholar] [CrossRef]
- Sun, W.L.; Chen, J.; Wang, Y.P.; Zheng, H. Autophagy Protects Breast Cancer Cells from Epirubicin-Induced Apoptosis and Facilitates Epirubicin-Resistance Development. Autophagy 2011, 7, 1035–1044. [Google Scholar] [CrossRef]
- Vitale, D.L.; Caon, I.; Parnigoni, A.; Sevic, I.; Spinelli, F.M.; Icardi, A.; Passi, A.; Vigetti, D.; Alaniz, L. Initial Identification of UDP-Glucose Dehydrogenase as a Prognostic Marker in Breast Cancer Patients, Which Facilitates Epirubicin Resistance and Regulates Hyaluronan Synthesis in MDA-MB-231 Cells. Biomolecules 2021, 11, 246. [Google Scholar] [CrossRef] [PubMed]
- Oyinlade, O.; Wei, S.; Lal, B.; Laterra, J.; Zhu, H.; Goodwin, C.R.; Wang, S.; Ma, D.; Wan, J.; Xia, S. Targeting UDP-α-d-Glucose 6-Dehydrogenase Inhibits Glioblastoma Growth and Migration. Oncogene 2018, 37, 2615–2629. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Chanukuppa, V.; Reddy, P.J.; Taunk, K.; Adhav, R.; Srivastava, S.; Santra, M.K.; Rapole, S. Global Proteomic Profiling Identifies Etoposide Chemoresistance Markers in Non-Small Cell Lung Carcinoma. J. Proteom. 2016, 138, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Xu, X.; Shao, M.; Yang, X.; He, G.; Qi, K.; Gu, J.; Wang, L. UDP-Glucose 6-Dehydrogenase Lessens Sorafenib Sensitivity via Modulating Unfolded Protein Response. Biochem. Biophys. Res. Commun. 2022, 613, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Harrington, B.S.; Kamdar, R.; Ning, F.; Korrapati, S.; Caminear, M.W.; Hernandez, L.F.; Butcher, D.; Edmondson, E.F.; Traficante, N.; Hendley, J.; et al. UGDH Promotes Tumor-Initiating Cells and a Fibroinflammatory Tumor Microenvironment in Ovarian Cancer. J. Exp. Clin. Cancer Res. 2023, 42, 270. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.P.; Zheng, C.C.; Huang, Y.N.; He, M.L.; Xu, W.W.; Li, B. Molecular Mechanisms of Chemo- and Radiotherapy Resistance and the Potential Implications for Cancer Treatment. MedComm 2021, 2, 315–340. [Google Scholar] [CrossRef] [PubMed]
- Frąszczak, K.; Barczyński, B. The Role of Cancer Stem Cell Markers in Ovarian Cancer. Cancers 2024, 16, 40. [Google Scholar] [CrossRef]
- Mazerska, Z.; Mróz, A.; Pawłowska, M.; Augustin, E. The Role of Glucuronidation in Drug Resistance. Pharmacol. Ther. 2016, 159, 35. [Google Scholar] [CrossRef]
- Fraunhoffer, N.A.; Abuelafia, A.M.; Chanez, B.; Bigonnet, M.; Gayet, O.; Roques, J.; Chuluyan, E.; Dusetti, N.; Iovanna, J. Inhibition of Glucuronidation in Pancreatic Cancer Improves Gemcitabine Anticancer Activity. Cancer Commun. 2022, 42, 1212–1216. [Google Scholar] [CrossRef]
- Cummings, J.; Ethell, B.T.; Jardine, L.; Boyd, G.; Macpherson, J.S.; Burchell, B.; Smyth, J.F.; Jodrell, D.I. Glucuronidation as a Mechanism of Intrinsic Drug Resistance in Human Colon Cancer: Reversal of Resistance by Food Additives. Cancer Res. 2003, 63, 8443–8450. [Google Scholar]
- Toole, B.P.; Slomiany, M.G. Hyaluronan: A Constitutive Regulator of Chemoresistance and Malignancy in Cancer Cells. Semin. Cancer Biol. 2008, 18, 244–250. [Google Scholar] [CrossRef]
- Liu, Z.; Hou, P.; Fang, J.; Shao, C.; Shi, Y.; Melino, G.; Peschiaroli, A. Hyaluronic Acid Metabolism and Chemotherapy Resistance: Recent Advances and Therapeutic Potential. Mol. Oncol. 2023; early view. [Google Scholar]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The Biology and Role of CD44 in Cancer Progression: Therapeutic Implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S. CD44 Intracellular Domain: A Long Tale of a Short Tail. Cancers 2023, 15, 5041. [Google Scholar] [CrossRef]
- Iczkowski, K.A. Cell Adhesion Molecule CD44: Its Functional Roles in Prostate Cancer. Am. J. Transl. Res. 2011, 3, 1–7. [Google Scholar]
- Vuorio, J.; Škerlová, J.; Fábry, M.; Veverka, V.; Vattulainen, I.; Řezáčová, P.; Martinez-Seara, H. N-Glycosylation Can Selectively Block or Foster Different Receptor–Ligand Binding Modes. Sci. Rep. 2021, 11, 5239. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, K.; Hackert, T.; Zöller, M. CD44/CD44v6 a Reliable Companion in Cancer-Initiating Cell Maintenance and Tumor Progression. Front. Cell Dev. Biol. 2018, 6, 97. [Google Scholar] [CrossRef]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K.; Götte, M.; Vigetti, D.; Passi, A.; Karamanos, N.K.; Skandalis, S.S. Roles and Targeting of the HAS/Hyaluronan/CD44 Molecular System in Cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar] [CrossRef]
- Sionov, R.V.; Naor, D. Hyaluronan-Independent Lodgment of CD44+ Lymphoma Cells in Lymphoid Organs. Int. J. Cancer 1997, 71, 462–469. [Google Scholar] [CrossRef]
- Wang, S.J.; Bourguignon, L.Y.W. Role of Hyaluronan-Mediated CD44 Signaling in Head and Neck Squamous Cell Carcinoma Progression and Chemoresistance. Am. J. Pathol. 2011, 178, 956–963. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Spevak, C.C.; Wong, G.; Xia, W.; Gilad, E. Hyaluronan-CD44 Interaction with Protein Kinase Cε Promotes Oncogenic Signaling by the Stem Cell Marker Nanog and the Production of MicroRNA-21, Leading to down-Regulation of the Tumor Suppressor Protein PDCD4, Anti-Apoptosis, and Chemotherapy Resistance in Breast Tumor Cells. J. Biol. Chem. 2009, 284, 26533–26546. [Google Scholar] [CrossRef]
- Zhang, H.; Ren, L.; Ding, Y.; Li, F.; Chen, X.; Ouyang, Y.; Zhang, Y.; Zhang, D. Hyaluronan-Mediated Motility Receptor Confers Resistance to Chemotherapy via TGFβ/Smad2-Induced Epithelial-Mesenchymal Transition in Gastric Cancer. FASEB J. 2019, 33, 6365–6377. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.W.; Xia, W.; Wong, G. Hyaluronan-Mediated CD44 Interaction with P300 and SIRT1 Regulates β-Catenin Signaling and NFκB-Specific Transcription Activity Leading to MDR1 and Bcl-XL Gene Expression and Chemoresistance in Breast Tumor Cells. J. Biol. Chem. 2009, 284, 2657–2671. [Google Scholar] [CrossRef] [PubMed]
- Compagnone, M.; Gatti, V.; Presutti, D.; Ruberti, G.; Fierro, C.; Markert, E.K.; Vousden, K.H.; Zhou, H.; Mauriello, A.; Anemone, L.; et al. ΔNp63-Mediated Regulation of Hyaluronic Acid Metabolism and Signaling Supports HNSCC Tumorigenesis. Proc. Natl. Acad. Sci. USA 2017, 114, 13254–13259. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, R.; Takahashi, F.; Cui, R.; Yoshioka, M.; Gu, T.; Sasaki, S.; Tominaga, S.; Nishio, K.; Tanabe, K.K.; Takahashi, K. Interaction between CD44 and Hyaluronate Induces Chemoresistance in Non-Small Cell Lung Cancer Cell. Cancer Lett. 2007, 252, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Regulating Multidrug Resistance with Hyaluronan. J. Biol. Chem. 2005, 280, e99925. [CrossRef]
- Vitale, D.; Kumar Katakam, S.; Greve, B.; Jang, B.; Oh, E.-S.; Alaniz, L.; Götte, M. Proteoglycans and Glycosaminoglycans as Regulators of Cancer Stem Cell Function and Therapeutic Resistance. FEBS J. 2019, 286, 2870–2882. [Google Scholar] [CrossRef]
- Ryoo, I.-G.; Choi, B.-H.; Ku, S.K.; Kwak, M.K. High CD44 Expression Mediates P62-Associated NFE2L2/NRF2 Activation in Breast Cancer Stem Cell-like Cells: Implications for Cancer Stem Cell Resistance. Redox Biol. 2018, 17, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 Axis Orchestrates Cancer Stem Cell Functions. Cell. Signal. 2019, 63, 109377. [Google Scholar] [CrossRef]
- Shiina, M.; Bourguignon, L.Y.W. Selective Activation of Cancer Stem Cells by Size-Specific Hyaluronan in Head and Neck Cancer. Int. J. Cell Biol. 2015, 2015, 989070. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, D.L.; Parnigoni, A.; Viola, M.; Karousou, E.; Sevic, I.; Moretto, P.; Passi, A.; Alaniz, L.; Vigetti, D. Deciphering Drug Resistance: Investigating the Emerging Role of Hyaluronan Metabolism and Signaling and Tumor Extracellular Matrix in Cancer Chemotherapy. Int. J. Mol. Sci. 2024, 25, 7607. https://doi.org/10.3390/ijms25147607
Vitale DL, Parnigoni A, Viola M, Karousou E, Sevic I, Moretto P, Passi A, Alaniz L, Vigetti D. Deciphering Drug Resistance: Investigating the Emerging Role of Hyaluronan Metabolism and Signaling and Tumor Extracellular Matrix in Cancer Chemotherapy. International Journal of Molecular Sciences. 2024; 25(14):7607. https://doi.org/10.3390/ijms25147607
Chicago/Turabian StyleVitale, Daiana L., Arianna Parnigoni, Manuela Viola, Evgenia Karousou, Ina Sevic, Paola Moretto, Alberto Passi, Laura Alaniz, and Davide Vigetti. 2024. "Deciphering Drug Resistance: Investigating the Emerging Role of Hyaluronan Metabolism and Signaling and Tumor Extracellular Matrix in Cancer Chemotherapy" International Journal of Molecular Sciences 25, no. 14: 7607. https://doi.org/10.3390/ijms25147607
APA StyleVitale, D. L., Parnigoni, A., Viola, M., Karousou, E., Sevic, I., Moretto, P., Passi, A., Alaniz, L., & Vigetti, D. (2024). Deciphering Drug Resistance: Investigating the Emerging Role of Hyaluronan Metabolism and Signaling and Tumor Extracellular Matrix in Cancer Chemotherapy. International Journal of Molecular Sciences, 25(14), 7607. https://doi.org/10.3390/ijms25147607









