O-Sialoglycoprotein Endopeptidase Deficiency Impairs Proteostasis and Induces Autophagy in Human Embryonic Stem Cells
Abstract
:1. Introduction
2. Results
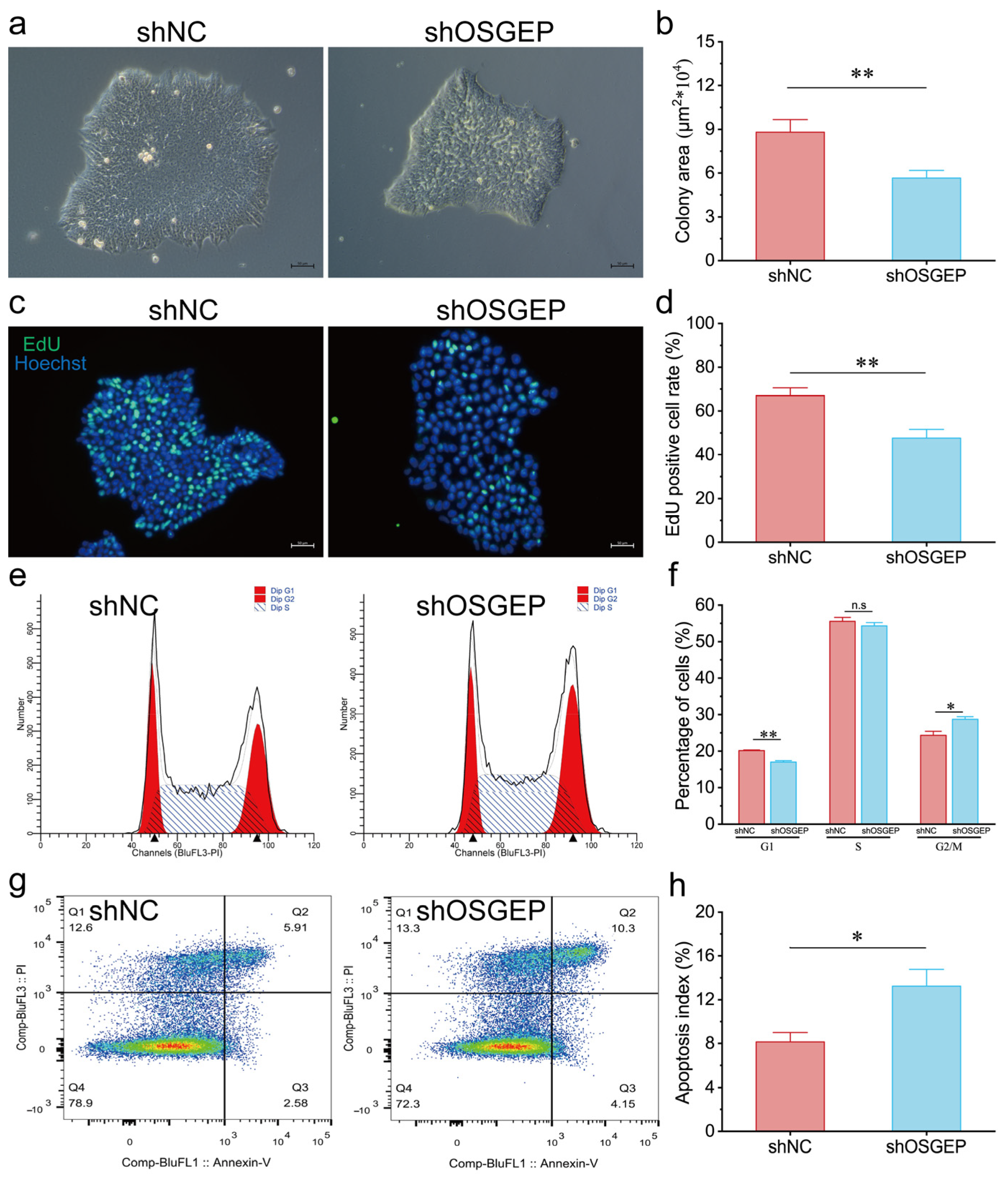
2.1. OSGEP Knockdown Results in Reduced Proliferation and Increased Apoptosis
2.2. OSGEP Knockdown Affects the Expression of Stemness Markers
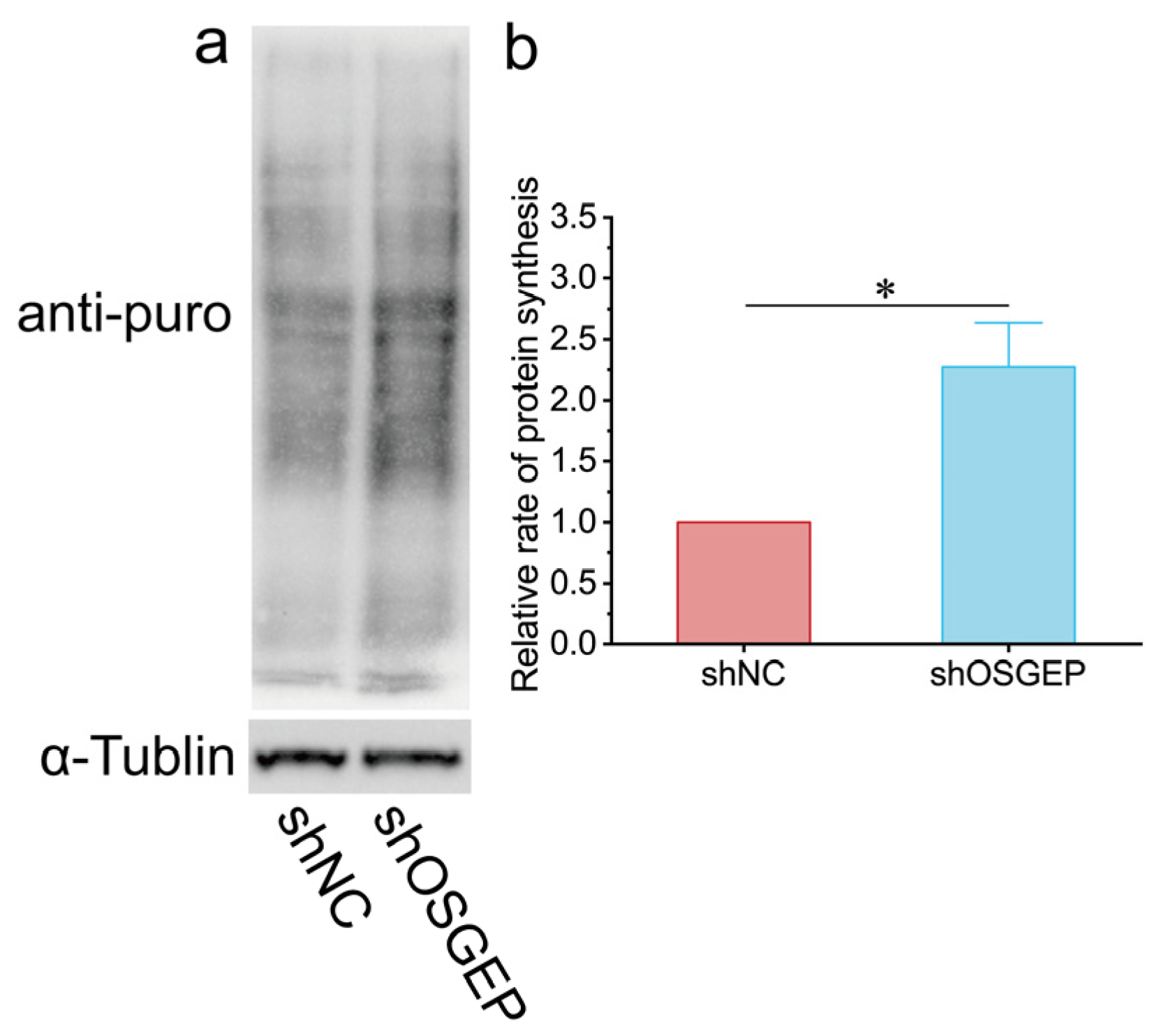
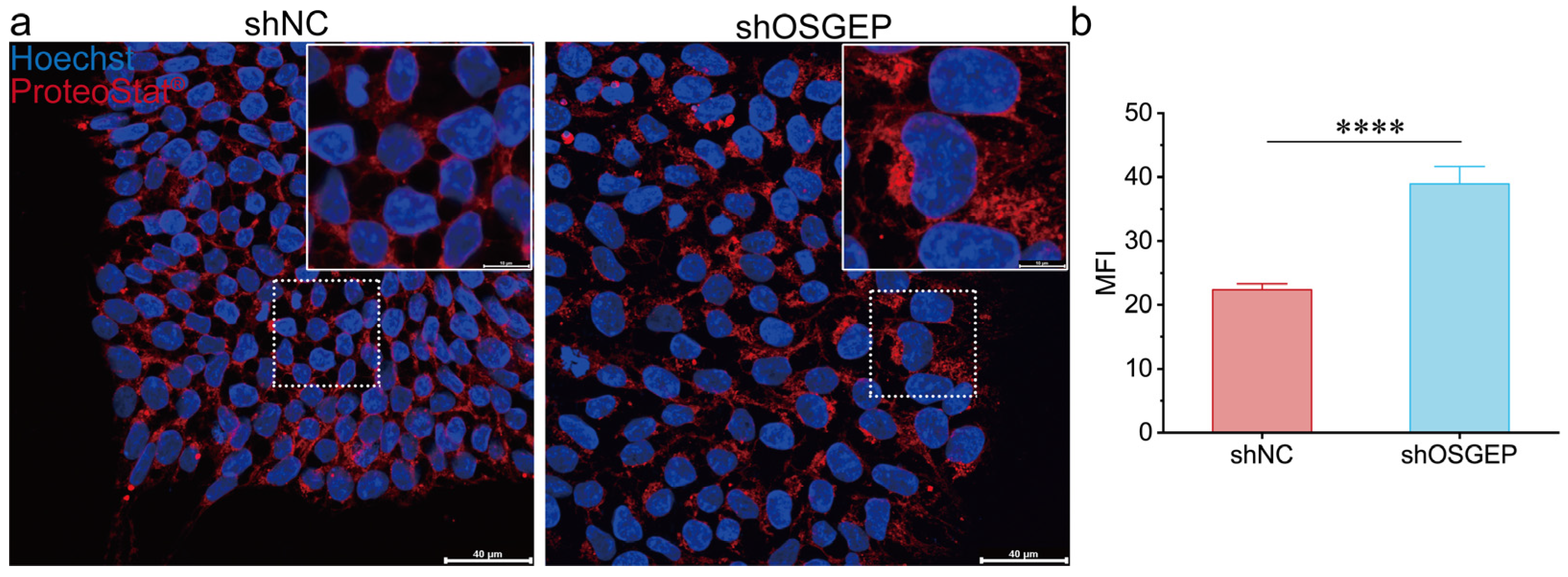
2.3. OSGEP Deficiency Perturbs Proteostasis
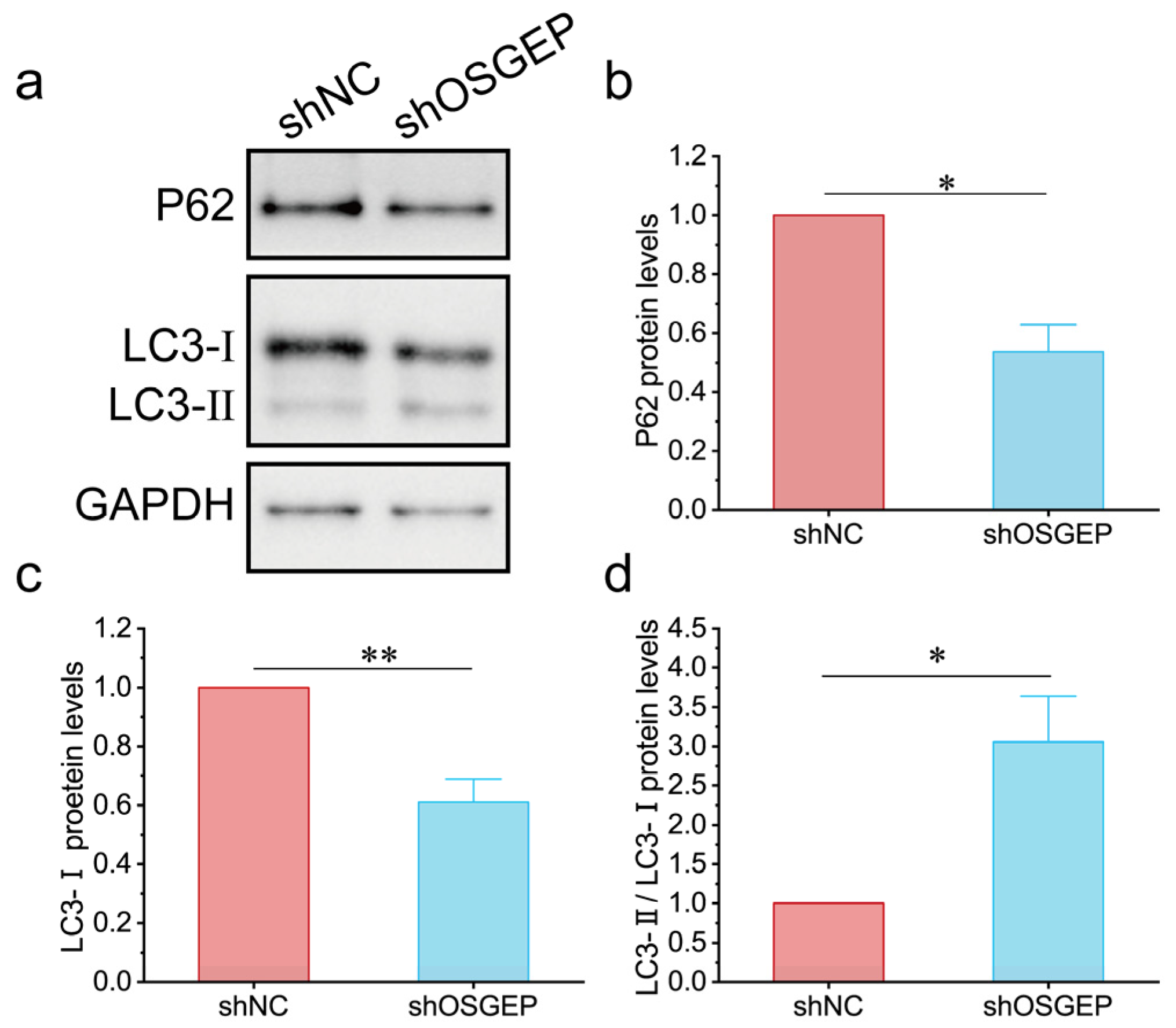
2.4. OSGEP Deficiency Induces Inappropriate Autophagy
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Lentivirus Production and Transduction of hESCs
4.3. HESC Self-Renewal and Proliferation Analysis
4.4. Flow Cytometry Analysis
4.5. Immunofluorescence Staining
4.6. Real-Time Quantitative PCR
4.7. Measurement of Protein Synthesis by Puromycin Incorporation
4.8. Western Blot Assay
4.9. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyer, O.; Mollet, G.; Dorval, G. Neurological involvement in monogenic podocytopathies. Pediatr. Nephrol. 2021, 36, 3571–3583. [Google Scholar] [CrossRef]
- Xu, S.; Hu, L.; Yang, L.; Wu, B.; Cao, Y.; Zhang, R.; Xu, X.; Ma, H.; Zhou, W.; Cheng, G.; et al. Galloway-Mowat Syndrome Type 3 Caused by OSGEP Gene Variants: A Case Report and Literature Review. Front. Pediatr. 2022, 10, 899991. [Google Scholar] [CrossRef]
- Mann, N.; Mzoughi, S.; Schneider, R.; Kühl, S.J.; Schanze, D.; Klämbt, V.; Lovric, S.; Mao, Y.; Shi, S.; Tan, W.; et al. Mutations in PRDM15 Are a Novel Cause of Galloway-Mowat Syndrome. J. Am. Soc. Nephrol. 2021, 32, 580–596. [Google Scholar] [CrossRef] [PubMed]
- Arrondel, C.; Missoury, S.; Snoek, R.; Patat, J.; Menara, G.; Collinet, B.; Liger, D.; Durand, D.; Gribouval, O.; Boyer, O.; et al. Defects in t(6)A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun. 2019, 10, 3967. [Google Scholar] [CrossRef]
- Braun, D.A.; Shril, S.; Sinha, A.; Schneider, R.; Tan, W.; Ashraf, S.; Hermle, T.; Jobst-Schwan, T.; Widmeier, E.; Majmundar, A.J.; et al. Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am. J. Med. Genet. A 2018, 176, 2460–2465. [Google Scholar] [CrossRef] [PubMed]
- Colin, E.; Huynh Cong, E.; Mollet, G.; Guichet, A.; Gribouval, O.; Arrondel, C.; Boyer, O.; Daniel, L.; Gubler, M.C.; Ekinci, Z.; et al. Loss-of-function mutations in WDR73 are responsible for microcephaly and steroid-resistant nephrotic syndrome: Galloway-Mowat syndrome. Am. J. Hum. Genet. 2014, 95, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A.; Tsukaguchi, H.; Koshimizu, E.; Nakazato, H.; Itoh, K.; Kuraoka, S.; Komohara, Y.; Shiina, M.; Nakamura, S.; Kitajima, M.; et al. Homozygous splicing mutation in NUP133 causes Galloway-Mowat syndrome. Ann. Neurol. 2018, 84, 814–828. [Google Scholar] [CrossRef] [PubMed]
- Rosti, R.O.; Sotak, B.N.; Bielas, S.L.; Bhat, G.; Silhavy, J.L.; Aslanger, A.D.; Altunoglu, U.; Bilge, I.; Tasdemir, M.; Yzaguirrem, A.D.; et al. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat syndrome. J. Med. Genet. 2017, 54, 399–403. [Google Scholar] [CrossRef]
- Braun, D.A.; Rao, J.; Mollet, G.; Schapiro, D.; Daugeron, M.C.; Tan, W.; Gribouval, O.; Boyer, O.; Revy, P.; Jobst-Schwan, T.; et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 2017, 49, 1529–1538. [Google Scholar] [CrossRef]
- Hong, S.Y.; Yang, J.J.; Li, S.Y.; Lee, I.C. A Wide Spectrum of Genetic Disorders Causing Severe Childhood Epilepsy in Taiwan: A Case Series of Ultrarare Genetic Cause and Novel Mutation Analysis in a Pilot Study. J. Pers. Med. 2020, 10, 281. [Google Scholar] [CrossRef]
- Yang, Y.; He, Y.; Zhen, L.; Li, D.Z. Fetal phenotype of Galloway-Mowat syndrome 3 caused by a specific OSGEP variant. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 242, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Gallego, A.; Furlano, M.; Pybus, M.; Barraca, D.; Martínez, A.B.; Mora Muñoz, E.; Torra, R.; Ars, E. Novel homozygous OSGEP gene pathogenic variants in two unrelated patients with Galloway-Mowat syndrome: Case report and review of the literature. BMC Nephrol. 2019, 20, 126. [Google Scholar] [CrossRef]
- Lin, P.Y.; Tseng, M.H.; Zenker, M.; Rao, J.; Hildebrandt, F.; Lin, S.H.; Lin, C.C.; Chang, J.H.; Hsu, C.H.; Lee, M.D.; et al. Galloway-Mowat syndrome in Taiwan: OSGEP mutation and unique clinical phenotype. Orphanet J. Rare Dis. 2018, 13, 226. [Google Scholar] [CrossRef]
- Wang, P.Z.T.; Prasad, C.; Rodriguez Cuellar, C.I.; Filler, G. Nephrological and urological complications of homozygous c.974G>A (p.Arg325Gln) OSGEP mutations. Pediatr. Nephrol. 2018, 33, 2201–2204. [Google Scholar] [CrossRef]
- Edvardson, S.; Prunetti, L.; Arraf, A.; Haas, D.; Bacusmo, J.M.; Hu, J.F.; Ta-Shma, A.; Dedon, P.C.; de Crécy-Lagard, V.; Elpeleg, O. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. 2017, 25, 545–551. [Google Scholar] [CrossRef]
- Teng, H.; Liang, C.; Liang, D.; Li, Z.; Wu, L. Novel variants in OSGEP leading to Galloway-Mowat syndrome by altering its subcellular localization. Clin. Chim. Acta 2021, 523, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Beenstock, J.; Sicheri, F. The structural and functional workings of KEOPS. Nucleic Acids Res. 2021, 49, 10818–10834. [Google Scholar] [CrossRef]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef]
- Klassen, R.; Ciftci, A.; Funk, J.; Bruch, A.; Butter, F.; Schaffrath, R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016, 44, 10946–10959. [Google Scholar] [CrossRef] [PubMed]
- Rezgui, V.A.; Tyagi, K.; Ranjan, N.; Konevega, A.L.; Mittelstaet, J.; Rodnina, M.V.; Peter, M.; Pedrioli, P.G. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. USA 2013, 110, 12289–12294. [Google Scholar] [CrossRef]
- Leeman, D.S.; Hebestreit, K.; Ruetz, T.; Webb, A.E.; McKay, A.; Pollina, E.A.; Dulken, B.W.; Zhao, X.; Yeo, R.W.; Ho, T.T.; et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 2018, 359, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Morrow, C.S.; Porter, T.J.; Xu, N.; Arndt, Z.P.; Ako-Asare, K.; Heo, H.J.; Thompson, E.A.N.; Moore, D.L. Vimentin Coordinates Protein Turnover at the Aggresome during Neural Stem Cell Quiescence Exit. Cell Stem Cell 2020, 26, 558–568.e559. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.D.; Ladha, S.; Ehrnhoefer, D.E.; Hayden, M.R. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 2015, 38, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Bordi, M.; Berg, M.J.; Mohan, P.S.; Peterhoff, C.M.; Alldred, M.J.; Che, S.; Ginsberg, S.D.; Nixon, R.A. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy 2016, 12, 2467–2483. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Sigrist, S. Autophagy and proteostasis in the control of synapse aging and disease. Curr. Opin. Neurobiol. 2018, 48, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Yue, Z. Autophagy and its normal and pathogenic states in the brain. Annu. Rev. Neurosci. 2014, 37, 55–78. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.L.; Lemmens, R.; Miskiewicz, K.; Broom, W.J.; Hansen, V.K.; van Vught, P.W.; Landers, J.E.; Sapp, P.; Van Den Bosch, L.; Knight, J.; et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 2009, 18, 472–481. [Google Scholar] [CrossRef] [PubMed]
- van Blitterswijk, M.; Mullen, B.; Wojtas, A.; Heckman, M.G.; Diehl, N.N.; Baker, M.C.; DeJesus-Hernandez, M.; Brown, P.H.; Murray, M.E.; Hsiung, G.Y.; et al. Genetic modifiers in carriers of repeat expansions in the C9ORF72 gene. Mol. Neurodegener. 2014, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Bruch, A.; Laguna, T.; Butter, F.; Schaffrath, R.; Klassen, R. Misactivation of multiple starvation responses in yeast by loss of tRNA modifications. Nucleic Acids Res. 2020, 48, 7307–7320. [Google Scholar] [CrossRef]
- Cocco, S.; Leone, A.; Roca, M.S.; Lombardi, R.; Piezzo, M.; Caputo, R.; Ciardiello, C.; Costantini, S.; Bruzzese, F.; Sisalli, M.J.; et al. Inhibition of autophagy by chloroquine prevents resistance to PI3K/AKT inhibitors and potentiates their antitumor effect in combination with paclitaxel in triple negative breast cancer models. J. Transl. Med. 2022, 20, 290. [Google Scholar] [CrossRef]
- Ruiz, S.; Panopoulos, A.D.; Herrerías, A.; Bissig, K.D.; Lutz, M.; Berggren, W.T.; Verma, I.M.; Izpisua Belmonte, J.C. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011, 21, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Smibert, P.; Zhao, X.; Hu, J.F.; Ramroop, J.; Kellner, S.M.; Benton, M.A.; Govind, S.; Dedon, P.C.; Sternglanz, R.; et al. An extensive allelic series of Drosophila kae1 mutants reveals diverse and tissue-specific requirements for t6A biogenesis. RNA 2015, 21, 2103–2118. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Nakazawa, N.; Wang, L.; Arakawa, O.; Yanagida, M. Negative Regulation of the Mis17-Mis6 Centromere Complex by mRNA Decay Pathway and EKC/KEOPS Complex in Schizosaccharomyces pombe. G3 2019, 9, 1815–1823. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.A.; Ellis, S.R.; True, H.L. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell Biol. 2010, 30, 354–363. [Google Scholar] [CrossRef]
- Daugeron, M.C.; Lenstra, T.L.; Frizzarin, M.; El Yacoubi, B.; Liu, X.; Baudin-Baillieu, A.; Lijnzaad, P.; Decourty, L.; Saveanu, C.; Jacquier, A.; et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. 2011, 39, 6148–6160. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, H.; Chen, S.; Liu, F.; Teng, Y.; Li, Y.; Liang, D.; Wu, L.; Li, Z. O-Sialoglycoprotein Endopeptidase Deficiency Impairs Proteostasis and Induces Autophagy in Human Embryonic Stem Cells. Int. J. Mol. Sci. 2024, 25, 7889. https://doi.org/10.3390/ijms25147889
Teng H, Chen S, Liu F, Teng Y, Li Y, Liang D, Wu L, Li Z. O-Sialoglycoprotein Endopeptidase Deficiency Impairs Proteostasis and Induces Autophagy in Human Embryonic Stem Cells. International Journal of Molecular Sciences. 2024; 25(14):7889. https://doi.org/10.3390/ijms25147889
Chicago/Turabian StyleTeng, Hua, Siyi Chen, Fang Liu, Yanling Teng, Yunlong Li, Desheng Liang, Lingqian Wu, and Zhuo Li. 2024. "O-Sialoglycoprotein Endopeptidase Deficiency Impairs Proteostasis and Induces Autophagy in Human Embryonic Stem Cells" International Journal of Molecular Sciences 25, no. 14: 7889. https://doi.org/10.3390/ijms25147889





