The Contrasting Effects of Two Distinct Exercise Training Modalities on Exhaustive Exercise-Induced Muscle Damage in Mice May Be Associated with Alterations in the Gut Microbiota
Abstract
:1. Introduction
2. Results
2.1. Effects of Two Different Training Modalities on Muscle Damage in Mice following Exhaustive Exercise
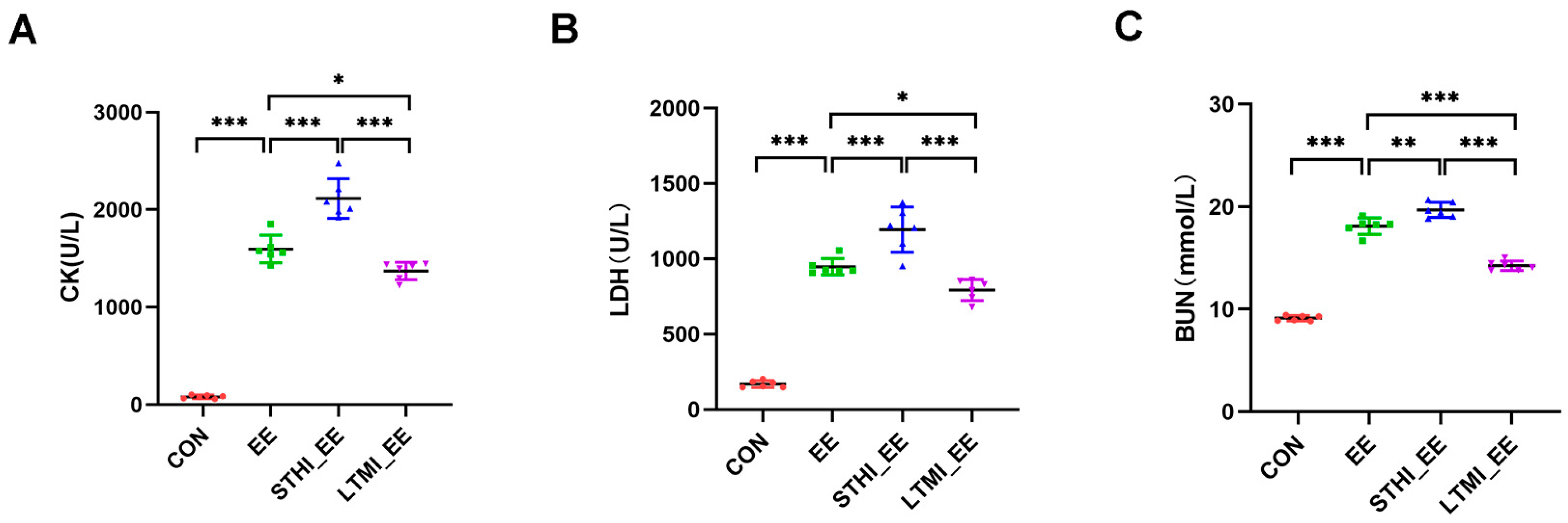
2.2. Effects of Two Different Training Modalities on the Levels of Serum CK, LDH, and BUN in Mice following Exhaustive Exercise
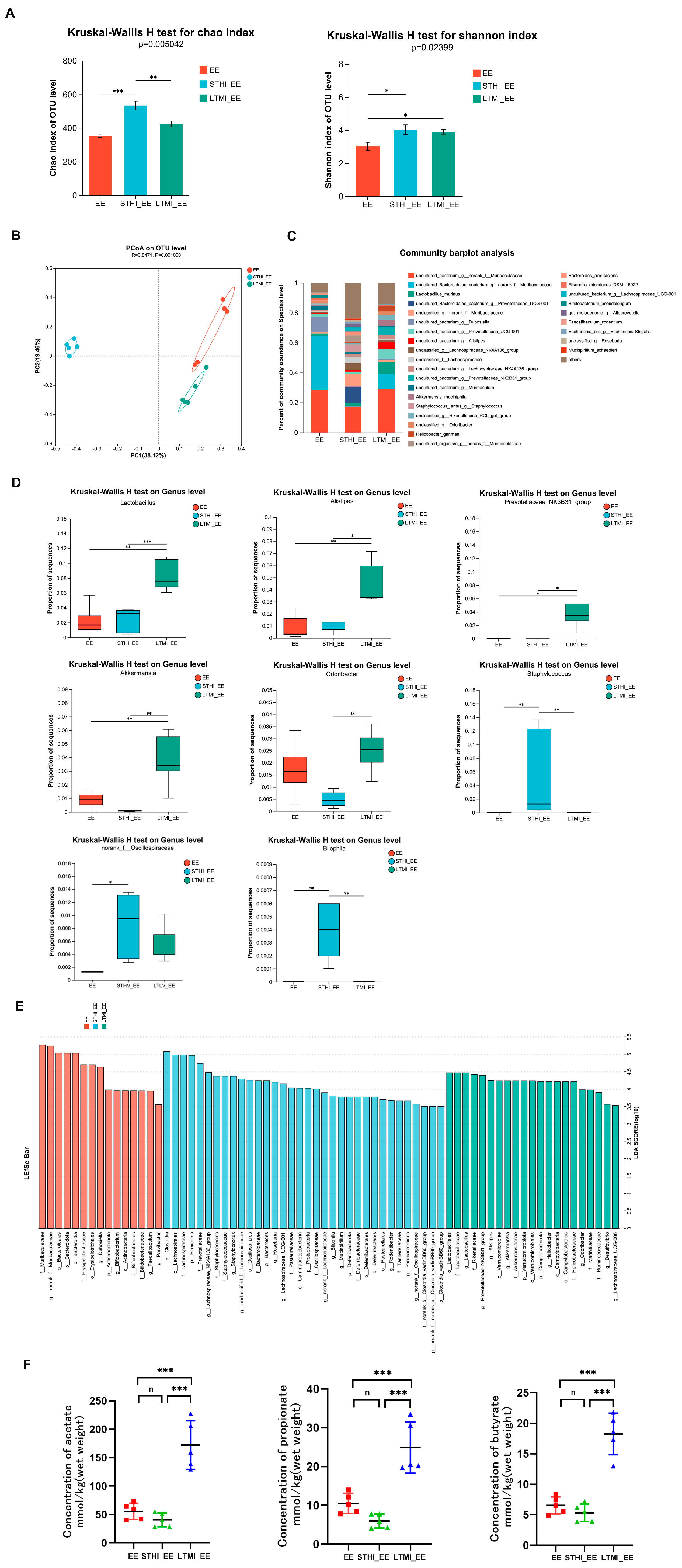
2.3. Effects of Two Different Training Modalities on Fecal Microbiota and Its SCFA Metabolites in Mice before Exhaustive Exercise
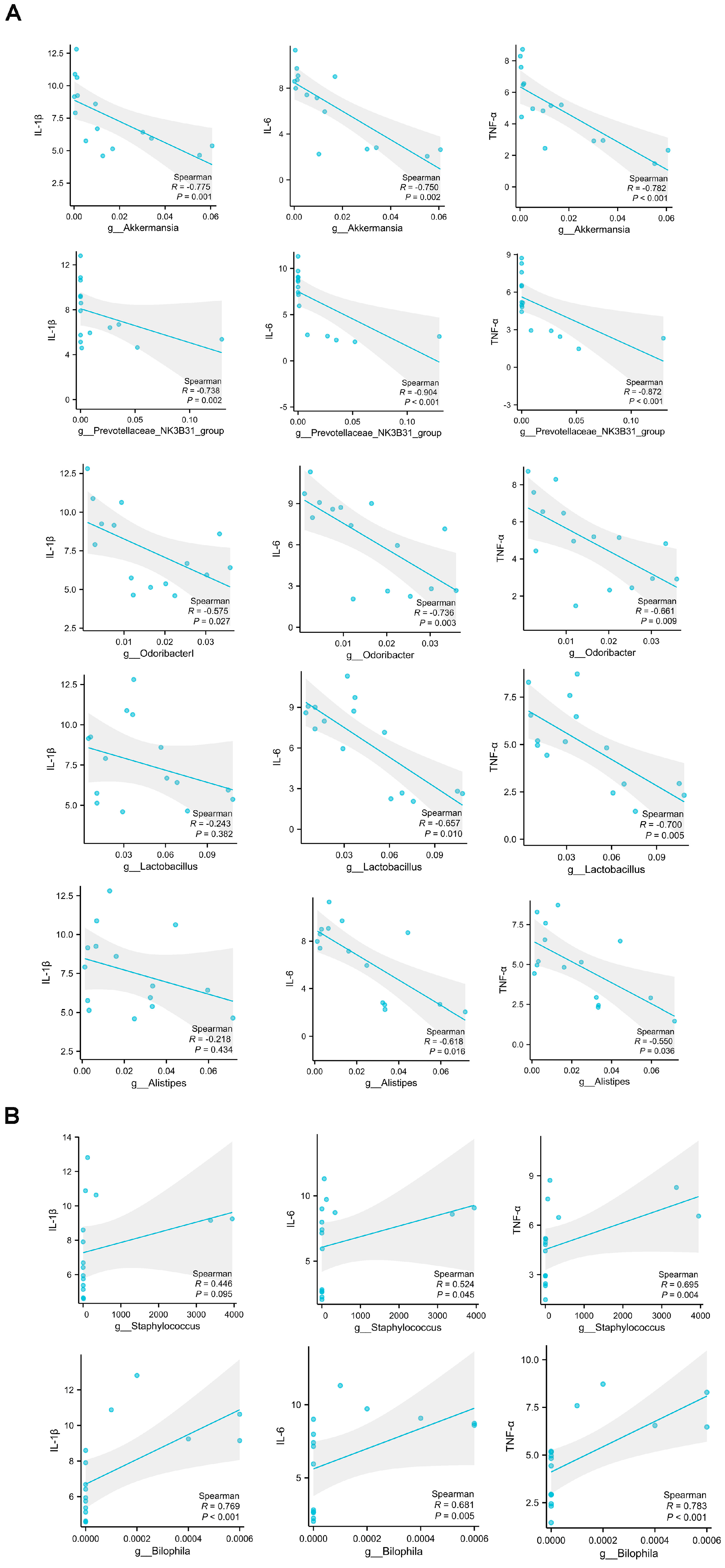
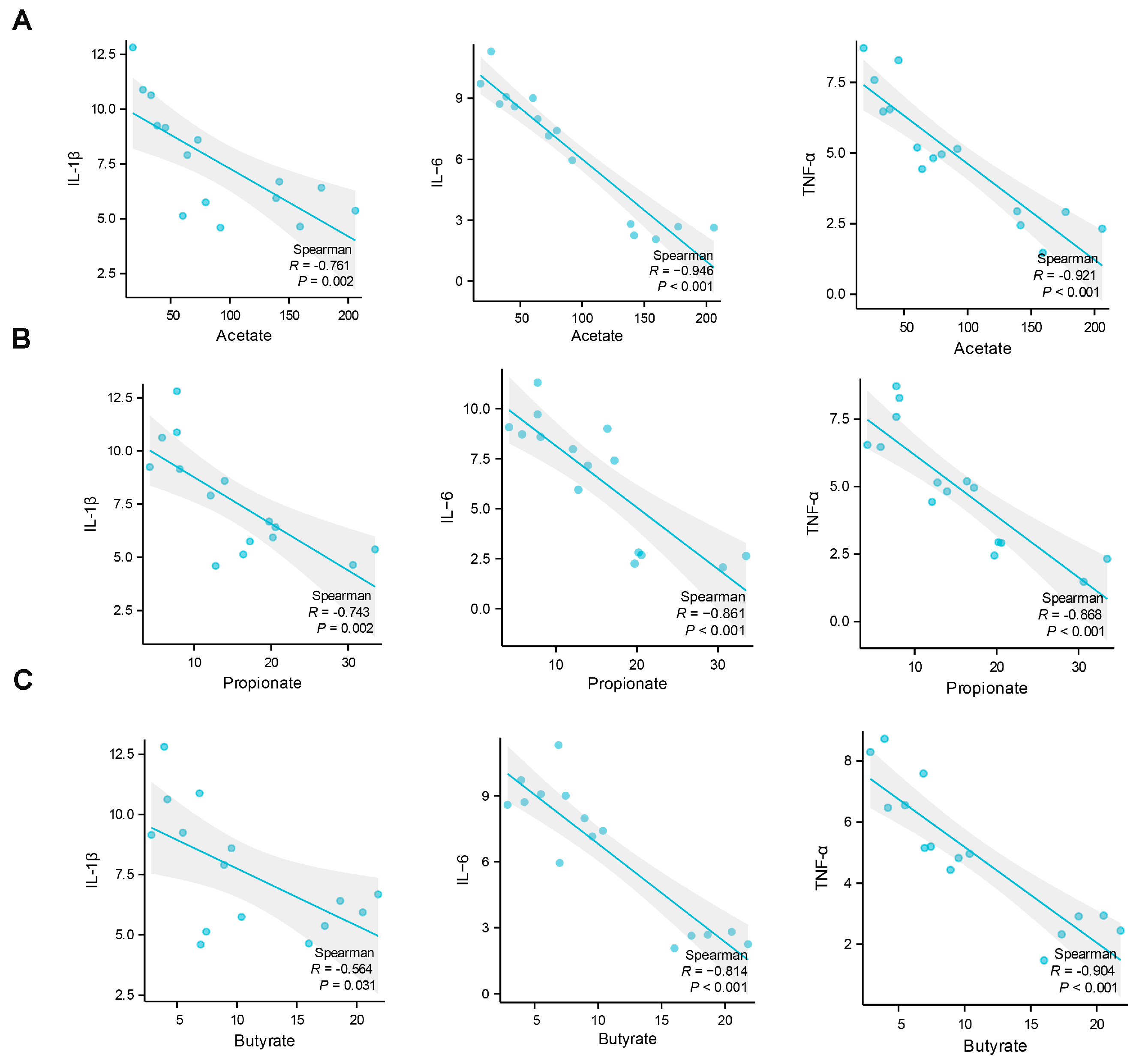
2.4. Analysis of the Correlations between Muscle Damage and Gut Microbiota at the Genus Level and Short-Chain Fatty Acids
2.5. Effects of Fecal Microbiota Transplantation from Mice That Underwent Different Exercise Training Modalities on Exhaustive Exercise-Induced Muscle Damage in Recipient Mice
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Ethics Statement
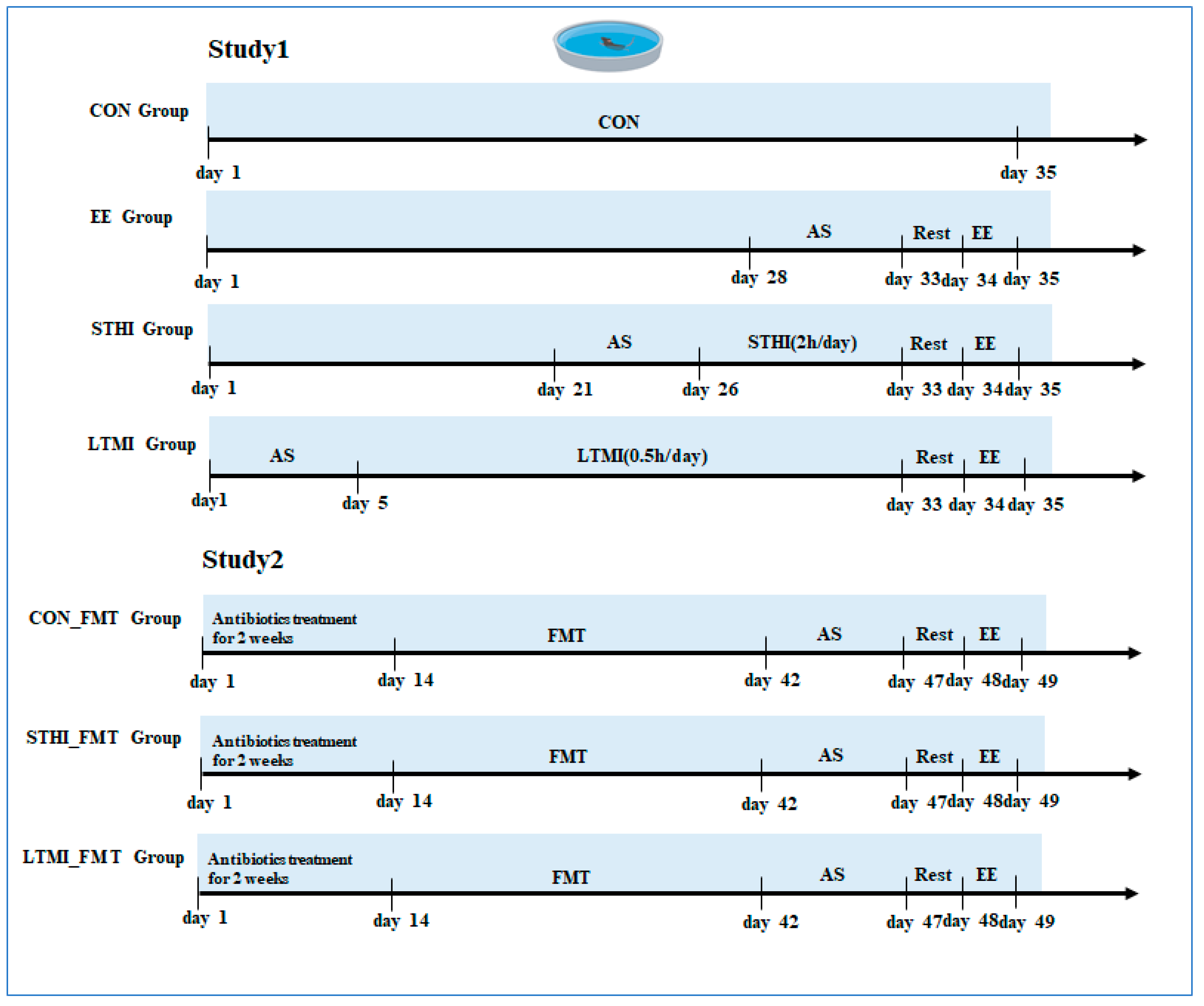
4.3. Experiment Design
4.4. Histopathological Analyses
4.5. Immunofluorescence Staining
4.6. Serum Biochemical Parameters
4.7. Real-Time Quantitative PCR
4.8. Western Blotting
4.9. Measurement of the Oxidative Stress Biomarkers
4.10. Fecal Microbiome Analysis
4.11. Measurement of Fecal SCFAs
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cuk, I.; Nikolaidis, P.T.; Markovic, S.; Knechtle, B. Age Differences in Pacing in Endurance Running: Comparison between Marathon and Half-Marathon Men and Women. Medicina 2019, 55, 479. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, C.A.Z.; Sierra, A.P.R.; Martínez Galán, B.S.; Maciel, J.F.S.; Manoel, R.; Barbeiro, H.V.; de Souza, H.P.; Cury-Boaventura, M.F. Time Course and Role of Exercise-Induced Cytokines in Muscle Damage and Repair After a Marathon Race. Front. Physiol. 2021, 12, 752144. [Google Scholar] [CrossRef] [PubMed]
- Kosowski, M.; Swoboda, K.; Chmura, J.; Kustrzycka-Kratochwil, D.; Todd, J.A.; Jankowska, E.A.; Reczuch, K.; Ponikowski, P. Inflammatory Activation Biomarker Profile after Marathon Running and Its Impact on Cardiovascular Stress in Amateur Middle-Aged Male Runners. Adv. Clin. Exp. Med. 2023, 32, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Clauss, M.; Gérard, P.; Mosca, A.; Leclerc, M. Interplay Between Exercise and Gut Microbiome in the Context of Human Health and Performance. Front. Nutr. 2021, 8, 637010. [Google Scholar] [CrossRef] [PubMed]
- Sardeli, A.V.; Araujo, R.B.; Woods, J.A.; Lord, J. Chacon-Mikahil MPT. Higher risk of upper respiratory tract infection post marathon running: When physical exercise becomes a threat to the immune system. Exerc. Immunol. Rev. 2024, in press. [Google Scholar]
- Fonseca, T.R.; Mendes, T.T.; Ramos, G.P.; Cabido, C.E.T.; Morandi, R.F.; Ferraz, F.O.; Miranda, A.S.; Mendonça, V.A.; Teixeira, A.L.; Silami-Garcia, E.; et al. Aerobic Training Modulates the Increase in Plasma Concentrations of Cytokines in Response to a Session of Exercise. J. Environ. Public. Health. 2021, 8, 1304139. [Google Scholar] [CrossRef] [PubMed]
- Roca, E.; Nescolarde, L.; Lupón, J.; Barallat, J.; Januzzi, J.L.; Liu, P.; Cruz Pastor, M.; Bayes-Genis, A. The Dynamics of Cardiovascular Biomarkers in Non-Elite Marathon Runners. J. Cardiovasc. Transl. Res. 2017, 10, 206–208. [Google Scholar] [CrossRef]
- Bull, F.C.; Al-Ansari, S.S.; Biddle, S.; Borodulin, K.; Buman, M.P.; Cardon, G.; Carty, C.; Chaput, J.P.; Chastin, S.; Chou, R.; et al. World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br. J. Sports Med. 2020, 54, 1451–1462. [Google Scholar] [CrossRef]
- O’Donovan, G.; Sarmiento, O.L.; Hamer, M. The Rise of the “Weekend Warrior”. J. Orthop. Sports Phys. Ther. 2018, 48, 604–606. [Google Scholar] [CrossRef]
- Dos Santos, M.; Ferrari, G.; Lee, D.H.; Rey-López, J.P.; Aune, D.; Liao, B.; Huang, W.; Nie, J.; Wang, Y.; Giovannucci, E.; et al. Association of the “Weekend Warrior” and Other Leisure-time Physical Activity Patterns with All-Cause and Cause-Specific Mortality: A Nationwide Cohort Study. JAMA Intern. Med. 2022, 182, 840–848. [Google Scholar] [CrossRef]
- O’Donovan, G.; Petermann-Rocha, F.; Ferrari, G.; Lee, I.M.; Hamer, M.; Stamatakis, E.; Sarmiento, O.L.; Ibáñez, A.; Lopez-Jaramillo, P. Associations of the ‘weekend warrior’ physical activity pattern with all-cause, cardiovascular disease, and cancer mortality: The Mexico City Prospective Study. Br. J. Sports Med. 2024, 58, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Kunutsor, S.K.; Jae, S.Y.; Laukkanen, J.A. ‘Weekend warrior’ and regularly active physical activity patterns confer similar cardiovascular and mortality benefits: A systematic meta-analysis. Eur. J. Prev. Cardiol. 2023, 30, e7–e10. [Google Scholar] [CrossRef]
- Wegierska, A.E.; Charitos, I.A.; Topi, S.; Potenza, M.A.; Montagnani, M.; Santacroce, L. The Connection Between Physical Exercise and Gut Microbiota: Implications for Competitive Sports Athletes. Sports Med. 2022, 52, 2355–2369. [Google Scholar] [CrossRef] [PubMed]
- Strasser, B.; Wolters, M.; Weyh, C.; Krüger, K.; Ticinesi, A. The Effects of Lifestyle and Diet on Gut Microbiota Composition, Inflammation and Muscle Performance in Our Aging Society. Nutrients 2021, 13, 2045. [Google Scholar] [CrossRef] [PubMed]
- Mailing, L.J.; Allen, J.M.; Buford, T.W.; Fields, C.J.; Woods, J.A. Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health. Exerc. Sport Sci. Rev. 2019, 47, 75–85. [Google Scholar] [CrossRef]
- Bonomini-Gnutzmann, R.; Plaza-Díaz, J.; Jorquera-Aguilera, C.; Rodríguez-Rodríguez, A.; Rodríguez-Rodríguez, F. Effect of Intensity and Duration of Exercise on Gut Microbiota in Humans: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 9518. [Google Scholar] [CrossRef]
- Donati Zeppa, S.; Agostini, D.; Gervasi, M.; Annibalini, G.; Amatori, S.; Ferrini, F.; Sisti, D.; Piccoli, G.; Barbieri, E.; Sestili, P.; et al. Mutual Interactions among Exercise, Sport Supplements and Microbiota. Nutrients 2019, 12, 17. [Google Scholar] [CrossRef]
- Marttinen, M.; Ala-Jaakkola, R.; Laitila, A.; Lehtinen, M.J. Gut Microbiota, Probiotics and Physical Performance in Athletes and Physically Active Individuals. Nutrients 2020, 12, 2936. [Google Scholar] [CrossRef] [PubMed]
- Bernat-Adell, M.D.; Collado-Boira, E.J.; Moles-Julio, P.; Panizo-González, N.; Martínez-Navarro, I.; Hernando-Fuster, B.; Hernando-Domingo, C. Recovery of Inflammation, Cardiac, and Muscle Damage Biomarkers After Running a Marathon. J. Strength Cond. Res. 2021, 35, 626–632. [Google Scholar] [CrossRef]
- Faria, F.R.; Gomes, A.C.; Antunes, A.; Rezende, K.R.; Pimentel, G.D.; Oliveira, C.L.P.; Antunes, B.M.; Lira, F.S.; Aoki, M.S.; Mota, J.F. Effects of turmeric extract supplementation on inflammation and muscle damage after a half-marathon race: A randomized, double-blind, placebo-controlled trial. Eur. J. Appl. Physiol. 2020, 120, 1531–1540. [Google Scholar] [CrossRef]
- Aoi, W.; Naito, Y.; Sakuma, K.; Kuchide, M.; Tokuda, H.; Maoka, T.; Toyokuni, S.; Oka, S.; Yasuhara, M.; Yoshikawa, T. Astaxanthin limits exercise-induced skeletal and cardiac muscle damage in mice. Antioxid. Redox Signal. 2003, 5, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, W.C.; Wu, Z.Y.; Fu, C.X.; Hui, A.L.; Gao, H.; Chen, P.P.; Du, B.; Zhang, H.W. Two macamide extracts relieve physical fatigue by attenuating muscle damage in mice. J. Sci. Food Agric. 2019, 99, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, S.; Al-Alusi, M.A.; Churchill, T.W.; Guseh, J.S.; Ellinor, P.T. Accelerometer-Derived “Weekend Warrior” Physical Activity and Incident Cardiovascular Disease. JAMA 2023, 330, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.H.; Huang, S.; Pu, Y.Q.; Zhao, Y.; Chen, Y.C.; Jiang, N.; Liu, M.L.; Bao, W.W.; Zhang, Y.S.; Hu, L.X.; et al. Whether weekend warrior activity and other leisure-time physical activity pattern reduce the risk of depression symptom in the representative adults? A population-based analysis of NHANES 2007–2020. J. Affect. Disord. 2023, 340, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yang, W.; Liu, F. The associations of weekend warrior and other physical activity patterns with the risk of all-cause and cardiovascular disease mortality in people with diabetes mellitus and chronic kidney disease: From NHANES 2007–2020. Int. Urol. Nephrol. 2024, 56, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, N.; Mizokami, T.; Niihara, H.; Yada, K.; Suzuki, K. Macrophage depletion by clodronate liposome attenuates muscle injury and inflammation following exhaustive exercise. Biochem. Biophys. Rep. 2015, 5, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zheng, J.; Qiu, J.; Wu, X.; Xu, Y.; Shen, W.; Sun, M. ALDH2 restores exhaustive exercise-induced mitochondrial dysfunction in skeletal muscle. Biochem. Biophys. Res. Commun. 2017, 485, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Ablitip, A.; Wang, R.; Luciana, T.; Wei, M.; Ma, X. Effects of Exercise on Gut Microbiota of Adults: A Systematic Review and Meta-Analysis. Nutrients 2024, 16, 1070. [Google Scholar] [CrossRef] [PubMed]
- Boytar, A.N.; Skinner, T.L.; Wallen, R.E.; Jenkins, D.G.; Dekker Nitert, M. The Effect of Exercise Prescription on the Human Gut Microbiota and Comparison between Clinical and Apparently Healthy Populations: A Systematic Review. Nutrients 2023, 15, 1534. [Google Scholar] [CrossRef]
- Allen, J.M.; Berg Miller, M.E.; Pence, B.D.; Whitlock, K.; Nehra, V.; Gaskins, H.R.; White, B.A.; Fryer, J.D.; Woods, J.A. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J. Appl. Physiol 2015, 118, 1059–1066. [Google Scholar] [CrossRef]
- Lensu, S.; Pekkala, S. Gut Microbiota, Microbial Metabolites and Human Physical Performance. Metabolites 2021, 11, 716. [Google Scholar] [CrossRef] [PubMed]
- Karl, J.P.; Margolis, L.M.; Madslien, E.H.; Murphy, N.E.; Castellani, J.W.; Gundersen, Y.; Hoke, A.V.; Levangie, M.W.; Kumar, R.; Chakraborty, N.; et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G559–G571. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Iglesias, P.; Massot-Cladera, M.; Rodríguez-Lagunas, M.J.; Franch, À.; Camps-Bossacoma, M.; Castell, M.; Pérez-Cano, F.J. A Cocoa Diet Can Partially Attenuate the Alterations in Microbiota and Mucosal Immunity Induced by a Single Session of Intensive Exercise in Rats. Front. Nutr. 2022, 9, 861533. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Huang, Y.; Yuan, D.; Liu, J. Atherosclerosis, gut microbiome, and exercise in a meta-omics perspective: A literature review. PeerJ 2024, 12, e17185. [Google Scholar] [CrossRef] [PubMed]
- Uchida, M.; Fujie, S.; Yano, H.; Iemitsu, M. Aerobic exercise training-induced alteration of gut microbiota composition affects endurance capacity. J. Physiol. 2023, 601, 2329–2344. [Google Scholar] [CrossRef] [PubMed]
- Scheiman, J.; Luber, J.M.; Chavkin, T.A.; MacDonald, T.; Tung, A.; Pham, L.D.; Wibowo, M.C.; Wurth, R.C.; Punthambaker, S.; Tierney, B.T.; et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 2019, 25, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Zmora, N.; Adolph, T.E.; Elinav, E. The intestinal microbiota fuelling metabolic inflammation. Nat. Rev. Immunol. 2020, 20, 40–54. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-Chain Fatty Acids and Their Association with Signalling Pathways in Inflammation, Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef]
- Brandsma, E.; Kloosterhuis, N.J.; Koster, M.; Dekker, D.C.; Gijbels, M.J.J.; van der Velden, S.; Ríos-Morales, M.; van Faassen, M.J.R.; Loreti, M.G.; de Bruin, A.; et al. A Proinflammatory Gut Microbiota Increases Systemic Inflammation and Accelerates Atherosclerosis. Circ. Res. 2019, 124, 94–100. [Google Scholar] [CrossRef]
- Wang, D.; Wang, L.; Han, L.; Wang, B.; Shi, R.; Ye, J.; Xia, B.; Zhao, Z.; Zhao, B.; Liu, X. Leucine-Restricted Diet Ameliorates Obesity-Linked Cognitive Deficits: Involvement of the Microbiota-Gut-Brain Axis. J. Agric. Food Chem. 2023, 71, 9404–9418. [Google Scholar] [CrossRef]
- Li, X.; Wang, Q.; Wu, D.; Zhang, D.W.; Li, S.C.; Zhang, S.W.; Chen, X.; Li, W. The effect of a novel anticonvulsant chemical Q808 on gut microbiota and hippocampus neurotransmitters in pentylenetetrazole-induced seizures in rats. BMC Neurosci. 2022, 23, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, W.; Wang, J.; Shi, J.; Sun, Y.; Wang, W.; Ning, G.; Liu, R.; Hong, J. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J. Mol. Endocrinol. 2017, 58, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Wang, X.; Liu, D.; Xu, A.; Cheng, H.; Li, L.; Zhang, C. Tolypocladium sinense Mycelium Polysaccharide Alleviates Obesity, Lipid Metabolism Disorder, and Inflammation Caused by High Fat Diet via Improving Intestinal Barrier and Modulating Gut Microbiota. Mol. Nutr. Food Res. 2024, 68, e2300759. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Wang, Y.; Zhang, S.; Zhang, X.; Du, Z.; Li, M.; Ding, K. Crataegus pinnatifida polysaccharide alleviates colitis via modulation of gut microbiota and SCFAs metabolism. Int. J. Biol. Macromol. 2021, 181, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yan, Y.; Wan, P.; Dong, W.; Huang, K.; Ran, L.; Mi, J.; Lu, L.; Zeng, X.; Cao, Y. Effects of long-term intake of anthocyanins from Lycium ruthenicum Murray on the organism health and gut microbiota in vivo. Food Res. Int. 2020, 130, 108952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, N.; Kan, J.; Sun, R.; Tang, S.; Wang, Z.; Chen, M.; Liu, J.; Jin, C. Anti-inflammatory activity of alkali-soluble polysaccharides from Arctium lappa L. and its effect on gut microbiota of mice with inflammation. Int. J. Biol. Macromol. 2020, 154, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tao, H.; Huang, H.; Xiao, Y.; Wu, X.; Li, M.; Shen, J.; Xiao, Z.; Zhao, Y.; Du, F.; et al. The dietary supplement Rhodiola crenulata extract alleviates dextran sulfate sodium-induced colitis in mice through anti-inflammation, mediating gut barrier integrity and reshaping the gut microbiome. Food Funct. 2021, 12, 3142–3158. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xiao, P.; Zhang, X.; Yang, Y.; Yang, M.; Wang, T.; Lu, H.; Tian, H.; Wang, H.; Liu, J. Inulin ameliorates schizophrenia via modulation of the gut microbiota and anti-inflammation in mice. Food Funct. 2021, 12, 1156–1175. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, Y.; Zhao, X.; Shang, C.; Xiang, M.; Li, L.; Cui, X. Microbiota-derived short-chain fatty acids: Implications for cardiovascular and metabolic disease. Front. Cardiovasc. Med. 2022, 9, 900381. [Google Scholar] [CrossRef]
- Niu, W.; Yang, F.; Fu, Z.; Dong, Y.; Zhang, Z.; Ju, J. The role of enteric dysbacteriosis and modulation of gut microbiota in the treatment of inflammatory bowel disease. Microb. Pathog. 2022, 165, 105381. [Google Scholar] [CrossRef]
- Astley, R.; Miller, F.C.; Mursalin, M.H.; Coburn, P.S.; Callegan, M.C. An Eye on Staphylococcus aureus Toxins: Roles in Ocular Damage and Inflammation. Toxins 2019, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Garrett, S.; Hong, W.; Zhang, J. Staphylococcus aureus Infections and Human Intestinal Microbiota. Pathogens 2024, 13, 276. [Google Scholar] [CrossRef]
- Olson, C.A.; Iñiguez, A.J.; Yang, G.E.; Fang, P.; Pronovost, G.N.; Jameson, K.G.; Rendon, T.K.; Paramo, J.; Barlow, J.T.; Ismagilov, R.F.; et al. Alterations in the gut microbiota contribute to cognitive impairment induced by the ketogenic diet and hypoxia. Cell Host Microbe 2021, 29, 1378–1392.e6. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Long, W.; Hao, B.; Ding, D.; Ma, X.; Zhao, L.; Pang, X. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog. 2017, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Agudelo-Ochoa, G.M.; Valdés-Duque, B.E.; Giraldo-Giraldo, N.A.; Jaillier-Ramírez, A.M.; Giraldo-Villa, A.; Acevedo-Castaño, I.; Yepes-Molina, M.A.; Barbosa-Barbosa, J.; Benítez-Paéz, A. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes 2020, 12, 1707610. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Vacharaksa, A.; Croxen, M.; Thanachayanont, T.; Finlay, B.B. Altering host resistance to infections through microbial transplantation. PLoS ONE 2011, 6, e26988. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yang, J.; Jian, Y.; Lei, Y.; Yao, S.; Hu, Z.; Liu, X.; Tang, C.; Liu, W. Treadmill Exercise Modulates Intestinal Microbes and Suppresses LPS Displacement to Alleviate Neuroinflammation in the Brains of APP/PS1 Mice. Nutrients 2022, 14, 4134. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Wang, S.; Tian, M.; Wang, L.; Gao, J.; Zhao, Q.; Li, Z.; Jia, X.; Yu, Y. Exercise Preconditioning Ameliorates Cognitive Impairment in Mice with Ischemic Stroke by Alleviating Inflammation and Modulating Gut Microbiota. Mediat. Inflamm. 2022, 2022, 2124230. [Google Scholar] [CrossRef]
- Lai, Z.L.; Tseng, C.H.; Ho, H.J.; Cheung, C.K.Y.; Lin, J.Y.; Chen, Y.J.; Cheng, F.C.; Hsu, Y.C.; Lin, J.T.; El-Omar, E.M.; et al. Fecal microbiota transplantation confers beneficial metabolic effects of diet and exercise on diet-induced obese mice. Sci. Rep. 2018, 8, 15625. [Google Scholar] [CrossRef]
- Vinolo, M.A.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Zhu, Y.; Wei, A.; Du, J.; Wang, Y.; Zheng, P.; Tu, M.; Wang, H.; Wen, L.; Yang, X. Traumatic Brain Injury Induces Gastrointestinal Dysfunction and Dysbiosis of Gut Microbiota Accompanied by Alterations of Bile Acid Profile. J. Neurotrauma 2022, 39, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Terzo, S.; Mulè, F.; Caldara, G.F.; Baldassano, S.; Puleio, R.; Vitale, M.; Cassata, G.; Ferrantelli, V.; Amato, A. Pistachio Consumption Alleviates Inflammation and Improves Gut Microbiota Composition in Mice Fed a High-Fat Diet. Int. J. Mol. Sci. 2020, 21, 365. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wang, J.; Kou, R.; Chen, M.; Zhang, B.; Zhang, Y.; Liu, J.; Xing, X.; Peng, B.; Wang, S. Polyphenol-rich oolong tea alleviates obesity and modulates gut microbiota in high-fat diet-fed mice. Front. Nutr. 2022, 9, 937279. [Google Scholar] [CrossRef] [PubMed]
- Bei, Y.; Wang, L.; Ding, R.; Che, L.; Fan, Z.; Gao, W.; Liang, Q.; Lin, S.; Liu, S.; Lu, X.; et al. Animal exercise studies in cardiovascular research: Current knowledge and optimal design-A position paper of the Committee on Cardiac Rehabilitation, Chinese Medical Doctors’ Association. J. Sport Health Sci. 2021, 10, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Bárcena, C.; Valdés-Mas, R.; Mayoral, P.; Garabaya, C.; Durand, S.; Rodríguez, F.; Fernández-García, M.T.; Salazar, N.; Nogacka, A.M.; Garatachea, N.; et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 2019, 25, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Li, X.; Li, X.; Wei, C.; Shi, C.; Hu, K.; Kong, D.; Luo, Q.; Xu, Y.; Shan, W.; et al. Fecal microbiota transplantation from young mice rejuvenates aged hematopoietic stem cells by suppressing inflammation. Blood 2023, 141, 1691–1707. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Species | Forward Primers Sequence (5′–3′) | Reverse Primers Sequence (5′–3′) |
|---|---|---|---|
| IL-1β | mouse | CCCAAGCAATACCCAAAGAA | TTGTGAGGTGCTGATGTACCA |
| IL6 | mouse | GAACAACGATGATGCACTTGC | CTTCATGTACTCCAGGTAGCTATGGT |
| TNF-α | mouse | CTTCTGTCTACTGAACTTCGGG | CACTTGGTGGTTTGCTACGAC |
| FoxO3a | mouse | TCACTGTATTCAGCTAGTGCAA | ATGATGGACTCCATGTCACATT |
| MAFbx | mouse | GACTGGACTTCTCGACTGCC | TCAGGGATGTGAGCTGTGAC |
| MuRF1 | mouse | ACCTGCTGGTGGAAAACATC | CTTCGTGTTCCTTGCACATC |
| GPADH | mouse | CATCACTGCCACCCAGAAGACTG | ATGCCAGTGAGCTTCCCGTTCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, C.; Lang, H.; Yu, H.; Zhou, M.; Rao, X.; Zhang, Q.; Yi, L.; Zhu, J.; Mi, M. The Contrasting Effects of Two Distinct Exercise Training Modalities on Exhaustive Exercise-Induced Muscle Damage in Mice May Be Associated with Alterations in the Gut Microbiota. Int. J. Mol. Sci. 2024, 25, 7837. https://doi.org/10.3390/ijms25147837
Zhang Y, Wang C, Lang H, Yu H, Zhou M, Rao X, Zhang Q, Yi L, Zhu J, Mi M. The Contrasting Effects of Two Distinct Exercise Training Modalities on Exhaustive Exercise-Induced Muscle Damage in Mice May Be Associated with Alterations in the Gut Microbiota. International Journal of Molecular Sciences. 2024; 25(14):7837. https://doi.org/10.3390/ijms25147837
Chicago/Turabian StyleZhang, Yong, Cong Wang, Hedong Lang, Hongtao Yu, Min Zhou, Xin Rao, Qianyong Zhang, Long Yi, Jundong Zhu, and Mantian Mi. 2024. "The Contrasting Effects of Two Distinct Exercise Training Modalities on Exhaustive Exercise-Induced Muscle Damage in Mice May Be Associated with Alterations in the Gut Microbiota" International Journal of Molecular Sciences 25, no. 14: 7837. https://doi.org/10.3390/ijms25147837




