Conserved Signaling Pathways in the Ciona robusta Gut
Abstract
1. Introduction
2. Results
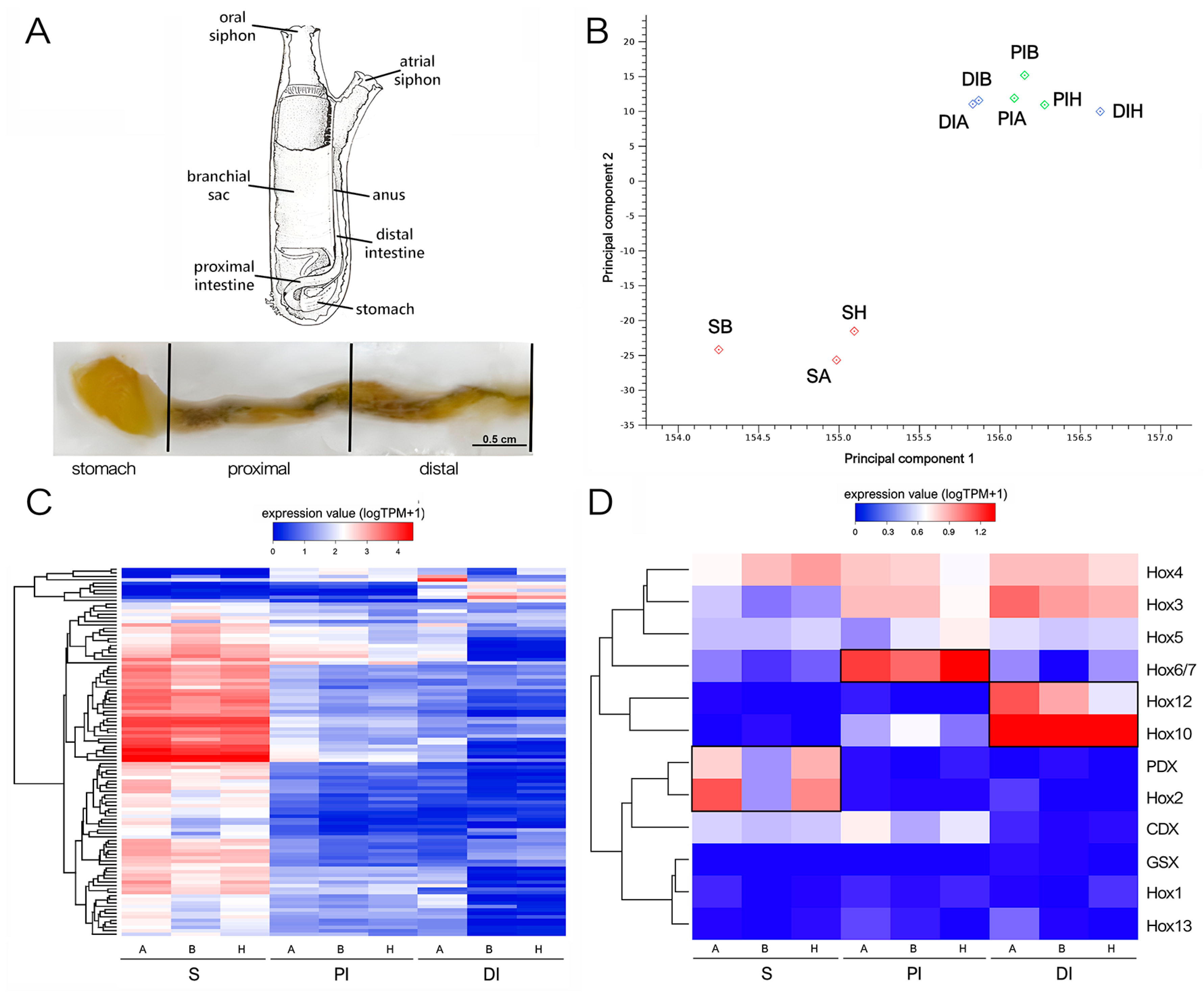
2.1. RNA-Seq Analysis and Validation of Differentially Expressed Genes (DEGs)
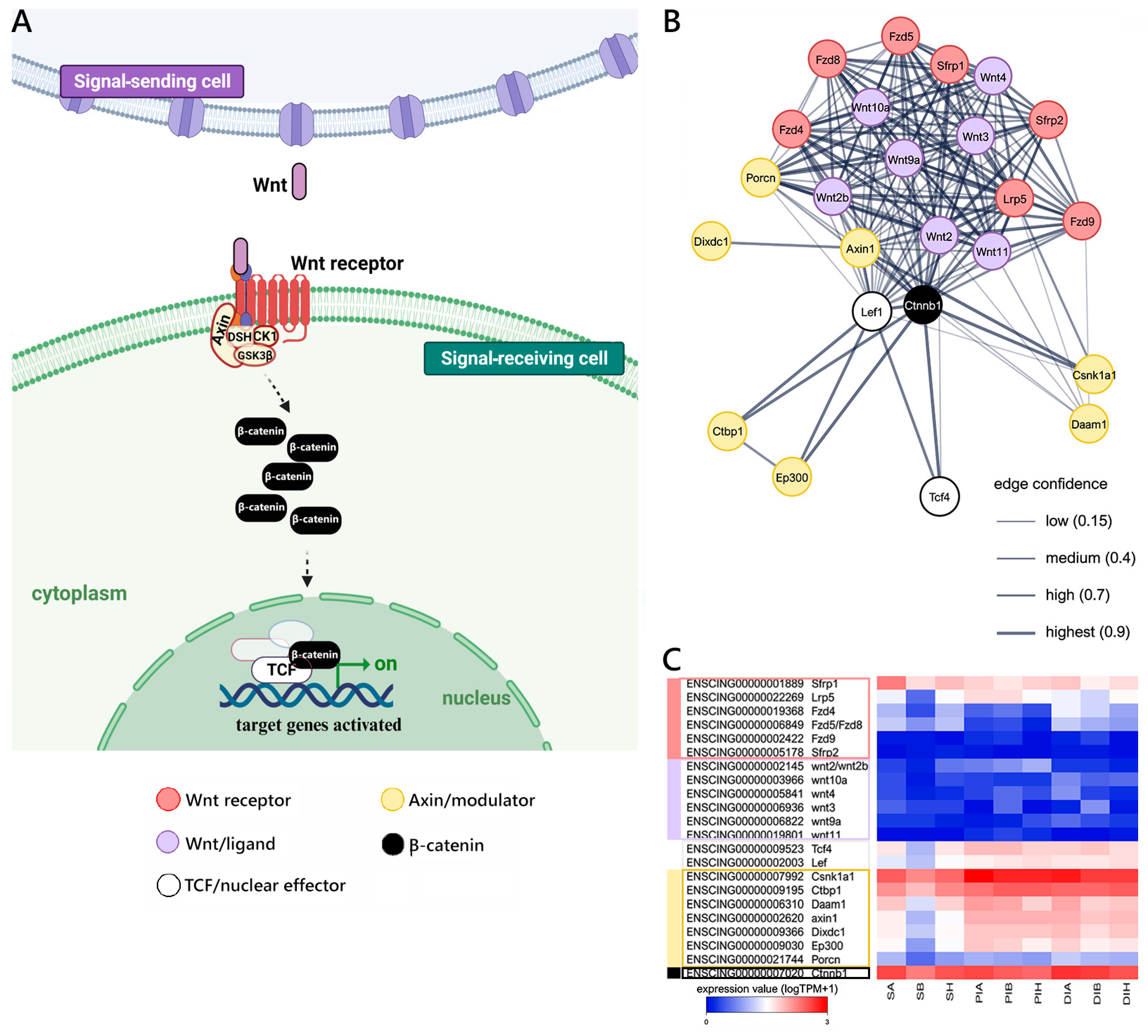
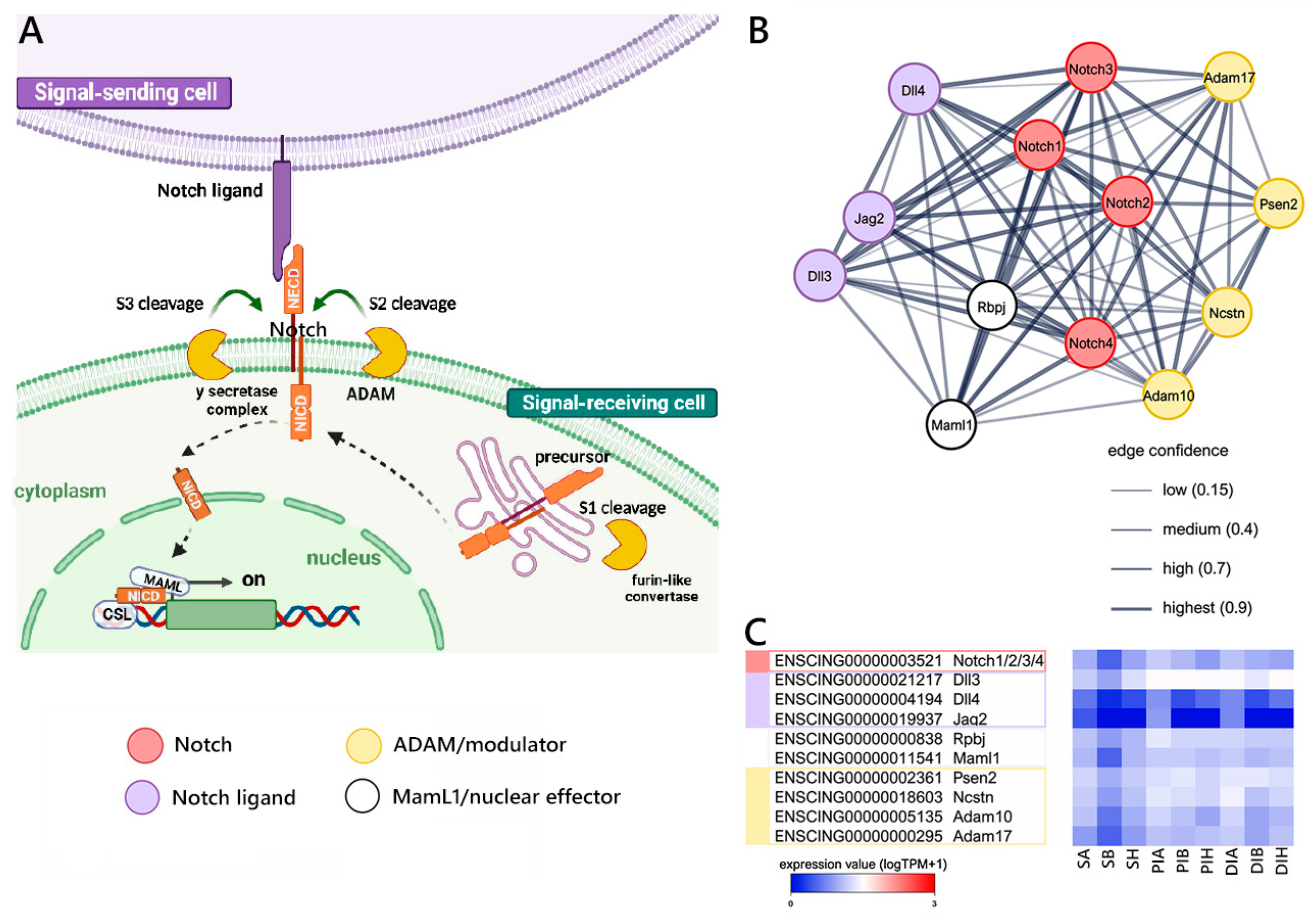
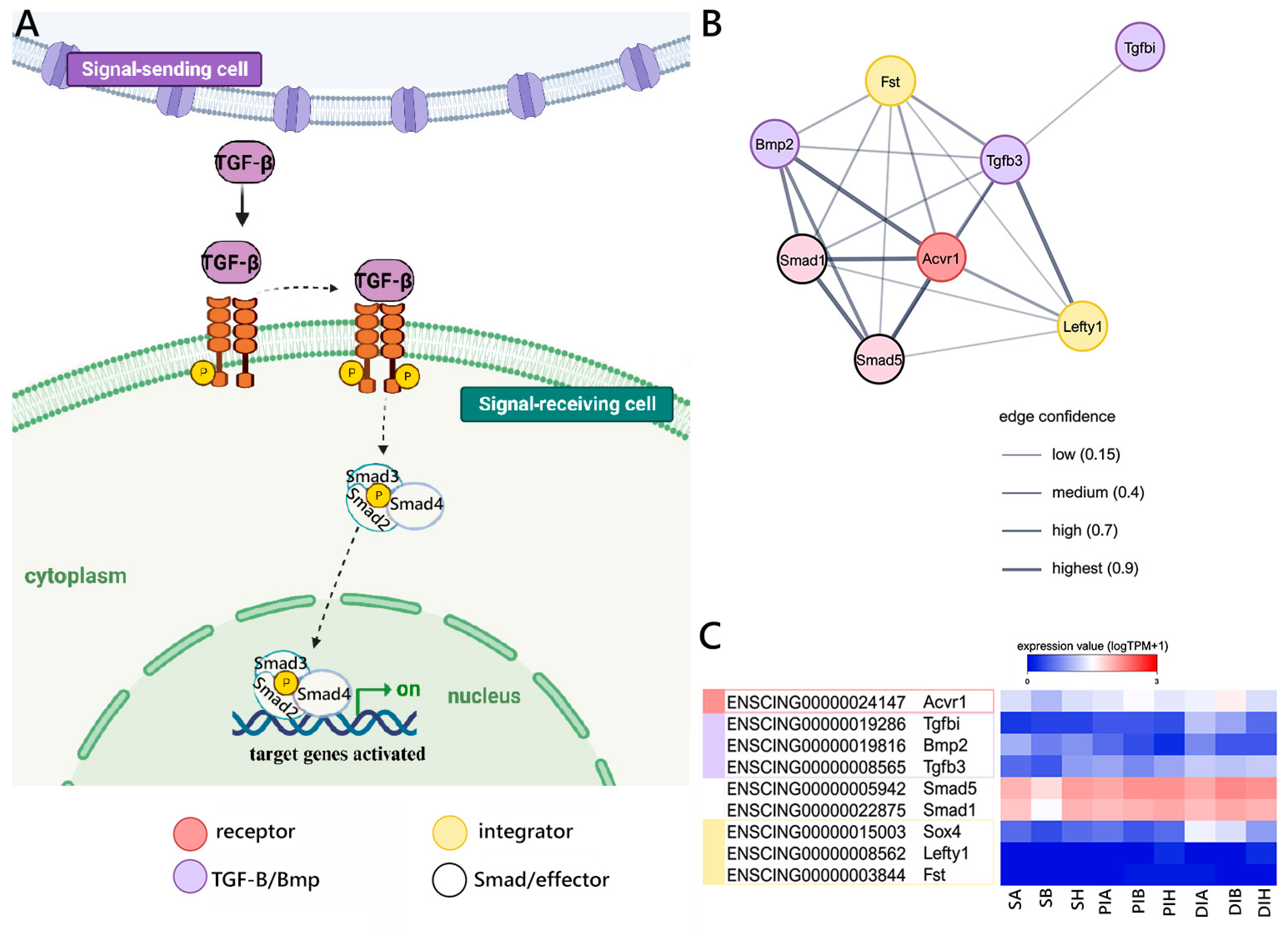
2.2. Evolutionarily Conserved Signaling Pathways
3. Discussion
4. Materials and Methods
4.1. Samples Collection
4.2. RNA-Seq
4.3. Bioinformatic Analyses
4.4. Real-Time Quantitative PCR (RT-qPCR)
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wanninger, A. Evolutionary Developmental Biology of Invertebrates 6: Deuterostomia; Springer: Berlin, Germany, 2015; ISBN 9783709118566. [Google Scholar]
- Burighel, P.; Cloney, R.A. Urochordata, Ascidiacea. In Microscopic Anatomy of Invertebrates, Volume 15: Hemichordata, Chaetognata, and Invertebrate Chordates; Wiley-Liss: New York, NY, USA, 1997; pp. 221–347. [Google Scholar]
- Liberti, A.; Bertocci, I.; Pollet, A.; Musco, L.; Locascio, A.; Ristoratore, F.; Spagnuolo, A.; Sordino, P. An Indoor Study of the Combined Effect of Industrial Pollution and Turbulence Events on the Gut Environment in a Marine Invertebrate: The Response of the Gut Environment to Polluted Sediment Reworking. Mar. Environ. Res. 2020, 158, 104950. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Toscano, F.; Arienzo, M.; Ferrara, L.; Procaccini, G.; Sordino, P. Temporal Correlation of Population Composition and Environmental Variables in the Marine Invader Ciona Robusta. Mar. Ecol. 2019, 40, e12543. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, Y.; Yi, C.; Fong, J.J.; Kim, W.; Rius, M.; Zhan, A. Genetic Signatures of Natural Selection in a Model Invasive Ascidian. Sci. Rep. 2017, 7, 44080. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Boni, R.; Buia, M.C.; Monfrecola, V.; Esposito, M.C.; Tosti, E. Ocean Acidification Impact on Ascidian Ciona Robusta Spermatozoa: New Evidence for Stress Resilience. Sci. Total Environ. 2019, 697, 134100. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Nakamura, R.; Yu, D.; Yoshida, R.; Hamada, M.; Fujie, M.; Hisata, K.; Takeda, H.; Satoh, N.; O’Neill, R. A Nearly Complete Genome of Ciona Intestinalis Type A (C. robusta) Reveals the Contribution of Inversion to Chromosomal Evolution in the Genus Ciona. Genome Biol. Evol. 2019, 11, 3144–3157. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, R.; Gissi, C.; Pennati, R.; Caicci, F.; Gasparini, F.; Manni, L. Morphological Evidence That the Molecularly Determined Ciona Intestinalis Type A and Type B Are Different Species: Ciona Robusta and Ciona Intestinalis. J. Zool. Syst. Evol. Res. 2015, 53, 186–193. [Google Scholar] [CrossRef]
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and Not Cephalochordates Are the Closest Living Relatives of Vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef]
- Tsagkogeorga, G.; Turon, X.; Hopcroft, R.R.; Tilak, M.K.; Feldstein, T.; Shenkar, N.; Loya, Y.; Huchon, D.; Douzery, E.J.; Delsuc, F. An Updated 18S RRNA Phylogeny of Tunicates Based on Mixture and Secondary Structure Models. BMC Evol. Biol. 2009, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Corbo, J.C.; Di Gregorio, A.; Levine, M. The Ascidian as a Model Organism in Developmental and Evolutionary Biology. Cell 2001, 106, 535–538. [Google Scholar] [CrossRef]
- Nakayama, S.; Sekiguchi, T.; Ogasawara, M. Molecular and Evolutionary Aspects of the Protochordate Digestive System. Cell Tissue Res. 2019, 377, 309–320. [Google Scholar] [CrossRef]
- Nakayama, S.; Ogasawara, M. Compartmentalized Expression Patterns of Pancreatic- and Gastric-Related Genes in the Alimentary Canal of the Ascidian Ciona Intestinalis: Evolutionary Insights into the Functional Regionality of the Gastrointestinal Tract in Olfactores. Cell Tissue Res. 2017, 370, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Satou, K.; Orito, W.; Ogasawara, M. Ordered Expression Pattern of Hox and ParaHox Genes along the Alimentary Canal in the Ascidian Juvenile. Cell Tissue Res. 2016, 365, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Liberti, A.; Natarajan, O.; Atkinson, C.G.F.; Sordino, P.; Dishaw, L.J. Reflections on the Use of an Invertebrate Chordate Model System for Studies of Gut Microbial Immune Interactions. Front. Immunol. 2021, 12, 642687. [Google Scholar] [CrossRef] [PubMed]
- Darras, S.; Nishida, H. The BMP/CHORDIN Antagonism Controls Sensory Pigment Cell Specification and Differentiation in the Ascidian Embryo. Dev. Biol. 2001, 236, 271–288. [Google Scholar] [CrossRef] [PubMed]
- Roure, A.; Chowdhury, R.; Darras, S. Regulation of Anterior Neurectoderm Specification and Differentiation by BMP Signaling in Ascidians. Development 2023, 150, dev201575. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Goricki, S.; Byerly, M.S.; Satoh, N.; Jeffery, W.R. Evolution of the Chordate Regeneration Blastema: Differential Gene Expression and Conserved Role of Notch Signaling during Siphon Regeneration in the Ascidian Ciona. Dev. Biol. 2015, 405, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Hino, K.; Satou, Y.; Yagi, K.; Satoh, N. A Genomewide Survey of Developmentally Relevant Genes in Ciona Intestinalis. VI. Genes for Wnt, TGFβ, Hedgehog and JAK/STAT Signalling Pathways. Dev. Genes Evol. 2003, 213, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Van Es, J.H.; Kuipers, J.; Kujala, P.; Van Den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of Stem Cells in Small Intestine and Colon by Marker Gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt Signals Are Essential for Homeostasis of the Intestinal Epithelium. Genes Dev. 2003, 17, 1709–1713. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-Catenin Signalling: Function, Biological Mechanisms, and Therapeutic Opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Gehart, H.; Clevers, H. Tales from the Crypt: New Insights into Intestinal Stem Cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Steinhart, Z.; Angers, S. Wnt Signaling in Development and Tissue Homeostasis. Development 2018, 145, dev146589. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch Signaling Pathway: Architecture, Disease, and Therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Schmierer, B.; Hill, C.S. TGFβ-SMAD Signal Transduction: Molecular Specificity and Functional Flexibility. Nat. Rev. Mol. Cell Biol. 2007, 8, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Millar, R.H.; Ciona, L.M.B.C. Memoirs on Typical British Marine Plants and Animals; Liverpool University Press: Liverpool, UK, 1953; Volume 35. [Google Scholar]
- Castelli-Gair Hombría, J.; Lovegrove, B. Beyond Homeosis—HOX Function in Morphogenesis and Organogenesis. Differentiation 2003, 71, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Grapin-Botton, A.; Melton, D.A. Endoderm Development: From Patterning to Organogenesis. Trends Genet. 2000, 16, 124–130. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, A.; Yu, H.; Dong, B. Comparative Transcriptomic Analysis Reveals the Functionally Segmented Intestine in Tunicate Ascidian. Int. J. Mol. Sci. 2023, 24, 6270. [Google Scholar] [CrossRef]
- Delsuc, F.; Philippe, H.; Tsagkogeorga, G.; Simion, P.; Tilak, M.K.; Turon, X.; López-Legentil, S.; Piette, J.; Lemaire, P.; Douzery, E.J.P. A Phylogenomic Framework and Timescale for Comparative Studies of Tunicates. BMC Biol. 2018, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, S.; Osugi, T.; Shiraishi, A.; Wada, A.; Satake, H. Comparative Analysis of Transcriptomic Profiles among Ascidians, Zebrafish, and Mice: Insights from Tissue-Specific Gene Expression. PLoS ONE 2021, 16, e0254308. [Google Scholar] [CrossRef]
- Sailaja, B.S.; He, X.C.; Li, L. The Regulatory Niche of Intestinal Stem Cells. J. Physiol. 2016, 594, 4827–4836. [Google Scholar] [CrossRef]
- Fre, S.; Pallavi, S.K.; Huyghe, M.; Laé, M.; Janssen, K.P.; Robine, S.; Artavanis-Tsakonas, S.; Louvard, D. Notch and Wnt Signals Cooperatively Control Cell Proliferation and Tumorigenesis in the Intestine. Proc. Natl. Acad. Sci. USA 2009, 106, 6309–6314. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, N.; Wang, W.; Christiaen, L. Initial characterization of Wnt-Tcf functions during Ciona heart development. Dev Biol. 2020, 448, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Yen, P.M.; Ando, S.; Feng, X.; Liu, Y.; Maruvada, P.; Xia, X. Thyroid Hormone Action at the Cellular, Genomic and Target Gene Levels. Mol. Cell. Endocrinol. 2006, 246, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Swalla, B.J. Molecular Phylogeny of the Protochordates: Chordate Evolution. Can. J. Zool. 2005, 83, 24–33. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of MRNA Abundance Using RNA-Seq Data: RPKM Measure Is Inconsistent among Samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Wright, R.M.; Aglyamova, G.V.; Meyer, E.; Matz, M.V. Gene Expression Associated with White Syndromes in a Reef Building Coral, Acropora Hyacinthus. BMC Genomics 2015, 16, 371. [Google Scholar] [CrossRef]
- Greco, S.; Gaetano, A.S.; Furlanis, G.; Capanni, F.; Manfrin, C.; Giulianini, P.G.; Santovito, G.; Edomi, P.; Pallavicini, A.; Gerdol, M. Gene Expression Profiling of Trematomus Bernacchii in Response to Thermal and Stabling Stress. Fishes 2022, 7, 387. [Google Scholar] [CrossRef]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2023, 51, D933–D941. [Google Scholar] [CrossRef]
- Muñoz, J.; Stange, D.E.; Schepers, A.G.; Van De Wetering, M.; Koo, B.K.; Itzkovitz, S.; Volckmann, R.; Kung, K.S.; Koster, J.; Radulescu, S.; et al. The Lgr5 Intestinal Stem Cell Signature: Robust Expression of Proposed Quiescent ‘+4’ Cell Markers. EMBO J. 2012, 31, 3079–3091. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Kapustin, Y.; Souvorov, A.; Tatusova, T.; Lipman, D. Splign: Algorithms for Computing Spliced Alignments with Identification of Paralogs. Biol. Direct 2008, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerdol, M.; Greco, S.; Marino, R.; Locascio, A.; Plateroti, M.; Sirakov, M. Conserved Signaling Pathways in the Ciona robusta Gut. Int. J. Mol. Sci. 2024, 25, 7846. https://doi.org/10.3390/ijms25147846
Gerdol M, Greco S, Marino R, Locascio A, Plateroti M, Sirakov M. Conserved Signaling Pathways in the Ciona robusta Gut. International Journal of Molecular Sciences. 2024; 25(14):7846. https://doi.org/10.3390/ijms25147846
Chicago/Turabian StyleGerdol, Marco, Samuele Greco, Rita Marino, Annamaria Locascio, Michelina Plateroti, and Maria Sirakov. 2024. "Conserved Signaling Pathways in the Ciona robusta Gut" International Journal of Molecular Sciences 25, no. 14: 7846. https://doi.org/10.3390/ijms25147846
APA StyleGerdol, M., Greco, S., Marino, R., Locascio, A., Plateroti, M., & Sirakov, M. (2024). Conserved Signaling Pathways in the Ciona robusta Gut. International Journal of Molecular Sciences, 25(14), 7846. https://doi.org/10.3390/ijms25147846









