Non-Hereditary Obesity Type Networks and New Drug Targets: An In Silico Approach
Abstract
1. Introduction
2. Results
2.1. Stress-Induced Obesidome
2.2. Autonomic Nervous System/Inflammation Obesidome
2.3. Virtual Screening on Natural Compounds for Drug Discovery
3. Discussion
3.1. Non-Hereditary Obesity Interactions Networks
3.2. Potent Natural Compounds Targeting FOXO1
4. Methods
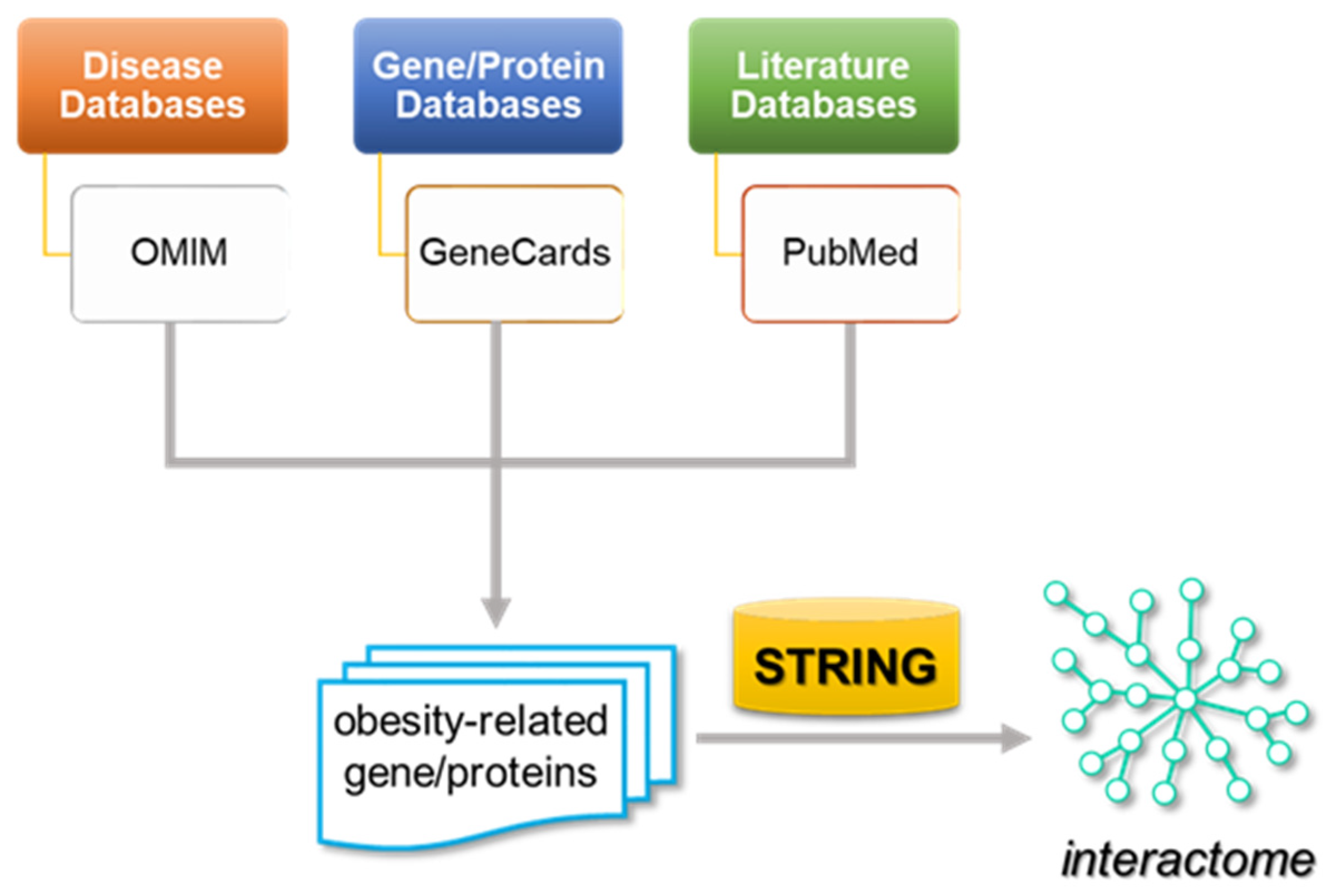
4.1. Network Construction
4.2. Drug Discovery
4.2.1. High-Throughput Virtual Screening of FOXO1 Protein
Target Protein
Chemical Compound Screening
Molecular Docking Simulations
Molecular Visualization
4.2.2. Protein Motif Detection
5. Conclusions
- Discovering new chronopharmacological drugs against obesity;
- Anticipating possible toxic effects of such natural products due to long-term taking;
- Ultradian and circadian rhythmicity identification and contribution to obesity pathogenesis as well as control;
- Through reverse translational research, repurposing not only drugs, but also pertinent biomarkers.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hruby, A.; Hu, F.B. The Epidemiology of Obesity: A Big Picture. PharmacoEconomics 2015, 33, 673–689. [Google Scholar] [CrossRef]
- Glass, T.A.; McAtee, M.J. Behavioral science at the crossroads in public health: Extending horizons, envisioning the future. Soc. Sci. Med. 2006, 62, 1650–1671. [Google Scholar] [CrossRef]
- Balke, H.; Nocito, A. A trip through the history of obesity. Praxis 2013, 102, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Christopoulou-Aletra, H.; Papavramidou, N. Methods used by the hippocratic physicians for weight reduction. World J. Surg. 2004, 28, 513–517. [Google Scholar] [CrossRef]
- Garawi, F.; Devries, K.; Thorogood, N.; Uauy, R. Global differences between women and men in the prevalence of obesity: Is there an association with gender inequality? Eur. J. Clin. Nutr. 2014, 68, 1101–1106. [Google Scholar] [CrossRef]
- Faka, A.; Chalkias, C.; Georgousopoulou, E.N.; Tripitsidis, A.; Pitsavos, C.; Panagiotakos, D.B. Identifying determinants of obesity in Athens, Greece through global and local statistical models. Spat. Spatio-Temporal Epidemiol. 2019, 29, 31–41. [Google Scholar] [CrossRef]
- OECD Publishing; European Observatory on Health Systems and Policies (Eds.) Greece: Country Health Profile 2023, State of Health in the EU; OECD Publishing: Paris, France; European Observatory on Health Systems and Policies: Brussels, Belgium, 2023. [Google Scholar]
- Ezzati, M. Excess weight and multimorbidity: Putting people’s health experience in risk factor epidemiology. Lancet Public Health 2017, 2, e252–e253. [Google Scholar] [CrossRef] [PubMed]
- NCD Risk Factor Collaboration. Trends in adult body-mass index in 200 countries from 1975 to 2014: A pooled analysis of 1698 population-based measurement studies with 19.2 million participants. Lancet 2016, 387, 1377–1396. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Xiong, Y.; Borck, P.C.; Jang, C.; Doulias, P.T.; Papazyan, R.; Fang, B.; Jiang, C.; Zhang, Y.; Briggs, E.R.; et al. Diet-Induced Circadian Enhancer Remodeling Synchronizes Opposing Hepatic Lipid Metabolic Processes. Cell 2018, 174, 831–842.e12. [Google Scholar] [CrossRef]
- Zee, P.C.; Attarian, H.; Videnovic, A. Circadian rhythm abnormalities. Continuum 2013, 19, 132–147. [Google Scholar] [CrossRef]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; van Hees, V.T.; Tyrrell, J.; Beaumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Arab, A.; Karimi, E.; Garaulet, M.; Scheer, F. Social jetlag and obesity: A systematic review and meta-analysis. Obes. Rev. 2024, 25, e13664. [Google Scholar] [CrossRef] [PubMed]
- Kolaitis, G.; Olff, M. Psychotraumatology in Greece. Eur. J. Psychotraumatol. 2017, 8, 135175. [Google Scholar] [CrossRef] [PubMed]
- Chrousos, G.P. The role of stress and the hypothalamic-pituitary-adrenal axis in the pathogenesis of the metabolic syndrome: Neuro-endocrine and target tissue-related causes. Int. J. Obes. 2000, 24 (Suppl. S2), S50–S55. [Google Scholar] [CrossRef] [PubMed]
- Manfredi-Lozano, M.; Roa, J.; Tena-Sempere, M. Connecting metabolism and gonadal function: Novel central neuropeptide pathways involved in the metabolic control of puberty and fertility. Front. Neuroendocrinol. 2018, 48, 37–49. [Google Scholar] [CrossRef]
- Rhie, Y.J. Kisspeptin/G protein-coupled receptor-54 system as an essential gatekeeper of pubertal development. Ann. Pediatr. Endocrinol. Metab. 2013, 18, 55–59. [Google Scholar] [CrossRef]
- Rupérez, A.I.; Gil, A.; Aguilera, C.M. Genetics of oxidative stress in obesity. Int. J. Mol. Sci. 2014, 15, 3118–3144. [Google Scholar] [CrossRef] [PubMed]
- Geronikolou, S.A.; Pavlopoulou, A.; Cokkinos, D.; Chrousos, G. Interactome of Obesity: Obesidome: Genetic Obesity, Stress Induced Obesity, Pathogenic Obesity Interaction. Adv. Exp. Med. Biol. 2017, 987, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Geronikolou, S.; Pavlopoulou, A.; Lambrou, G.I.; Koutelekos, J.; Cokkinos, D.; Albanopoulos, K.; Chrousos, G.P. Kisspeptin and the Genetic Obesity Interactome. Adv. Exp. Med. Biol. 2021, 1339, 111–117. [Google Scholar] [CrossRef]
- Summerbell, C.D.; Waters, E.; Edmunds, L.D.; Kelly, S.; Brown, T.; Campbell, K.J. Interventions for preventing obesity in children. Cochrane Database Syst. Rev. 2005, 3, Cd001871. [Google Scholar] [CrossRef]
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017, 25, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, H. Obesity: Epidemiology, Pathophysiology, and Therapeutics. Front. Endocrinol. 2021, 12, 706978. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Sierra, A.; Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Martinez, J.A. Diet, Gut Microbiota, and Obesity: Links with Host Genetics and Epigenetics and Potential Applications. Adv. Nutr. 2019, 10, S17–S30. [Google Scholar] [CrossRef] [PubMed]
- Bartelt, A.; Heeren, J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 2014, 10, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.T.; Drewnosksi, A.; Kumanyika, S.; Glass, T.A. A systems-oriented multilevel framework for addressing obesity in the 21st century. Prev. Chronic Dis. 2009, 6, A82. [Google Scholar] [PubMed]
- Perna, S.; Giacosa, A.; Bonitta, G.; Bologna, C.; Isu, A.; Guido, D.; Rondanelli, M. Effects of Hazelnut Consumption on Blood Lipids and Body Weight: A Systematic Review and Bayesian Meta-Analysis. Nutrients 2016, 8, 747. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Sung, S.H. Platyphylloside Isolated from Betula platyphylla Inhibit Adipocyte Differentiation and Induce Lipolysis Via Regulating Adipokines Including PPARgamma in 3T3-L1 Cells. Pharmacogn. Mag. 2016, 12, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.R.; Lee, I.K.; Ly, S.Y.; Yang, K.J.; Gang, G.T.; Kim, Y.H.; Hwang, J.H.; Yun, B.S.; Lee, C.H. A Phellinus baumii extract reduces obesity in high-fat diet-fed mice and absorption of triglyceride in lipid-loaded mice. J. Med. Food 2011, 14, 209–218. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Wang, B.; Su, M.; Zhou, S.; Luo, P.; Chen, L. Prevention and control effects of edible fungi and their active ingredients on obesity: An updated review of research and mechanism. J. Funct. Foods 2023, 107, 105621. [Google Scholar] [CrossRef]
- Brent, M.M.; Anand, R.; Marmorstein, R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 2008, 16, 1407–1416. [Google Scholar] [CrossRef]
- Jouanna, J. Hippocrate: Oeuvres Complètes. Collection Budé; Les Belles Lettres: Paris, France, 1988; ISBN 978-2-251-00396-2. [Google Scholar]
- Cannon, W. Wisdom of the Body; W.W. Norton & Company: New York, NY, USA, 1932. [Google Scholar]
- Billman, G.E. Homeostasis: The Underappreciated and Far Too Often Ignored Central Organizing Principle of Physiology. Front. Physiol. 2020, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Marks, D.F. Homeostatic theory of obesity. Health Psychol. Open 2015, 2, 2055102915590692. [Google Scholar] [CrossRef] [PubMed]
- Sobradillo, P.; Pozo, F.; Agustí, Á. P4 Medicine: The Future Around the Corner. Arch. Bronconeumol. 2011, 47, 35–40. [Google Scholar] [CrossRef]
- Flores, M.; Glusman, G.; Brogaard, K.; Price, N.D.; Hood, L. P4 medicine: How systems medicine will transform the healthcare sector and society. Pers. Med. 2013, 10, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Brodie, E.D., III; Gregory, B.; Lisch, D.; Riddle, N.C. The Epigenome and Beyond: How Does Non-genetic Inheritance Change Our View of Evolution? Integr. Comp. Biol. 2021, 61, 2199–2207. [Google Scholar] [CrossRef] [PubMed]
- Kahan, L.G.; Mehrzad, R. Chapter 10—Environmental factors related to the obesity epidemic. In Obesity; Mehrzad, R., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 117–139. [Google Scholar]
- Grün, F.; Blumberg, B. Environmental obesogens: Organotins and endocrine disruption via nuclear receptor signaling. Endocrinology 2006, 147, S50–S55. [Google Scholar] [CrossRef] [PubMed]
- Baik, J.-H. Stress and the dopaminergic reward system. Exp. Mol. Med. 2020, 52, 1879–1890. [Google Scholar] [CrossRef] [PubMed]
- Odle, A.K.; MacNicol, M.C.; Childs, G.V.; MacNicol, A.M. Post-Transcriptional Regulation of Gnrhr: A Checkpoint for Metabolic Control of Female Reproduction. Int. J. Mol. Sci. 2021, 22, 3312. [Google Scholar] [CrossRef] [PubMed]
- Rosmond, R.; Dallman, M.F.; Björntorp, P. Stress-related cortisol secretion in men: Relationships with abdominal obesity and endocrine, metabolic and hemodynamic abnormalities. J. Clin. Endocrinol. Metab. 1998, 83, 1853–1859. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Lee, M.J.; Pramyothin, P.; Karastergiou, K.; Fried, S.K. Deconstructing the roles of glucocorticoids in adipose tissue biology and the development of central obesity. Biochim. Biophys. Acta 2014, 1842, 473–481. [Google Scholar] [CrossRef]
- Bonanni, A.; Basile, M.; Montone, R.A.; Crea, F. Impact of the exposome on cardiovascular disease. Eur. Heart J. Suppl. 2023, 25, B60–B64. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Trayhurn, P. Acute and prolonged effects of TNF-α on the expression and secretion of inflammation-related adipokines by human adipocytes differentiated in culture. Pflug. Arch. Eur. J. Physiol. 2006, 452, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Kern, P.A.; Saghizadeh, M.; Ong, J.M.; Bosch, R.J.; Deem, R.; Simsolo, R.B. The expression of tumor necrosis factor in human adipose tissue. Regulation by obesity, weight loss, and relationship to lipoprotein lipase. J. Clin. Investig. 1995, 95, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Röszer, T. Adipose Tissue Immunometabolism and Apoptotic Cell Clearance. Cells 2021, 10, 2288. [Google Scholar] [CrossRef] [PubMed]
- Rudic, R.D.; McNamara, P.; Curtis, A.M.; Boston, R.C.; Panda, S.; Hogenesch, J.B.; Fitzgerald, G.A. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004, 2, e377. [Google Scholar] [CrossRef] [PubMed]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Marcheva, B.; Ramsey, K.M.; Buhr, E.D.; Kobayashi, Y.; Su, H.; Ko, C.H.; Ivanova, G.; Omura, C.; Mo, S.; Vitaterna, M.H.; et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010, 466, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Shimba, S.; Ishii, N.; Ohta, Y.; Ohno, T.; Watabe, Y.; Hayashi, M.; Wada, T.; Aoyagi, T.; Tezuka, M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 12071–12076. [Google Scholar] [CrossRef]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef]
- Shimba, A.; Cui, G.; Tani-Ichi, S.; Ogawa, M.; Abe, S.; Okazaki, F.; Kitano, S.; Miyachi, H.; Yamada, H.; Hara, T.; et al. Glucocorticoids Drive Diurnal Oscillations in T Cell Distribution and Responses by Inducing Interleukin-7 Receptor and CXCR4. Immunity 2018, 48, 286–298.e6. [Google Scholar] [CrossRef] [PubMed]
- Bellet, M.M.; Deriu, E.; Liu, J.Z.; Grimaldi, B.; Blaschitz, C.; Zeller, M.; Edwards, R.A.; Sahar, S.; Dandekar, S.; Baldi, P.; et al. Circadian clock regulates the host response to Salmonella. Proc. Natl. Acad. Sci. USA 2013, 110, 9897–9902. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, S.; Dubeau-Laramée, G.; Ohm, H.; Labrecque, N.; Olivier, M.; Cermakian, N. The circadian clock in immune cells controls the magnitude of Leishmania parasite infection. Sci. Rep. 2017, 7, 10892. [Google Scholar] [CrossRef] [PubMed]
- Carvalho Cabral, P.; Tekade, K.; Stegeman, S.K.; Olivier, M.; Cermakian, N. The involvement of host circadian clocks in the regulation of the immune response to parasitic infections in mammals. Parasite Immunol. 2022, 44, e12903. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, T.W.; Hall, S.; Begley, N.; Forman, R.; Brown, S.; Vonslow, R.; Saer, B.; Little, M.C.; Murphy, E.A.; Hurst, R.J.; et al. The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci. Rep. 2018, 8, 3782. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, A.; Xie, W.; Agapov, E.; Brown, S.; Steinberg, D.; Tidwell, R.; Sajol, G.; Schutz, R.; Weaver, R.; Yu, H.; et al. BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal Immunol. 2018, 11, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, T.; Dhar, J.; Patel, S.; Kondratov, R.; Barik, S. Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate Immun. 2017, 23, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Fernández Alfonso, T.; Celentano, A.M.; Gonzalez Cappa, S.M.; Golombek, D.A. The circadian system of Trypanosoma cruzi-infected mice. Chronobiol. Int. 2003, 20, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Rijo-Ferreira, F.; Carvalho, T.; Afonso, C.; Sanches-Vaz, M.; Costa, R.M.; Figueiredo, L.M.; Takahashi, J.S. Sleeping sickness is a circadian disorder. Nat. Commun. 2018, 9, 62. [Google Scholar] [CrossRef]
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N.; et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015, 17, 681–689. [Google Scholar] [CrossRef]
- Cani, P.D.; Van Hul, M. Gut microbiota in overweight and obesity: Crosstalk with adipose tissue. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 164–183. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Khoshbakht, S.; Başkurt, D.; Vural, A.; Vural, S. Behçet’s Disease: A Comprehensive Review on the Role of HLA-B*51, Antigen Presentation, and Inflammatory Cascade. Int. J. Mol. Sci. 2023, 24, 16382. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Fagundes, C.T.; Yang, G.; Palsson-McDermott, E.M.; Wochal, P.; McGettrick, A.F.; Foley, N.H.; Early, J.O.; Chen, L.; Zhang, H.; et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl. Acad. Sci. USA 2015, 112, 7231–7236. [Google Scholar] [CrossRef] [PubMed]
- Sutton, C.E.; Finlay, C.M.; Raverdeau, M.; Early, J.O.; DeCourcey, J.; Zaslona, Z.; O’Neill, L.A.J.; Mills, K.H.G.; Curtis, A.M. Loss of the molecular clock in myeloid cells exacerbates T cell-mediated CNS autoimmune disease. Nat. Commun. 2017, 8, 1923. [Google Scholar] [CrossRef] [PubMed]
- Ruben, M.D.; Wu, G.; Smith, D.F.; Schmidt, R.E.; Francey, L.J.; Lee, Y.Y.; Anafi, R.C.; Hogenesch, J.B. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci. Transl. Med. 2018, 10, eaat8806. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Samblas, M.; Milagro, F.I.; Riezu-Boj, J.I.; Crujeiras, A.B.; Martinez, J.A.; Project, M. Circadian gene methylation profiles are associated with obesity, metabolic disturbances and carbohydrate intake. Chronobiol. Int. 2018, 35, 969–981. [Google Scholar] [CrossRef]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Zimanyi, I.A.; Fathi, Z.; Poindexter, G.S. Central control of feeding behavior by neuropeptide Y. Curr. Pharm. Des. 1998, 4, 349–366. [Google Scholar] [CrossRef]
- Guarino, D.; Nannipieri, M.; Iervasi, G.; Taddei, S.; Bruno, R.M. The Role of the Autonomic Nervous System in the Pathophysiology of Obesity. Front. Physiol. 2017, 8, 665. [Google Scholar] [CrossRef]
- Fidan-Yaylali, G.; Yaylali, Y.T.; Erdogan, Ç.; Can, B.; Senol, H.; Gedik-Topçu, B.; Topsakal, S. The Association between Central Adiposity and Autonomic Dysfunction in Obesity. Med. Princ. Pract. 2016, 25, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Lambert, E.A.; Straznicky, N.E.; Dixon, J.B.; Lambert, G.W. Should the sympathetic nervous system be a target to improve cardiometabolic risk in obesity? Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H244–H258. [Google Scholar] [CrossRef] [PubMed]
- Geronikolou, S.A.; Albanopoulos, K.; Chrousos, G.; Cokkinos, D. Evaluating the Homeostasis Assessment Model Insulin Resistance and the Cardiac Autonomic System in Bariatric Surgery Patients: A Meta-Analysis. Adv. Exp. Med. Biol. 2017, 988, 249–259. [Google Scholar] [PubMed]
- Mateus, K.; Bonfante, I.L.P.; Sardeli, A.V.; Duft, R.G.; Gáspari, A.F.; Trombeta, J.; Morari, J.; Rodrigues, B.; Torsoni, M.A.; Chacon-Mikahil, M.P.T.; et al. Effects of Different Exercise Types on Chrna7 and Chrfam7a Expression in Healthy Normal Weight and Overweight Type 2 Diabetic Adults. Biomedicines 2023, 11, 565. [Google Scholar] [CrossRef] [PubMed]
- Bukowiecki, L.; Collet, A.J.; Follea, N.; Guay, G.; Jahjah, L. Brown adipose tissue hyperplasia: A fundamental mechanism of adaptation to cold and hyperphagia. Am. J. Physiol. Metab. 1982, 242, E353–E359. [Google Scholar] [CrossRef]
- Marsili, A.; Aguayo-Mazzucato, C.; Chen, T.; Kumar, A.; Chung, M.; Lunsford, E.P.; Harney, J.W.; Van-Tran, T.; Gianetti, E.; Ramadan, W.; et al. Mice with a targeted deletion of the type 2 deiodinase are insulin resistant and susceptible to diet induced obesity. PLoS ONE 2011, 6, e20832. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.W.; Monemdjou, S.; Gavrilova, O.; Leon, L.R.; Marcus-Samuels, B.; Chou, C.J.; Everett, C.; Kozak, L.P.; Li, C.; Deng, C.; et al. Lack of obesity and normal response to fasting and thyroid hormone in mice lacking uncoupling protein-3. J. Biol. Chem. 2000, 275, 16251–16257. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.H.; Cypess, A.M.; Kahn, C.R. Cellular bioenergetics as a target for obesity therapy. Nat. Rev. Drug Discov. 2010, 9, 465–482. [Google Scholar] [CrossRef] [PubMed]
- Jalba, M.S.; Rhoads, G.G.; Demissie, K. Association of codon 16 and codon 27 β 2-adrenergic receptor gene polymorphisms with obesity: A meta-analysis. Obesity 2008, 16, 2096–2106. [Google Scholar] [CrossRef]
- Kurylowicz, A.; Jonas, M.; Lisik, W.; Jonas, M.; Wicik, Z.A.; Wierzbicki, Z.; Chmura, A.; Puzianowska-Kuznicka, M. Obesity is associated with a decrease in expression but not with the hypermethylation of thermogenesis-related genes in adipose tissues. J. Transl. Med. 2015, 13, 31. [Google Scholar] [CrossRef]
- Navarro, V.M. Metabolic regulation of kisspeptin—The link between energy balance and reproduction. Nat. Rev. Endocrinol. 2020, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Hudson, A.D.; Kauffman, A.S. Metabolic actions of kisspeptin signaling: Effects on body weight, energy expenditure, and feeding. Pharmacol. Ther. 2022, 231, 107974. [Google Scholar] [CrossRef] [PubMed]
- Romere, C.; Duerrschmid, C.; Bournat, J.; Constable, P.; Jain, M.; Xia, F.; Saha, P.K.; Del Solar, M.; Zhu, B.; York, B.; et al. Asprosin, a Fasting-Induced Glucogenic Protein Hormone. Cell 2016, 165, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Yun, S.; Jeong, J.H.; Jung, T.W. Asprosin impairs insulin secretion in response to glucose and viability through TLR4/JNK-mediated inflammation. Mol. Cell. Endocrinol. 2019, 486, 96–104. [Google Scholar] [CrossRef]
- Pîrsean, C.; Neguț, C.; Stefan-van Staden, R.I.; Dinu-Pirvu, C.E.; Armean, P.; Udeanu, D.I. The salivary levels of leptin and interleukin-6 as potential inflammatory markers in children obesity. PLoS ONE 2019, 14, e0210288. [Google Scholar] [CrossRef] [PubMed]
- Duerrschmid, C.; He, Y.; Wang, C.; Li, C.; Bournat, J.C.; Romere, C.; Saha, P.K.; Lee, M.E.; Phillips, K.J.; Jain, M.; et al. Asprosin is a centrally acting orexigenic hormone. Nat. Med. 2017, 23, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, J.; Liu, Y. The extracellular matrix glycoprotein fibrillin-1 in health and disease. Front. Cell Dev. Biol. 2023, 11, 1302285. [Google Scholar] [CrossRef]
- Tsoutsouki, J.; Abbara, A.; Dhillo, W. Novel therapeutic avenues for kisspeptin. Curr. Opin. Pharmacol. 2022, 67, 102319. [Google Scholar] [CrossRef]
- Cerulli, A.; Lauro, G.; Masullo, M.; Cantone, V.; Olas, B.; Kontek, B.; Nazzaro, F.; Bifulco, G.; Piacente, S. Cyclic Diarylheptanoids from Corylus avellana Green Leafy Covers: Determination of Their Absolute Configurations and Evaluation of Their Antioxidant and Antimicrobial Activities. J. Nat. Prod. 2017, 80, 1703–1713. [Google Scholar] [CrossRef]
- Esposito, T.; Sansone, F.; Franceschelli, S.; Del Gaudio, P.; Picerno, P.; Aquino, R.P.; Mencherini, T. Hazelnut (Corylus avellana L.) Shells Extract: Phenolic Composition, Antioxidant Effect and Cytotoxic Activity on Human Cancer Cell Lines. Int. J. Mol. Sci. 2017, 18, 392. [Google Scholar] [CrossRef]
- Arruda, H.S.; Borsoi, F.T.; Andrade, A.C.; Pastore, G.M.; Marostica Junior, M.R. Scientific Advances in the Last Decade on the Recovery, Characterization, and Functionality of Bioactive Compounds from the Araticum Fruit (Annona crassiflora Mart.). Plants 2023, 12, 1536. [Google Scholar] [CrossRef]
- Mateos, R.; Salvador, M.D.; Fregapane, G.; Goya, L. Why Should Pistachio Be a Regular Food in Our Diet? Nutrients 2022, 14, 3207. [Google Scholar] [CrossRef]
- Lin, H.; Li, J.; Sun, M.; Wang, X.; Zhao, J.; Zhang, W.; Lv, G.; Wang, Y.; Lin, Z. Effects of hazelnut soluble dietary fiber on lipid-lowering and gut microbiota in high-fat-diet-fed rats. Int. J. Biol. Macromol. 2024, 256, 128538. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of Open Natural Products database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef]
- 2020 News Releases. Available online: https://www.cancer.gov/news-events/press-releases/2020 (accessed on 7 June 2021).
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]




| Gene/Protein Symbol | Description | Network |
|---|---|---|
| ACE | angiotensin-converting enzyme | 2 |
| ACE2 | angiotensin-converting enzyme 2 | 2 |
| ADIPOQ | adiponectin, C1Q, and collagen domain containing | 1, 2 |
| ADRB2 | adrenoceptor beta 2 | 1, 2 |
| ADRB3 | adrenoceptor beta 3 | 1, 2 |
| AGRP | agouti-related neuropeptide | 1,2 |
| AGT | Angiotensinogen | 2 |
| ANK3 | ankyrin 3 | 1, 2 |
| ARNTL | basic helix–loop–helix ARNT-like 1 | 1,2 |
| ARRB1 | arrestin beta 1 | 1, 2 |
| ARRB2 | arrestin beta 2 | 1, 2 |
| AVP | arginine vasopressin | 2 |
| CARTPT | cocaine- and amphetamine-regulated transcript | 2 |
| CCK | Cholecystokinin | 1, 2 |
| CCRN4L | nocturnin | 1, 2 |
| CD14 | CD14 molecule | 1, 2 |
| CD163 | CD163 molecule | 1, 2 |
| CHRNA7 | neuronal acetylcholine receptor subunit alpha | 2 |
| CLOCK | nocturnin | 1, 2 |
| CREB1 | cAMP responsive element binding protein 1 | 1, 2 |
| CRH | corticotropin-releasing hormone | 1, 2 |
| CRP | C-reactive protein | 1, 2 |
| CSNK1A1 | casein kinase 1 alpha 1 | 2 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 1, 2 |
| CXCL2 | C-X-C motif chemokine ligand 2 | 1, 2 |
| CXCL8 | C-X-C motif chemokine ligand 8 | 1, 2 |
| DBP | D-box binding PAR bZIP transcription factor | 1, 2 |
| FBN1 | fibrillin 1 | 1,2 |
| FOS | Fos proto-oncogene, AP-1 transcription factor subunit | 1, 2 |
| FOXO1 | forkhead box O1 | 2 |
| FTO | FTO alpha-ketoglutarate-dependent dioxygenase | 1, 2 |
| GCG | Glucagon | 1, 2 |
| GDF15 | growth differentiation factor 15 | 1, 2 |
| GHRL | Ghrelin | 1, 2 |
| GHSR | growth hormone secretagogue receptor | 1 |
| GLP1R | glucagon-like peptide 1 receptor | 1, 2 |
| GLP2R | glucagon-like peptide 2 receptor | 1, 2 |
| GNAL | G protein subunit alpha L | 1,2 |
| GNRH1 | gonadotropin-releasing hormone 1 | 1, 2 |
| GOT1 | glutamic-oxaloacetic transaminase 1 | 1, 2 |
| GPT | glutamic-pyruvic transaminase | 1, 2 |
| GSK3B | glycogen synthase kinase 3 beta | 1, 2 |
| H2AX | H2A.X variant histone | 1, 2 |
| HSD11B1 | hydroxysteroid 11-Beta Dehydrogenase 1 | 1 |
| HSD11B2 | hydroxysteroid 11-Beta Dehydrogenase 2 | 1 |
| HSD3B1 | hydroxy-delta-5-steroid dehydrogenase, 3 beta-, and steroid delta-isomerase 1 | 1 |
| HTR2A | 5-HT2A receptor | 2 |
| IFNG | interferon gamma | 2 |
| IGF1 | insulin-like growth factor 1 | 1, 2 |
| IGFBP1 | insulin-like growth factor binding protein 1 | 1 |
| IKBKG | inhibitor of nuclear factor kappa B kinase regulatory subunit gamma | 2 |
| IL10 | interleukin 10 | 1, 2 |
| IL18 | interleukin 18 | 1, 2 |
| IL1B | interleukin 1 beta | 1, 2 |
| IL6 | interleukin 6 | 2 |
| IL6R | interleukin 6 receptor | 2 |
| IL7 | interleukin 7 | 1, 2 |
| INS | insulin | 1, 2 |
| JAK2 | janus kinase 2 | 2 |
| KCNQ2 | potassium voltage-gated channel subfamily Q member 2 | 1, 2 |
| KISS1 | KiSS-1 metastasis-suppressor | 1, 2 |
| KISS1R | KISS1 receptor | 1, 2 |
| LEP | Leptin | 1, 2 |
| LEPR | leptin receptor | 1, 2 |
| LHCGR | luteinizing hormone/choriogonadotropin receptor | 1, 2 |
| LIPN | lipase family member N | 1, 2 |
| LRP5 | LDL receptor related protein 5 | 1, 2 |
| MC1R | melanocortin 1 receptor | 1, 2 |
| MC2R | melanocortin 2 receptor | 1, 2 |
| MC3R | melanocortin 3 receptor | 1, 2 |
| MC4R | melanocortin 4 receptor | 1, 2 |
| MC5R | melanocortin 5 receptor | 1, 2 |
| MTRNR2L5 | MT-RNR2-like 5 (pseudogene) | 1, 2 |
| NCOR1 | nuclear receptor corepressor 1 | 1, 2 |
| NFIL3 | nuclear factor, interleukin 3 regulated | 2 |
| NFKB1 | nuclear factor kappa b | 1 |
| NPPB | natriuretic peptide B | 1, 2 |
| NPY | neuropeptide Y | 1, 2 |
| NR1D1 | nuclear receptor subfamily 1 group D member 1 | 2 |
| NR1D2 | nuclear receptor subfamily 1 group D member 2 | 2 |
| NR3C1 | nuclear receptor subfamily 3 group C member 1 | 1 |
| NR3C2 | nuclear receptor subfamily 3 group C member 2 | 1 |
| OR4M1 | olfactory receptor family 4 subfamily M member 1 | 1, 2 |
| PCK1 | phosphoenolpyruvate carboxykinase 1 | 1, 2 |
| PCSK1 | proprotein convertase subtilisin/kexin type 1 | 1, 2 |
| PLCB1 | phospholipase C beta 1 | 1, 2 |
| PLCB2 | phospholipase C beta 2 | 1, 2 |
| PLCB3 | phospholipase C beta 3 | 1, 2 |
| PLCB4 | phospholipase C beta 4 | 1, 2 |
| PLCE1 | phospholipase C epsilon 1 | 1 |
| PLIN1 | perilipin 1 | 1, 2 |
| POMC | proopiomelanocortin | 1, 2 |
| PPARG2 | peroxisome proliferator-activated receptor gamma | 1, 2 |
| PPY | pancreatic polypeptide | 1, 2 |
| RBP4 | retinol-binding protein 4 | 1, 2 |
| THRA | thyroid hormone receptor alpha | 1, 2 |
| THRB | thyroid hormone receptor beta | 1, 2 |
| TNF | tumor necrosis factor | 2 |
| TP53 | tumor protein p53 | 1 |
| TRPC6 | transient receptor potential cation channel subfamily C member 6 | 1, 2 |
| UCP1 | uncoupling protein 1 | 1, 2 |
| UCP2 | uncoupling protein 2 | 1, 2 |
| UCP3 | uncoupling protein 3 | 1, 2 |
| Stress Induced | ANS/Inflammation Induced | ||
|---|---|---|---|
| Hub | Interactions | Hub | Interactions |
| GCG | 32 | INS | 37 |
| POMC | 32 | GCG | 34 |
| INS | 31 | LEP | 34 |
| LEP | 29 | POMC | 32 |
| NPY | 25 | IL6 | 29 |
| CRH | 22 | AVP | 28 |
| FOS | 22 | NPY | 27 |
| ADIPOQ | 19 | AGT | 25 |
| MC4R | 19 | CRH | 24 |
| ADRB2 | 18 | FOS | 22 |
| CXCL8 | 18 | ADIPOQ | 21 |
| ADRB3 | 17 | CXCL8 | 21 |
| CCK | 17 | ADRB2 | 20 |
| GHRL | 17 | CCK | 20 |
| GNRH1 | 17 | MC4R | 20 |
| IGF1 | 17 | GNRH1 | 19 |
| IL10 | 19 | ||
| ADRB3 | 18 | ||
| IL1B | 18 | ||
| TNF | 18 | ||
| IGF1 | 17 | ||
| COCONUT CID | Docking Scores (kcal/mol) | Compound Name | Source | 2D Image |
|---|---|---|---|---|
| CNP0087111 | −14.7 | carpinontriol B | Corylus avellana (hazelnut); Carpinus cordata (hornbeam) | 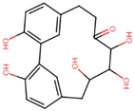 |
| CNP0191412 | −13.5 | alnusonol | Alnus japonica (Japanese alder) |  |
| CNP0307211 | −13 | acerogenin E | Betula spp. (birch) | 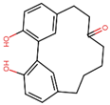 |
| CNP0276542 | −9.3 | phelligridin E | Phellinus igniarius (willow bracket fungus) | 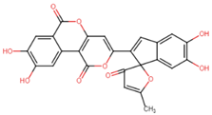 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geronikolou, S.A.; Pavlopoulou, A.; Uça Apaydin, M.; Albanopoulos, K.; Cokkinos, D.V.; Chrousos, G. Non-Hereditary Obesity Type Networks and New Drug Targets: An In Silico Approach. Int. J. Mol. Sci. 2024, 25, 7684. https://doi.org/10.3390/ijms25147684
Geronikolou SA, Pavlopoulou A, Uça Apaydin M, Albanopoulos K, Cokkinos DV, Chrousos G. Non-Hereditary Obesity Type Networks and New Drug Targets: An In Silico Approach. International Journal of Molecular Sciences. 2024; 25(14):7684. https://doi.org/10.3390/ijms25147684
Chicago/Turabian StyleGeronikolou, Styliani A., Athanasia Pavlopoulou, Merve Uça Apaydin, Konstantinos Albanopoulos, Dennis V. Cokkinos, and George Chrousos. 2024. "Non-Hereditary Obesity Type Networks and New Drug Targets: An In Silico Approach" International Journal of Molecular Sciences 25, no. 14: 7684. https://doi.org/10.3390/ijms25147684
APA StyleGeronikolou, S. A., Pavlopoulou, A., Uça Apaydin, M., Albanopoulos, K., Cokkinos, D. V., & Chrousos, G. (2024). Non-Hereditary Obesity Type Networks and New Drug Targets: An In Silico Approach. International Journal of Molecular Sciences, 25(14), 7684. https://doi.org/10.3390/ijms25147684








