Comparative Analysis of Breast Cancer Metabolomes Highlights Fascin’s Central Role in Regulating Key Pathways Related to Disease Progression
Abstract
1. Introduction
2. Results
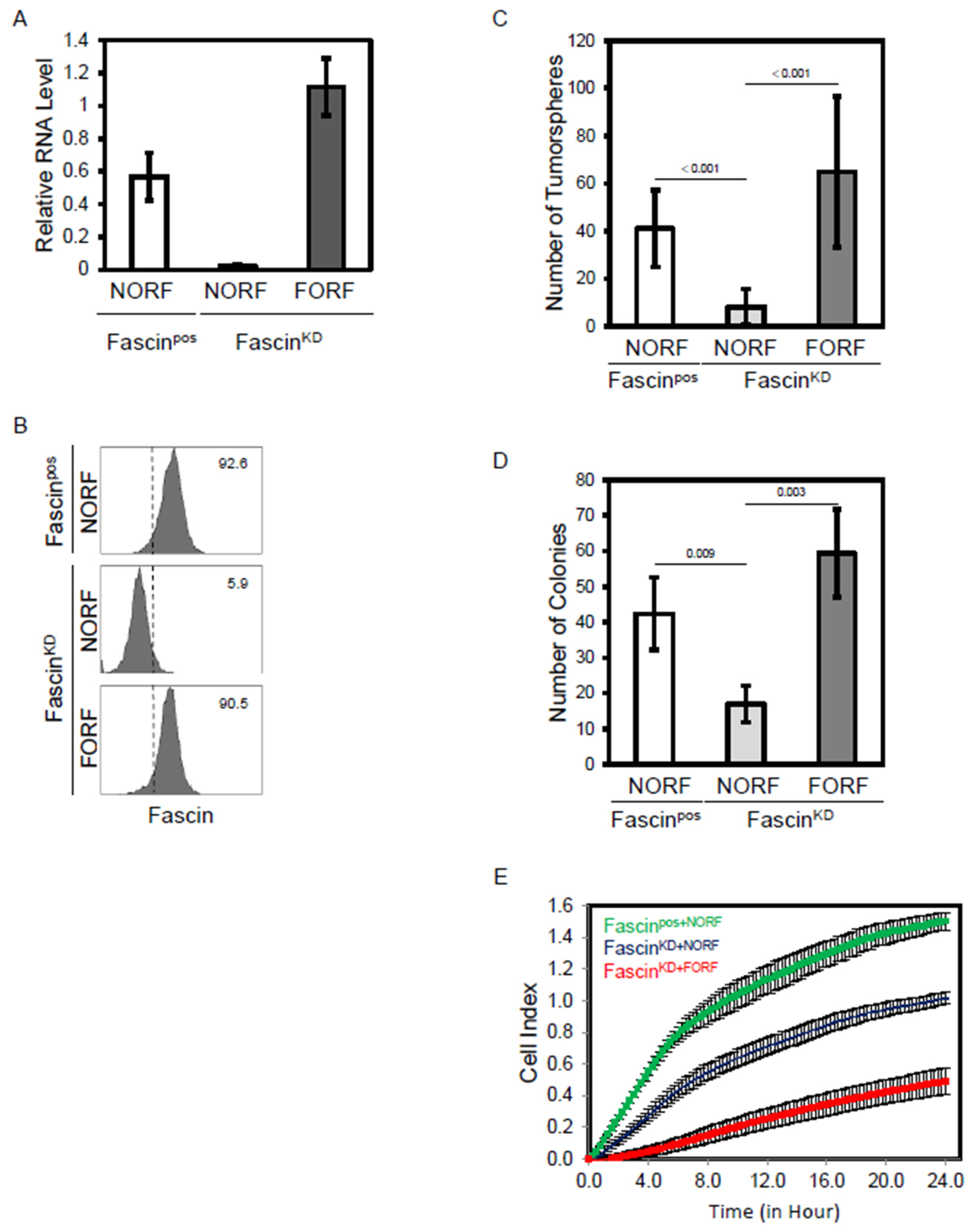
2.1. Fascin Expression Regulates Functions of MDA-MB-231 Cells Linked to Breast Cancer Progression
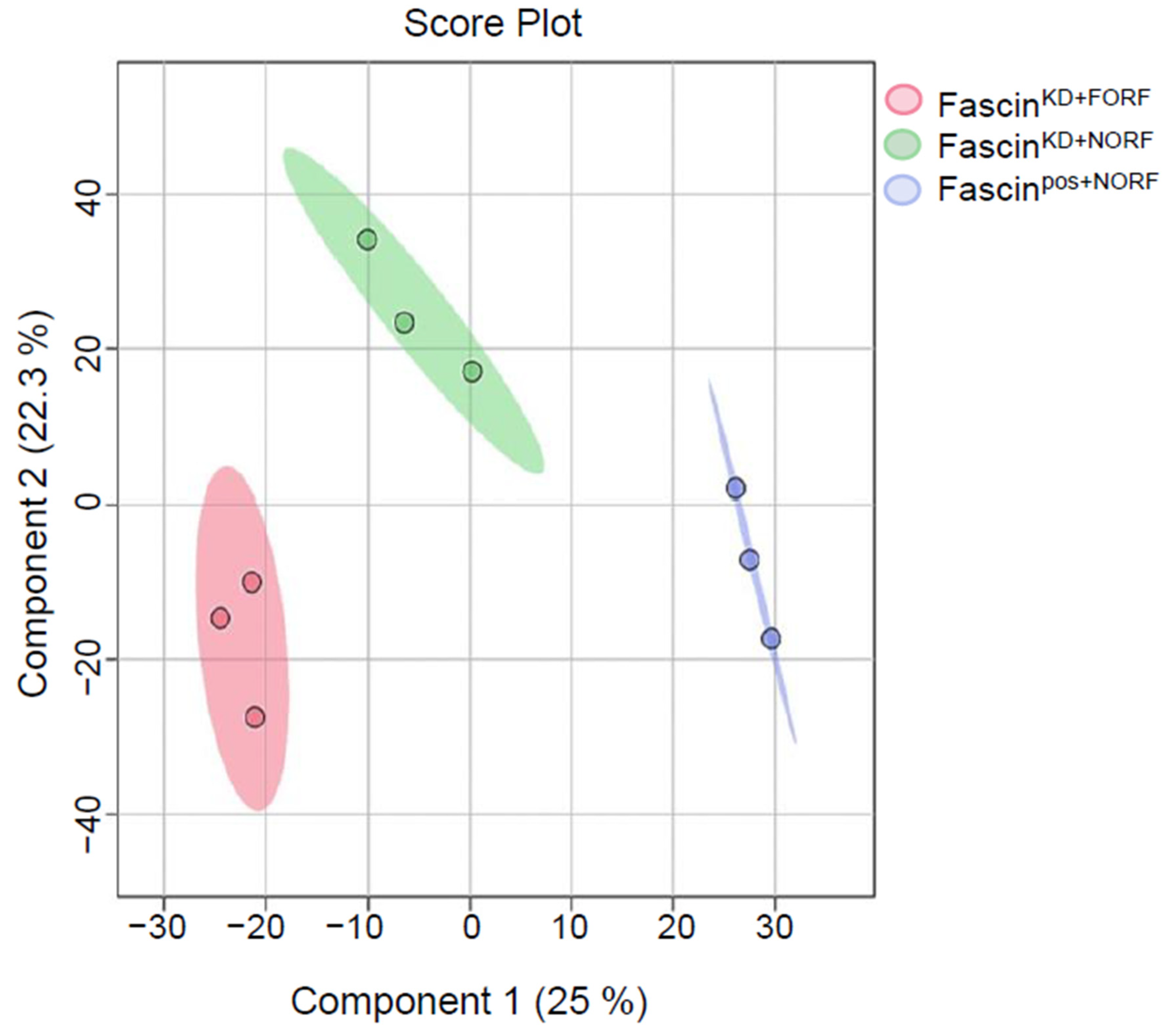
2.2. Fascin Expression Dysregulates the Metabolomics Profile of MDA-MB-231 Cells
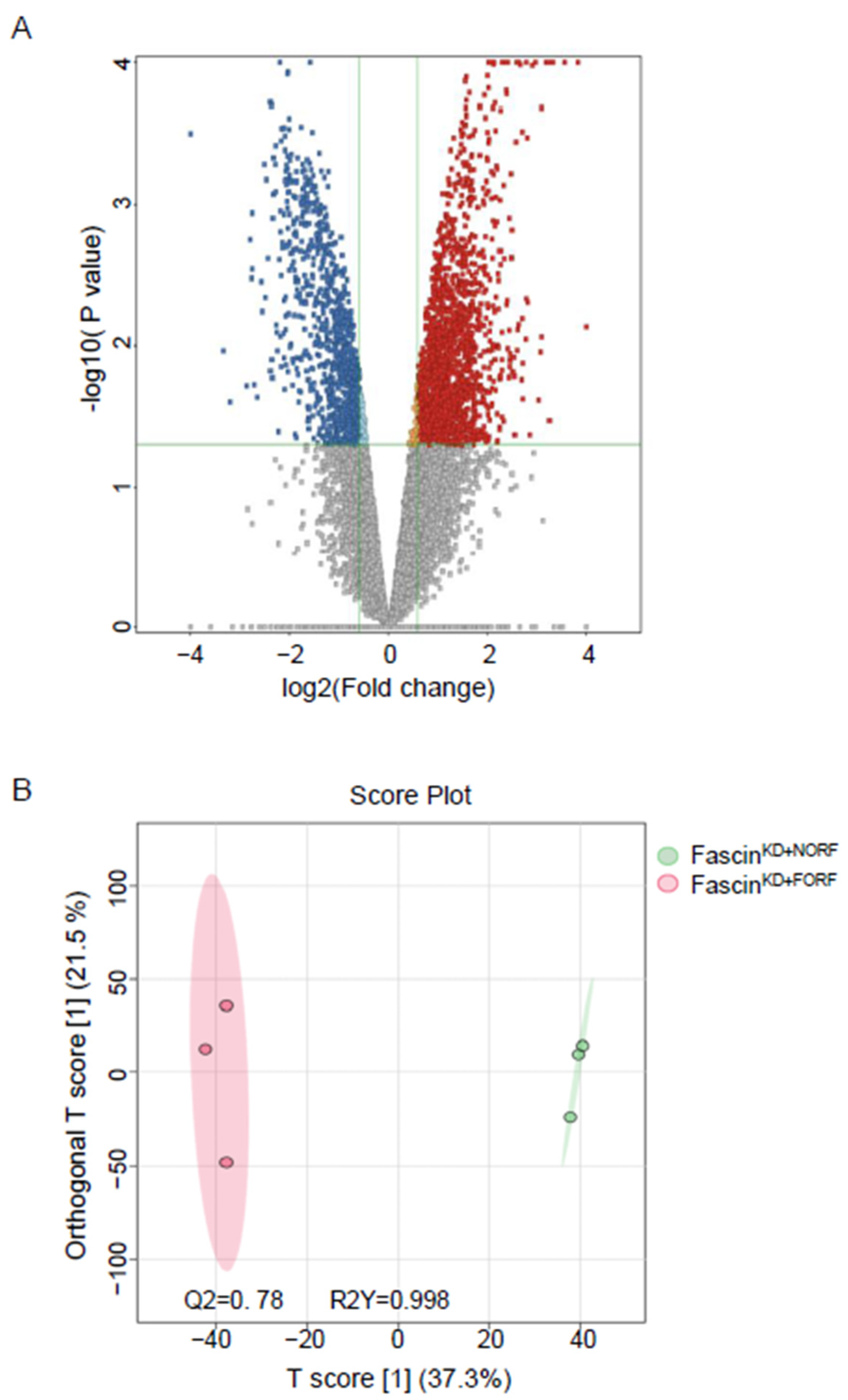
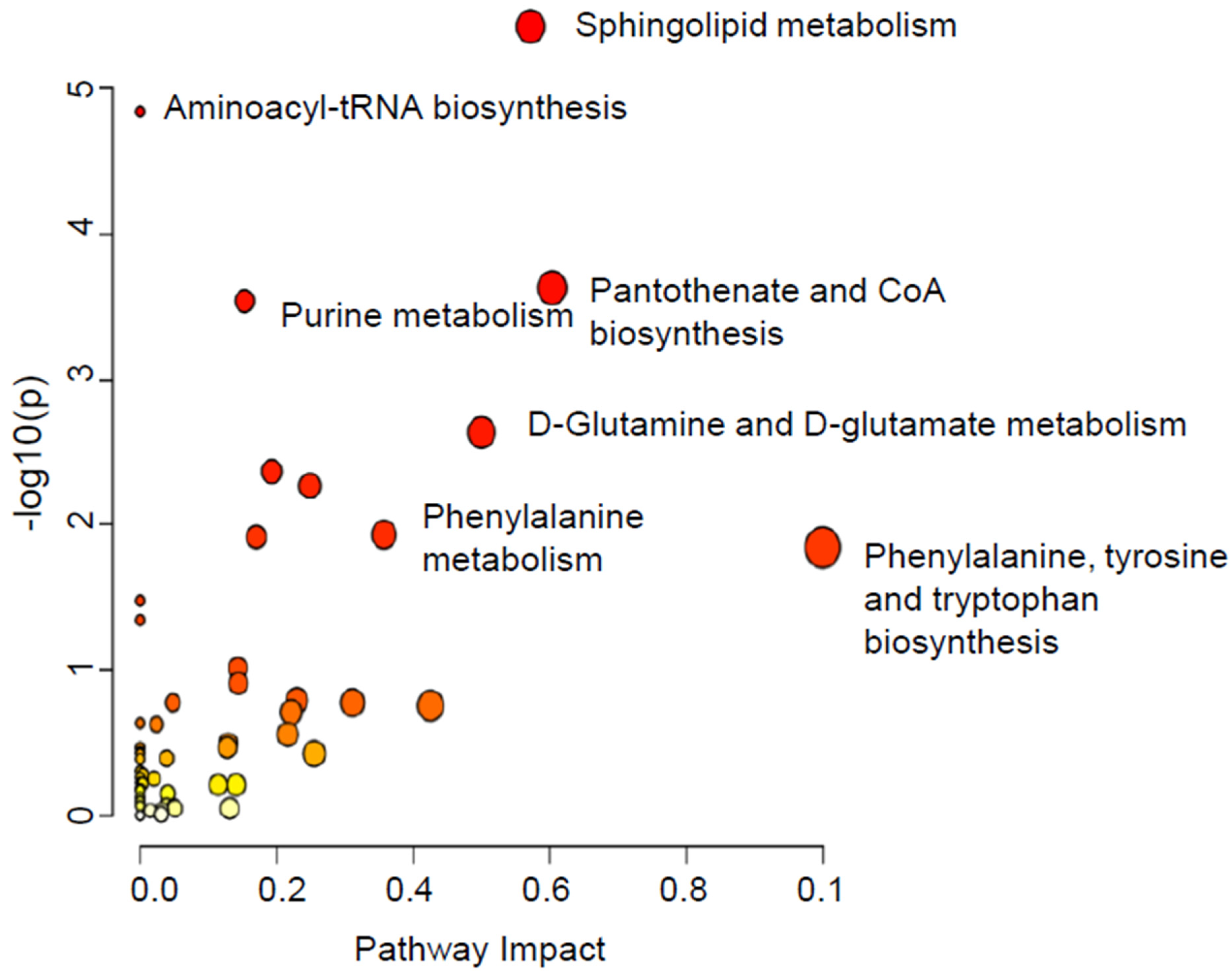
2.2.1. A-Dysregulated Metabolomics Profile and Pathway Analysis of MDA-MB-231 Cell Pellet by Fascin Expression
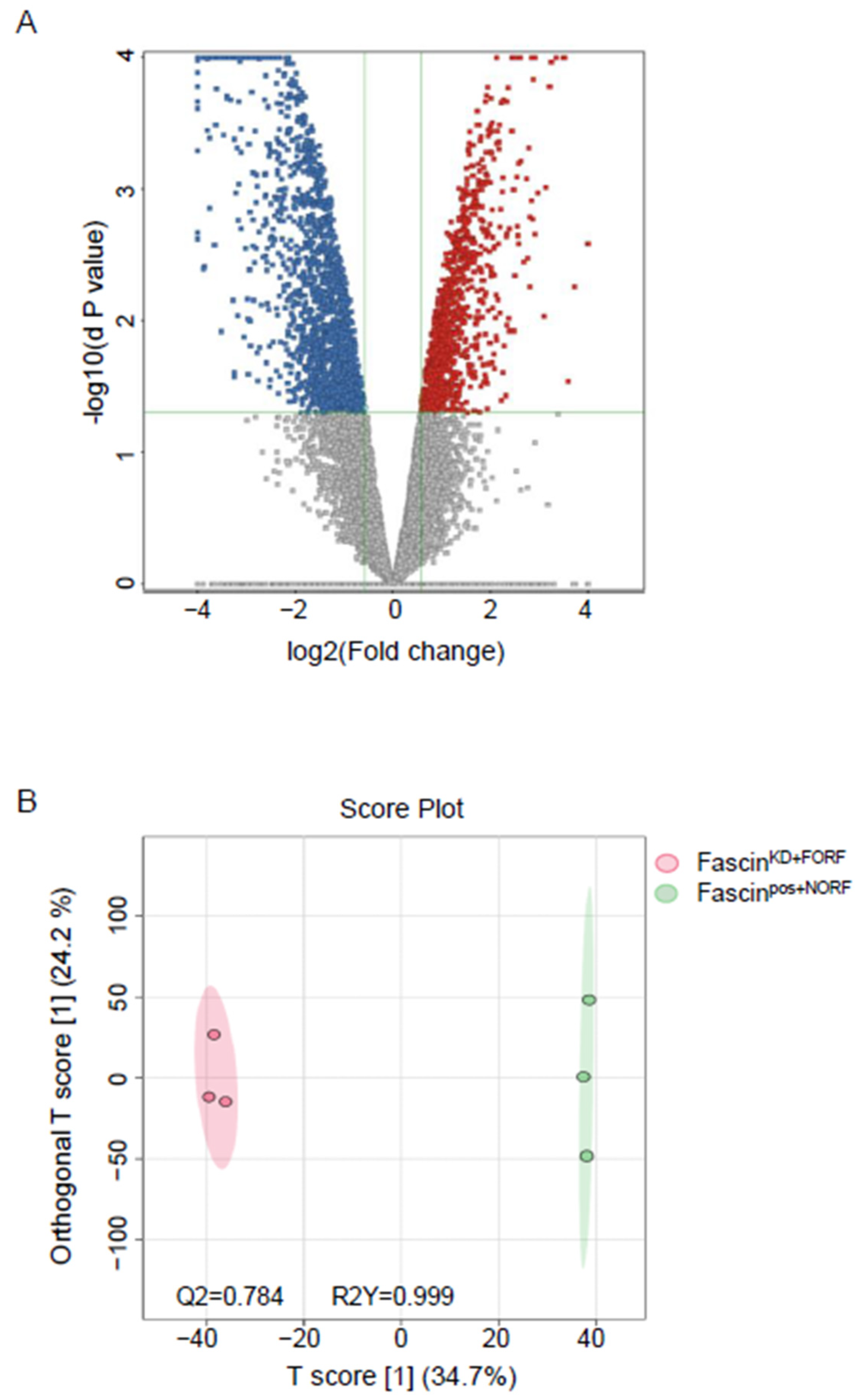
2.2.2. B-Dysregulated Metabolomics Profile and Pathway Analysis of MDA-MB-231 Secretomes by Fascin Expression
3. Discussion
4. Materials and Methods
4.1. Cells and Reagents
4.2. Gene Knockdown and Restoration
4.3. RT-qPCR and Flow Cytometry
4.4. Functional Assays
4.5. Sample Preparation
4.6. LC-MS Metabolomics
4.7. Data and Statistical Analyses
4.8. Metabolite Identification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orrantia-Borunda, E.; Anchondo-Nunez, P.; Acuna-Aguilar, L.E.; Gomez-Valles, F.O.; Ramirez-Valdespino, C.A. Subtypes of Breast Cancer. In Breast Cancer; Mayrovitz, H.N., Ed.; Exon Publications: Brisbane, Australia, 2022. [Google Scholar]
- Dellapasqua, S.; Bagnardi, V.; Balduzzi, A.; Iorfida, M.; Rotmensz, N.; Santillo, B.; Viale, G.; Ghisini, R.; Veronesi, P.; Luini, A.; et al. Outcomes of patients with breast cancer who present with ipsilateral supraclavicular or internal mammary lymph node metastases. Clin. Breast Cancer 2014, 14, 53–60. [Google Scholar] [CrossRef][Green Version]
- Hudis, C.A.; Gianni, L. Triple-negative breast cancer: An unmet medical need. Oncologist 2011, 16 (Suppl. S1), 1–11. [Google Scholar] [CrossRef]
- Kinross, J.M.; Holmes, E.; Darzi, A.W.; Nicholson, J.K. Metabolic phenotyping for monitoring surgical patients. Lancet 2011, 377, 1817–1819. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Han, Y.; Yuan, Y.; Wang, P.; Song, G.; Yuan, X.; Zhang, M.; Xie, N.; Wang, X. Exploratory urinary metabolic biomarkers and pathways using UPLC-Q-TOF-HDMS coupled with pattern recognition approach. Analyst 2012, 137, 4200–4208. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Barnett, D.A.; Culf, A.S.; Chute, I. Cell culture metabolomics: Applications and future directions. Drug Discov. Today 2010, 15, 610–621. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, A.; Han, Y.; Wang, P.; Sun, H.; Song, G.; Dong, T.; Yuan, Y.; Yuan, X.; Zhang, M.; et al. Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Mol. Cell Proteom. 2012, 11, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, A.; Sun, H. Future perspectives of Chinese medical formulae: Chinmedomics as an effector. OMICS 2012, 16, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Alakwaa, F.M.; Chaudhary, K.; Garmire, L.X. Deep Learning Accurately Predicts Estrogen Receptor Status in Breast Cancer Metabolomics Data. J. Proteome Res. 2018, 17, 337–347. [Google Scholar] [CrossRef]
- Dougan, M.M.; Li, Y.; Chu, L.W.; Haile, R.W.; Whittemore, A.S.; Han, S.S.; Moore, S.C.; Sampson, J.N.; Andrulis, I.L.; John, E.M.; et al. Metabolomic profiles in breast cancer:a pilot case-control study in the breast cancer family registry. BMC Cancer 2018, 18, 532. [Google Scholar] [CrossRef]
- Kanaan, Y.M.; Sampey, B.P.; Beyene, D.; Esnakula, A.K.; Naab, T.J.; Ricks-Santi, L.J.; Dasi, S.; Day, A.; Blackman, K.W.; Frederick, W.; et al. Metabolic profile of triple-negative breast cancer in African-American women reveals potential biomarkers of aggressive disease. Cancer Genom. Proteom. 2014, 11, 279–294. [Google Scholar]
- Al-Ansari, M.M.; AlMalki, R.H.; Dahabiyeh, L.A.; Abdel Rahman, A.M. Metabolomics-Microbiome Crosstalk in the Breast Cancer Microenvironment. Metabolites 2021, 11, 758. [Google Scholar] [CrossRef] [PubMed]
- Al-Alwan, M.; Olabi, S.; Ghebeh, H.; Barhoush, E.; Tulbah, A.; Al-Tweigeri, T.; Ajarim, D.; Adra, C. Fascin is a key regulator of breast cancer invasion that acts via the modification of metastasis-associated molecules. PLoS ONE 2011, 6, e27339. [Google Scholar] [CrossRef]
- Ghebeh, H.; Al-Khaldi, S.; Olabi, S.; Al-Dhfyan, A.; Al-Mohanna, F.; Barnawi, R.; Tulbah, A.; Al-Tweigeri, T.; Ajarim, D.; Al-Alwan, M. Fascin is involved in the chemotherapeutic resistance of breast cancer cells predominantly via the PI3K/Akt pathway. Br. J. Cancer 2014, 111, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Barnawi, R.; Al-Khaldi, S.; Majid, S.; Qattan, A.; Bakheet, T.; Fallatah, M.; Ghebeh, H.; Alajez, N.M.; Al-Alwan, M. Comprehensive Transcriptome and Pathway Analyses Revealed Central Role for Fascin in Promoting Triple-Negative Breast Cancer Progression. Pharmaceuticals 2021, 14, 1228. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Huang, C.; Gunda, V.; Sun, J.; Chellappan, S.P.; Li, Z.; Izumi, V.; Fang, B.; Koomen, J.; Singh, P.K.; et al. Fascin Controls Metastatic Colonization and Mitochondrial Oxidative Phosphorylation by Remodeling Mitochondrial Actin Filaments. Cell Rep. 2019, 28, 2824–2836.e8. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yu, P.; Tang, J. Characterization of Triple-Negative Breast Cancer MDA-MB-231 Cell Spheroid Model. Onco. Targets Ther. 2020, 13, 5395–5405. [Google Scholar] [CrossRef]
- Barnawi, R.; Al-Khaldi, S.; Colak, D.; Tulbah, A.; Al-Tweigeri, T.; Fallatah, M.; Monies, D.; Ghebeh, H.; Al-Alwan, M. beta1 Integrin is essential for fascin-mediated breast cancer stem cell function and disease progression. Int. J. Cancer 2019, 145, 830–841. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Li, L.; Cao, J.; Guo, Y.; Wu, Y.; Gao, W. Fascin actin-bundling protein 1 in human cancer: Promising biomarker or therapeutic target? Mol. Ther. Oncolytics 2021, 20, 240–264. [Google Scholar] [CrossRef]
- Min, K.W.; Chae, S.W.; Kim, D.H.; Do, S.I.; Kim, K.; Lee, H.J.; Sohn, J.H.; Pyo, J.S.; Kim, D.H.; Oh, S.; et al. Fascin expression predicts an aggressive clinical course in patients with advanced breast cancer. Oncol. Lett. 2015, 10, 121–130. [Google Scholar] [CrossRef]
- Rodriguez-Pinilla, S.M.; Sarrio, D.; Honrado, E.; Hardisson, D.; Calero, F.; Benitez, J.; Palacios, J. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin. Cancer Res. 2006, 12, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Tang, C.H.; Chang, H.T.; Li, X.N.; Zhao, Y.M.; Su, C.M.; Hu, G.N.; Zhang, T.; Sun, X.X.; Zeng, Y.; et al. Fascin-1 as a novel diagnostic marker of triple-negative breast cancer. Cancer Med. 2016, 5, 1983–1988. [Google Scholar] [CrossRef] [PubMed]
- Yoder, B.J.; Tso, E.; Skacel, M.; Pettay, J.; Tarr, S.; Budd, T.; Tubbs, R.R.; Adams, J.C.; Hicks, D.G. The expression of fascin, an actin-bundling motility protein, correlates with hormone receptor-negative breast cancer and a more aggressive clinical course. Clin. Cancer Res. 2005, 11, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Arnone, A.A.; Cline, J.M.; Soto-Pantoja, D.R.; Cook, K.L. Investigating the role of endogenous estrogens, hormone replacement therapy, and blockade of estrogen receptor-alpha activity on breast metabolic signaling. Breast Cancer Res. Treat. 2021, 190, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ba-Alawi, W.; Deblois, G.; Cruickshank, J.; Duan, S.; Lima-Fernandes, E.; Haight, J.; Tonekaboni, S.A.M.; Fortier, A.M.; Kuasne, H.; et al. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer. Nat. Commun. 2020, 11, 4205. [Google Scholar] [CrossRef] [PubMed]
- Kisanga, E.R.; Mellgren, G.; Lien, E.A. Excretion of hydroxylated metabolites of tamoxifen in human bile and urine. Anticancer. Res. 2005, 25, 4487–4492. [Google Scholar] [PubMed]
- Spiegel, S.; Milstien, S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 2011, 11, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Homaidan, F.R.; Chakroun, I.; El-Sabban, M.E. Regulation of nuclear factor-kappaB in intestinal epithelial cells in a cell model of inflammation. Mediat. Inflamm. 2003, 12, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.; Huang, J.; Huang, X.Y.; Zhang, J.J. A signal transducer and activator of transcription 3.Nuclear Factor kappaB (Stat3.NFkappaB) complex is necessary for the expression of fascin in metastatic breast cancer cells in response to interleukin (IL)-6 and tumor necrosis factor (TNF)-alpha. J. Biol. Chem. 2014, 289, 30082–30089. [Google Scholar] [CrossRef]
- Acharya, S.; Yao, J.; Li, P.; Zhang, C.; Lowery, F.J.; Zhang, Q.; Guo, H.; Qu, J.; Yang, F.; Wistuba, I.I.; et al. Sphingosine Kinase 1 Signaling Promotes Metastasis of Triple-Negative Breast Cancer. Cancer Res. 2019, 79, 4211–4226. [Google Scholar] [CrossRef]
- Canals, D.; Salamone, S.; Santacreu, B.J.; Nemeth, E.; Aguilar, D.; Hernandez-Corbacho, M.J.; Adada, M.; Staquicini, D.I.; Arap, W.; Pasqualini, R.; et al. Ceramide launches an acute anti-adhesion pro-migration cell signaling program in response to chemotherapy. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 7610–7630. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, S.; Selvam, S.P.; Mehrotra, S.; Kawamori, T.; Snider, A.J.; Obeid, L.M.; Shao, Y.; Sabbadini, R.; Ogretmen, B. Communication between host organism and cancer cells is transduced by systemic sphingosine kinase 1/sphingosine 1-phosphate signalling to regulate tumour metastasis. EMBO Mol. Med. 2012, 4, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Fujiwara, A.; Nakamura, K.; Sadzuka, Y. The effects of vitamin B(6) compounds on cell proliferation and melanogenesis in B16F10 melanoma cells. Oncol. Lett. 2018, 15, 5181–5184. [Google Scholar] [CrossRef]
- Zhang, P.; Suidasari, S.; Hasegawa, T.; Yanaka, N.; Kato, N. Vitamin B(6) activates p53 and elevates p21 gene expression in cancer cells and the mouse colon. Oncol. Rep. 2014, 31, 2371–2376. [Google Scholar] [CrossRef] [PubMed]
- Lurie, G.; Wilkens, L.R.; Shvetsov, Y.B.; Ollberding, N.J.; Franke, A.A.; Henderson, B.E.; Kolonel, L.N.; Goodman, M.T. Prediagnostic plasma pyridoxal 5′-phosphate (vitamin b6) levels and invasive breast carcinoma risk: The multiethnic cohort. Cancer Epidemiol. Biomark. Prev. A Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2012, 21, 1942–1948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.M.; Cook, N.R.; Albert, C.M.; Gaziano, J.M.; Buring, J.E.; Manson, J.E. Effect of combined folic acid, vitamin B6, and vitamin B12 on cancer risk in women: A randomized trial. Jama 2008, 300, 2012–2021. [Google Scholar] [CrossRef]
- Christopher, S.A.; Diegelman, P.; Porter, C.W.; Kruger, W.D. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 2002, 62, 6639–6644. [Google Scholar] [PubMed]
- Su, C.Y.; Chang, Y.C.; Chan, Y.C.; Lin, T.C.; Huang, M.S.; Yang, C.J.; Hsiao, M. MTAP is an independent prognosis marker and the concordant loss of MTAP and p16 expression predicts short survival in non-small cell lung cancer patients. Eur. J. Surg. Oncol. 2014, 40, 1143–1150. [Google Scholar] [CrossRef]
- Shibuya, H.; Hamamura, K.; Hotta, H.; Matsumoto, Y.; Nishida, Y.; Hattori, H.; Furukawa, K.; Ueda, M.; Furukawa, K. Enhancement of malignant properties of human osteosarcoma cells with disialyl gangliosides GD2/GD3. Cancer Sci. 2012, 103, 1656–1664. [Google Scholar] [CrossRef]
- Ohkawa, Y.; Miyazaki, S.; Hamamura, K.; Kambe, M.; Miyata, M.; Tajima, O.; Ohmi, Y.; Yamauchi, Y.; Furukawa, K.; Furukawa, K. Ganglioside GD3 enhances adhesion signals and augments malignant properties of melanoma cells by recruiting integrins to glycolipid-enriched microdomains. J. Biol. Chem. 2010, 285, 27213–27223. [Google Scholar] [CrossRef]
- Heng, B.; Lim, C.K.; Lovejoy, D.B.; Bessede, A.; Gluch, L.; Guillemin, G.J. Understanding the role of the kynurenine pathway in human breast cancer immunobiology. Oncotarget 2016, 7, 6506–6520. [Google Scholar] [CrossRef] [PubMed]
- Uyttenhove, C.; Pilotte, L.; Theate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.J.; DuHadaway, J.B.; Donover, P.S.; Sutanto-Ward, E.; Prendergast, G.C. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat. Med. 2005, 11, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Bonifacio, V.D.B.; Pereira, S.A.; Serpa, J.; Vicente, J.B. Cysteine metabolic circuitries: Druggable targets in cancer. Br. J. Cancer 2021, 124, 862–879. [Google Scholar] [CrossRef] [PubMed]
- Neurauter, G.; Grahmann, A.V.; Klieber, M.; Zeimet, A.; Ledochowski, M.; Sperner-Unterweger, B.; Fuchs, D. Serum phenylalanine concentrations in patients with ovarian carcinoma correlate with concentrations of immune activation markers and of isoprostane-8. Cancer Lett. 2008, 272, 141–147. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.J.; Huang, X.Y. Anti-Metastasis Fascin Inhibitors Decrease the Growth of Specific Subtypes of Cancers. Cancers 2020, 12, 2287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, Y.; Zhang, J.J.; Huang, X.Y. Fascin Inhibitors Decrease Cell Migration and Adhesion While Increase Overall Survival of Mice Bearing Bladder Cancers. Cancers 2021, 13, 2698. [Google Scholar] [CrossRef]
- Barnawi, R.; Al-Khaldi, S.; Majed Sleiman, G.; Sarkar, A.; Al-Dhfyan, A.; Al-Mohanna, F.; Ghebeh, H.; Al-Alwan, M. Fascin Is Critical for the Maintenance of Breast Cancer Stem Cell Pool Predominantly via the Activation of the Notch Self-Renewal Pathway. Stem. Cells 2016, 34, 2799–2813. [Google Scholar] [CrossRef] [PubMed]
- AlMalki, R.H.; Sebaa, R.; Al-Ansari, M.M.; Al-Alwan, M.; Alwehaibi, M.A.; Rahman, A.M.A.E. coli Secretome Metabolically Modulates MDA-MB-231 Breast Cancer Cells’ Energy Metabolism. Int. J. Mol. Sci. 2023, 24, 4219. [Google Scholar] [CrossRef]
- Abdel Rahman, A.M.; Pawling, J.; Ryczko, M.; Caudy, A.A.; Dennis, J.W. Targeted metabolomics in cultured cells and tissues by mass spectrometry: Method development and validation. Anal. Chim. Acta 2014, 845, 53–61. [Google Scholar] [CrossRef]
- Aleidi, S.M.; Alnehmi, E.A.; Alshaker, M.; Masood, A.; Benabdelkamel, H.; Al-Ansari, M.M.; Abdel Rahman, A.M. A Distinctive Human Metabolomics Alteration Associated with Osteopenic and Osteoporotic Patients. Metabolites 2021, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [PubMed]
- Gu, X.; Al Dubayee, M.; Alshahrani, A.; Masood, A.; Benabdelkamel, H.; Zahra, M.; Li, L.; Abdel Rahman, A.M.; Aljada, A. Distinctive metabolomics patterns associated with insulin resistance and type 2 diabetes mellitus. Front. Mol. Biosci. 2020, 7, 411. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L. HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AlMalki, R.H.; Al-Nasrallah, H.K.; Aldossry, A.; Barnawi, R.; Al-Khaldi, S.; Almozyan, S.; Al-Ansari, M.M.; Ghebeh, H.; Abdel Rahman, A.M.; Al-Alwan, M. Comparative Analysis of Breast Cancer Metabolomes Highlights Fascin’s Central Role in Regulating Key Pathways Related to Disease Progression. Int. J. Mol. Sci. 2024, 25, 7891. https://doi.org/10.3390/ijms25147891
AlMalki RH, Al-Nasrallah HK, Aldossry A, Barnawi R, Al-Khaldi S, Almozyan S, Al-Ansari MM, Ghebeh H, Abdel Rahman AM, Al-Alwan M. Comparative Analysis of Breast Cancer Metabolomes Highlights Fascin’s Central Role in Regulating Key Pathways Related to Disease Progression. International Journal of Molecular Sciences. 2024; 25(14):7891. https://doi.org/10.3390/ijms25147891
Chicago/Turabian StyleAlMalki, Reem H., Huda K. Al-Nasrallah, Alanoud Aldossry, Rayanah Barnawi, Samiyah Al-Khaldi, Sheema Almozyan, Mysoon M. Al-Ansari, Hazem Ghebeh, Anas M. Abdel Rahman, and Monther Al-Alwan. 2024. "Comparative Analysis of Breast Cancer Metabolomes Highlights Fascin’s Central Role in Regulating Key Pathways Related to Disease Progression" International Journal of Molecular Sciences 25, no. 14: 7891. https://doi.org/10.3390/ijms25147891
APA StyleAlMalki, R. H., Al-Nasrallah, H. K., Aldossry, A., Barnawi, R., Al-Khaldi, S., Almozyan, S., Al-Ansari, M. M., Ghebeh, H., Abdel Rahman, A. M., & Al-Alwan, M. (2024). Comparative Analysis of Breast Cancer Metabolomes Highlights Fascin’s Central Role in Regulating Key Pathways Related to Disease Progression. International Journal of Molecular Sciences, 25(14), 7891. https://doi.org/10.3390/ijms25147891





