In Pursuit of Novel Markers: Unraveling the Potential of miR-106, CEA and CA 19-9 in Gastric Adenocarcinoma Diagnosis and Staging
Abstract
1. Introduction
2. Results
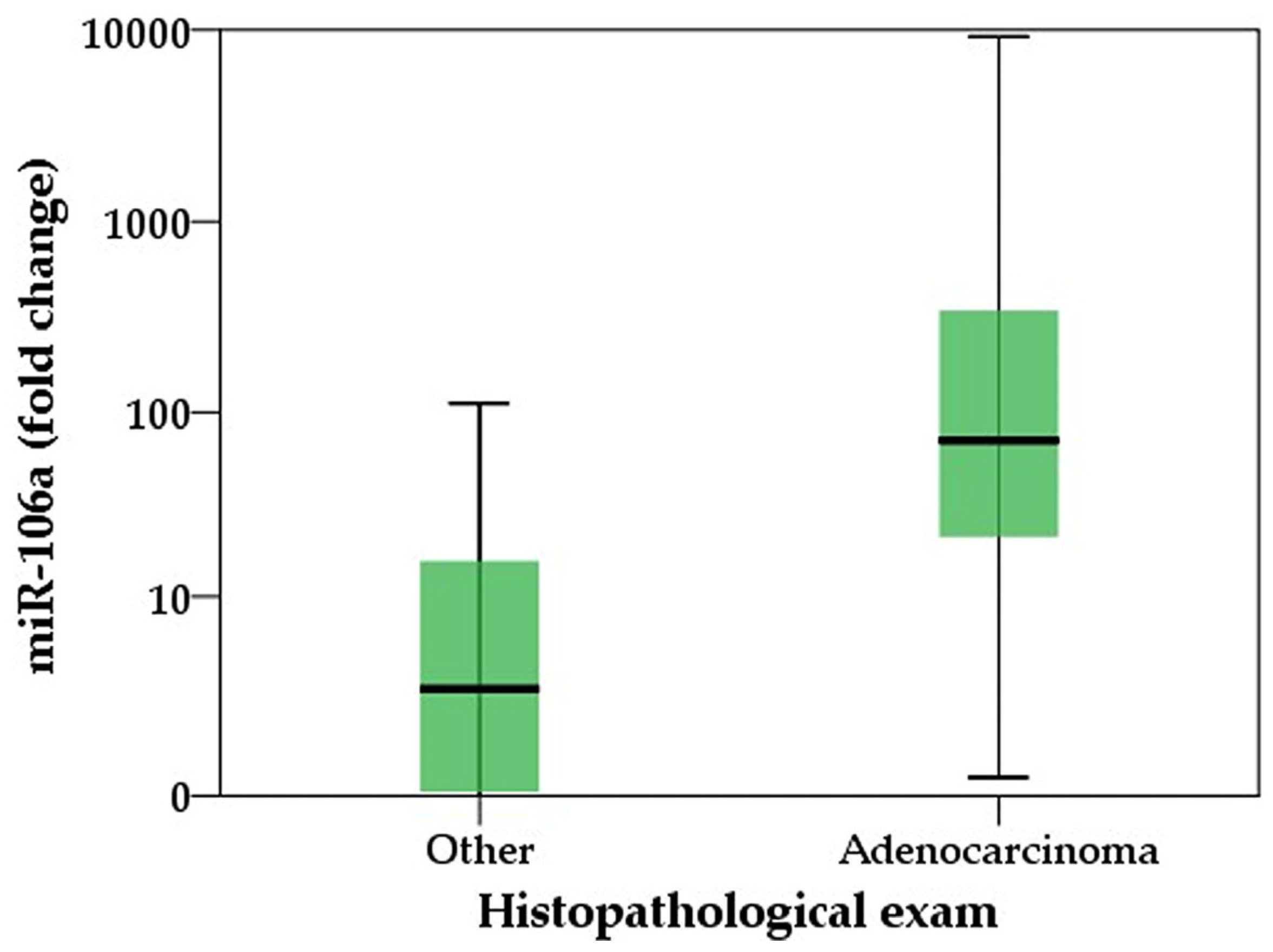
2.1. Higher Expressions Levels of miR-106 in Gastric Juice Are Corelated with CEA Levels
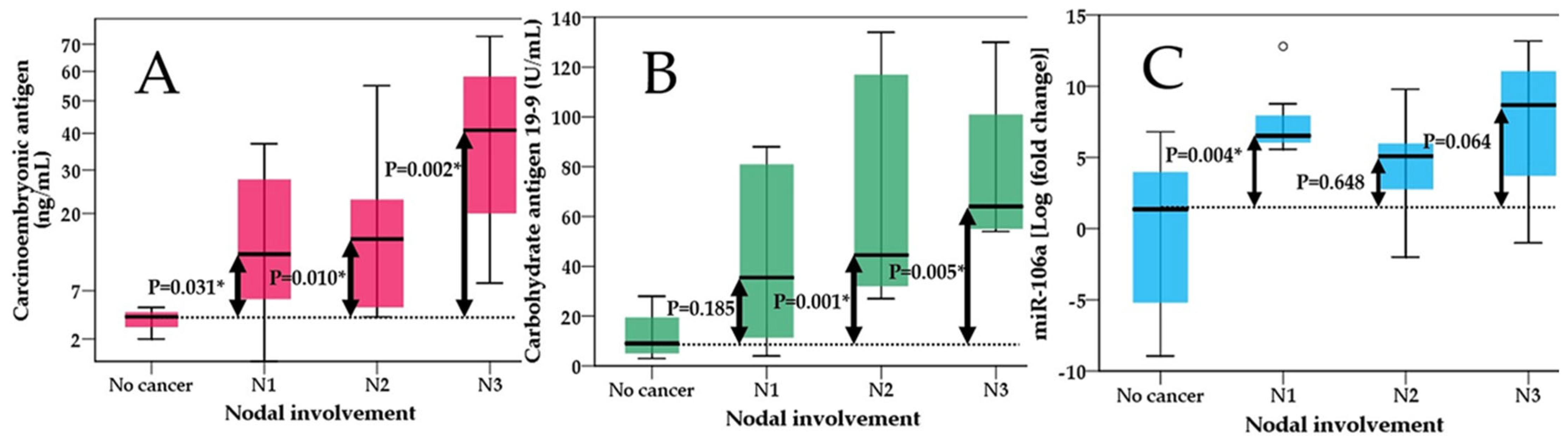
2.2. Comparisons between miR-106 Levels, CEA, CA 19-9 in Predicting Tumor Extension, Nodal Involvement and Metastases
2.3. Comparative Analysis of miR-106, CEA, and CA 19-9 as Predictive Biomarkers for Positive Histopathological Examination in Gastric Cancer
3. Discussions
4. Materials and Methods
- Adult participants, aged 18 years and older;
- Patients diagnosed with adenocarcinoma at various stages, with histopathological biopsy results confirming the diagnosis;
- Patients with an endoscopic description of the lesion.
- Patients with other associated malignancies;
- Prior chemotherapy for gastric cancer.
4.1. MiRNA Extraction and Quantification
4.2. Statistical Analysis
5. Conclusions
6. Limitation of the Study
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Azarbarzin, S.; Safaralizadeh, R.; Khojasteh, M.B.; Baghbanzadeh, A.; Baradaran, B. Current Perspectives on the Dysregulated microRNAs in Gastric Cancer. Mol. Biol. Rep. 2020, 47, 7253–7264. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tang, X.; Liu, Y.; Zhu, J.; Liu, J. Induction/Reversal of Drug Resistance in Gastric Cancer by Non-Coding RNAs (Review). Int. J. Oncol. 2019, 54, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Shen, H.; Li, H.; Cao, Y.; Qin, R.; Long, L.; Zhu, X.; Xie, C.; Xu, W. miR-106a Confers Cisplatin Resistance by Regulating PTEN/Akt Pathway in Gastric Cancer Cells. Acta Biochim. Biophys. Sin. 2013, 45, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhu, M.; Liao, B.; Tian, T.; Li, M.; Wang, Z.; Chen, G. Upregulation of miR-552 Predicts Unfavorable Prognosis of Gastric Cancer and Promotes the Proliferation, Migration, and Invasion of Gastric Cancer Cells. Oncol. Res. Treat. 2020, 43, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Liu, X.; Lin, F.; Li, P.; Liu, K.; Geng, R.; Dai, C.; Lin, Y.; Tang, W.; Wu, Z.; et al. MicroRNA-421 Regulated by HIF-1α Promotes Metastasis, Inhibits Apoptosis, and Induces Cisplatin Resistance by Targeting E-Cadherin and Caspase-3 in Gastric Cancer. Oncotarget 2016, 7, 24466–24482. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.-B.; He, Y.-F.; Li, X.-Q.; Wang, K.; Wang, R.-L. The Role of miRNA and lncRNA in Gastric Cancer. Oncotarget 2017, 8, 81572–81582. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-K. Circulating microRNAs and Long Non-Coding RNAs in Gastric Cancer Diagnosis: An Update and Review. World J. Gastroenterol. 2015, 21, 9863. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.-L.; Wang, T.; Zhang, K.-H. MicroRNAs as Potential Biomarkers for Diagnosis, Therapy and Prognosis of Gastric Cancer. OncoTargets Ther. 2018, 11, 3891–3900. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Jiang, J.; Fu, Y.; Liu, T.; Yu, Y.; Zhang, X. MiR-129-5p Functions as a Tumor Suppressor in Gastric Cancer Progression through Targeting ADAM9. Biomed. Pharmacother. 2018, 105, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Kupcinskas, J. MicroRNAs as Non-Invasive Diagnostic Biomarkers for Gastric Cancer: Current Insights and Future Perspectives. World J. Gastroenterol. 2018, 24, 3313–3329. [Google Scholar] [CrossRef]
- Ishiguro, H. Role of microRNAs in Gastric Cancer. World J. Gastroenterol. 2014, 20, 5694. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Min, B.-H.; Jang, J.; Kang, S.Y.; Bae, H.; Jang, S.S.; Kim, J.-I.; Kim, K.-M. MicroRNA Expression Profiles in Gastric Carcinogenesis. Sci. Rep. 2018, 8, 14393. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, E.; Giarnieri, E.; Giovagnoli, M.R.; Montagnini, M.; Proietti, A.; D’urso, R.; Mercantini, P.; Balducci, G.; Cavallini, M. Gastric Juice MicroRNAs as Potential Biomarkers for Screening Gastric Cancer: A Systematic Review. Anticancer Res. 2018, 38, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, P.; Yang, W.; Mi, H.; Pan, J.; Huang, Y.; Hou, Z.; Hou, Q.; Luo, Q.; Liu, F. Identification of Non-Invasive Biomarkers for Chronic Atrophic Gastritis from Serum Exosomal microRNAs. BMC Cancer 2019, 19, 129. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Kupcinskas, J.; Wex, T.; Malfertheiner, P. Macro-Role of MicroRNA in Gastric Cancer. Dig. Dis. 2012, 30, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA Expression across Human Tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, J. MiR-129-5p Suppresses Gastric Cancer Cell Invasion and Proliferation by Inhibiting COL1A1. Biochem. Cell Biol. 2018, 96, 19–25. [Google Scholar] [CrossRef]
- Sagar, S.K. miR-106b as an Emerging Therapeutic Target in Cancer. Genes Dis. 2022, 9, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Sun, Z.; Wang, D.; Du, T. MiR-106b-5p Regulates Esophageal Squamous Cell Carcinoma Progression by Binding to HPGD. BMC Cancer 2022, 22, 308. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Xie, Z.; Lei, X.; Tang, G.; Gan, R.; Yang, X. Clinical Crosstalk between microRNAs and Gastric Cancer (Review). Int. J. Oncol. 2021, 58, 7. [Google Scholar] [CrossRef] [PubMed]
- Daneshpour, M.; Ghadimi-Daresajini, A. Overview of miR-106a Regulatory Roles: From Cancer to Aging. Bioengineering 2023, 10, 892. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Guo, J.; Miao, Y.; Jiang, Z.; Huan, R.; Zhang, Y.; Li, D.; Zhong, J. Detection of miR-106a in Gastric Carcinoma and Its Clinical Significance. Clin. Chim. Acta 2009, 400, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, N.; He, S.; Yan, R.; Zhang, J. MicroRNA-106a Functions as an Oncogene in Human Gastric Cancer and Contributes to Proliferation and Metastasis in Vitro and in Vivo. Clin. Exp. Metastasis 2016, 33, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Sun, J.; Zhang, N.; Zheng, Y.; Wang, X.; Lv, L.; Liu, J.; Xu, Y.; Shen, Y.; Yang, M. Noncoding RNAs in Gastric Cancer: Implications for Drug Resistance. Mol. Cancer 2020, 19, 62. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-S. MicroRNAs as Potential Biomarkers for Gastric Cancer. World J. Gastroenterol. 2014, 20, 12007. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.; Kitsukawa, Y.; Itoh, K. Carcinoembryonic Antigen (CEA) in Gastric Juice or Feces as an Aid in the Diagnosis of Gastrointestinal Cancer. Ann. Surg. 1979, 189, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Kurokawa, Y.; Miyazaki, Y.; Makino, T.; Takahashi, T.; Yamasaki, M.; Nakajima, K.; Takiguchi, S.; Mori, M.; Doki, Y. The Characteristics of the Serum Carcinoembryonic Antigen and Carbohydrate Antigen 19-9 Levels in Gastric Cancer Cases. Surg. Today 2017, 47, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, Z.; Lu, N. Molecular Targeted Therapy for the Treatment of Gastric Cancer. J. Exp. Clin. Cancer Res. 2016, 35, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-Z.; Cheng, T.-T.; He, Q.-J.; Lei, Z.-Y.; Chi, J.; Tang, Z.; Liao, Q.-X.; Zhang, H.; Zeng, L.-S.; Cui, S.-Z. LINC01133 as ceRNA Inhibits Gastric Cancer Progression by Sponging miR-106a-3p to Regulate APC Expression and the Wnt/β-Catenin Pathway. Mol. Cancer 2018, 17, 126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, N.; He, S.; Lui, Y.; Lu, G.; Zhao, L. MicroRNA-106a Targets TIMP2 to Regulate Invasion and Metastasis of Gastric Cancer. FEBS Lett. 2014, 588, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Shen, Y.; Lin, K.; Zou, L.; Shen, Y.; Zhu, Y. Comprehensive and Integrative Analysis Identifies microRNA-106 as a Novel Non-Invasive Biomarker for Detection of Gastric Cancer. J. Transl. Med. 2018, 16, 127. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A. A Systematic Review of microRNAs as Potential Biomarkers for Diagnosis and Prognosis of Gastric Cancer. Immunogenetics 2021, 73, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Guo, J.-M.; Lou, Y.-R.; Zhang, X.-J.; Zhong, F.-D.; Jiang, Z.; Cheng, J.; Xiao, B.-X. Detection of Circulating Tumor Cells in Peripheral Blood from Patients with Gastric Cancer Using microRNA as a Marker. J. Mol. Med. 2010, 88, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Petrocca, F.; Vecchione, A.; Croce, C.M. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008, 68, 8191–8194. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, L.; Liu, B.; Yang, L.; Meng, X.; Zhang, W.; Ma, Y.; Xiao, H. Genome-Wide microRNA Profiles Identify miR-378 as a Serum Biomarker for Early Detection of Gastric Cancer. Cancer Lett. 2012, 316, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Gan, N.; Jia, L.; Zheng, L. A Sandwich Electrochemical Immunosensor Using Magnetic DNA Nanoprobes for Carcinoembryonic Antigen. Int. J. Mol. Sci. 2011, 12, 7410–7423. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.W.; Kibbey, W.E.; Divecchia, L.; Anderson, G.; Catalano, P.; Minton, J.P. Carcinoembryonic Antigen. Clinical and Historical Aspects. Cancer 1976, 37, 62–81. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.; Snyder, J.; Miller, E.; Vandevoorde, J.; Miller, O.; Hines, L.; Burns, J. Carcinoembryonic Antigen (CEA) assayA Laboratory Adjunct in the Diagnosis and Management of Cancer. Hum. Pathol. 1974, 5, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.C. Effect of Age and Cigarette Smoking on Carcinoembryonic Antigen Levels. JAMA J. Am. Med. Assoc. 1976, 235, 1975. [Google Scholar] [CrossRef]
- Park, S.-H.; Ku, K.-B.; Chung, H.-Y.; Yu, W. Prognostic Significance of Serum and Tissue Carcinoembryonic Antigen in Patients with Gastric Adenocarcinomas. Cancer Res. Treat. 2008, 40, 16. [Google Scholar] [CrossRef] [PubMed]
- Sisik, A.; Kaya, M.; Bas, G.; Basak, F.; Alimoglu, O. CEA and CA 19-9 Are Still Valuable Markers for the Prognosis of Colorectal and Gastric Cancer Patients. Asian Pac. J. Cancer Prev. 2013, 14, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.L.; Slater, E.D.; Sanders, G.K.; Prichard, J.G. Serum Tumor Markers. Am. Fam. Physician 2003, 68, 1075–1082. [Google Scholar] [PubMed]
- Shibata, C.; Nakano, T.; Yasumoto, A.; Mitamura, A.; Sawada, K.; Ogawa, H.; Miura, T.; Ise, I.; Takami, K.; Yamamoto, K.; et al. Comparison of CEA and CA19-9 as a Predictive Factor for Recurrence after Curative Gastrectomy in Gastric Cancer. BMC Surg. 2022, 22, 213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xu, C.-W.; Shao, Y.; Wang, H.-T.; Wu, Y.-F.; Song, Y.-Y.; Li, X.-B.; Liang, W.-B. Evaluation and Identification of microRNA-106 in the Diagnosis of Cancer: A Meta-Analysis. Int. J. Clin. Exp. Med. 2014, 7, 3746. [Google Scholar] [PubMed]
- Cheng, H.; Zhang, L.; Cogdell, D.E.; Zheng, H.; Schetter, A.J.; Nykter, M.; Harris, C.C.; Chen, K.; Hamilton, S.R.; Zhang, W. Circulating Plasma MiR-141 Is a Novel Biomarker for Metastatic Colon Cancer and Predicts Poor Prognosis. PLoS ONE 2011, 6, e17745. [Google Scholar] [CrossRef] [PubMed]



| Variable | Normal Values | Control Group | Adenocarcinoma Group | p-Value | |
|---|---|---|---|---|---|
| Demographic characteristics | |||||
| Gender | Male | 7 (31.8%) | 15 (68.2%) | 0.132 | |
| Female | 9 (56.2%) | 7 (43.8%) | |||
| Age | 54 (±12.8) | 67 (±11.2) | 0.002 | ||
| Significant comorbidities | |||||
| Diabetes | 2 (12.5%) | 5 (22.7%) | 0.645 | ||
| Cardiovascular disease | 3 (18.7%) | 10 (45.4%) | 0.086 | ||
| No comorbidities | 0 | 2 (9.1%) | 0.215 | ||
| Laboratory assessment | |||||
| Hemoglobin levels g/dL | 12–15 | 13.5 (±0.8) | 8.1 (±1.9) | <0.001 | |
| Hematocrit % | 40–47 | 44 (±1.5) | 33(±5) | <0.001 | |
| MCHC, g/dL | 32–36 | 34 (±0.9) | 28 (±4) | <0.001 | |
| MCV, fL | 80–100 | 88 (±6) | 73 (±6) | <0.001 | |
| CRP, mg/L | <5 | 2.6 (±1.1) | 24.6 (±14.3) | <0.001 | |
| LDH, U/L | 130–230 | 153 (±23) | 265 (±64) | <0.001 | |
| ALP, U/L | 20–140 | 130 (±18) | 142 (±49) | 0.324 | |
| CEA, ng/mL | <3 | 3.8 (±0.9) | 21 (±19) | <0.001 | |
| miR-106a (fold change) | 2.7 (0.04–15.9) | 71.3 (21.5–343.7) | <0.001 | ||
| CA 19-9 U/mL | 0–27 | 11.7 (±8.7) | 57.9 (±40) | <0.001 | |
| 95.0% Confidence Interval for B | ||||
|---|---|---|---|---|
| Variable | B | p-Value | Lower Bound | Upper Bound |
| Gender | 2.274 | 0.236 | −1.561 | 6.11 |
| Age | 0.008 | 0.918 | −0.147 | 0.163 |
| Carcinoembryonic antigen (CEA) | 0.175 | 0.024 | 0.025 | 0.326 |
| Carbohydrate antigen 19-9 (CA 19-9) | −0.01 | 0.779 | −0.078 | 0.059 |
| Univariable Regression | Multivariable Regression | |||||||
|---|---|---|---|---|---|---|---|---|
| p-Value | Odds Ratio (OR) | 95% CI for OR | p-Value | Odds Ratio (OR) | 95% CI for OR | |||
| Variable | Lower | Upper | Lower | Upper | ||||
| miR-106 | 0.001 | 14,733 | 2,965 | 73,208 | 0.007 | 12,032 | 1948 | 74,305 |
| CEA | 0.001 | 19 | 3604 | 100,154 | 0.003 | 30 | 3141 | 286,574 |
| CA 19-9 | <0.001 | 67.5 | 6795 | 670,529 | 0.002 | 55,866 | 4512 | 691,687 |
| Variable | Area | Std. Error | p-Value | Confidence Interval 95% | |
|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||
| miR-106 model | 0.892 | 0.052 | <0.001 | 0.789 | 0.995 |
| CEA model | 0.919 | 0.043 | <0.001 | 0.836 | 1.000 |
| CA 19-9 model | 0.936 | 0.041 | <0.001 | 0.856 | 1.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boicean, A.; Boeras, I.; Birsan, S.; Ichim, C.; Todor, S.B.; Onisor, D.M.; Brusnic, O.; Bacila, C.; Dura, H.; Roman-Filip, C.; et al. In Pursuit of Novel Markers: Unraveling the Potential of miR-106, CEA and CA 19-9 in Gastric Adenocarcinoma Diagnosis and Staging. Int. J. Mol. Sci. 2024, 25, 7898. https://doi.org/10.3390/ijms25147898
Boicean A, Boeras I, Birsan S, Ichim C, Todor SB, Onisor DM, Brusnic O, Bacila C, Dura H, Roman-Filip C, et al. In Pursuit of Novel Markers: Unraveling the Potential of miR-106, CEA and CA 19-9 in Gastric Adenocarcinoma Diagnosis and Staging. International Journal of Molecular Sciences. 2024; 25(14):7898. https://doi.org/10.3390/ijms25147898
Chicago/Turabian StyleBoicean, Adrian, Ioana Boeras, Sabrina Birsan, Cristian Ichim, Samuel Bogdan Todor, Danusia Maria Onisor, Olga Brusnic, Ciprian Bacila, Horatiu Dura, Corina Roman-Filip, and et al. 2024. "In Pursuit of Novel Markers: Unraveling the Potential of miR-106, CEA and CA 19-9 in Gastric Adenocarcinoma Diagnosis and Staging" International Journal of Molecular Sciences 25, no. 14: 7898. https://doi.org/10.3390/ijms25147898
APA StyleBoicean, A., Boeras, I., Birsan, S., Ichim, C., Todor, S. B., Onisor, D. M., Brusnic, O., Bacila, C., Dura, H., Roman-Filip, C., Ognean, M. L., Tanasescu, C., Hasegan, A., Bratu, D., Porr, C., Roman-Filip, I., Neamtu, B., & Fleaca, S. R. (2024). In Pursuit of Novel Markers: Unraveling the Potential of miR-106, CEA and CA 19-9 in Gastric Adenocarcinoma Diagnosis and Staging. International Journal of Molecular Sciences, 25(14), 7898. https://doi.org/10.3390/ijms25147898








