Vitamin D Receptor Regulates the Expression of the Grainyhead-Like 1 Gene
Abstract
1. Introduction
2. Results
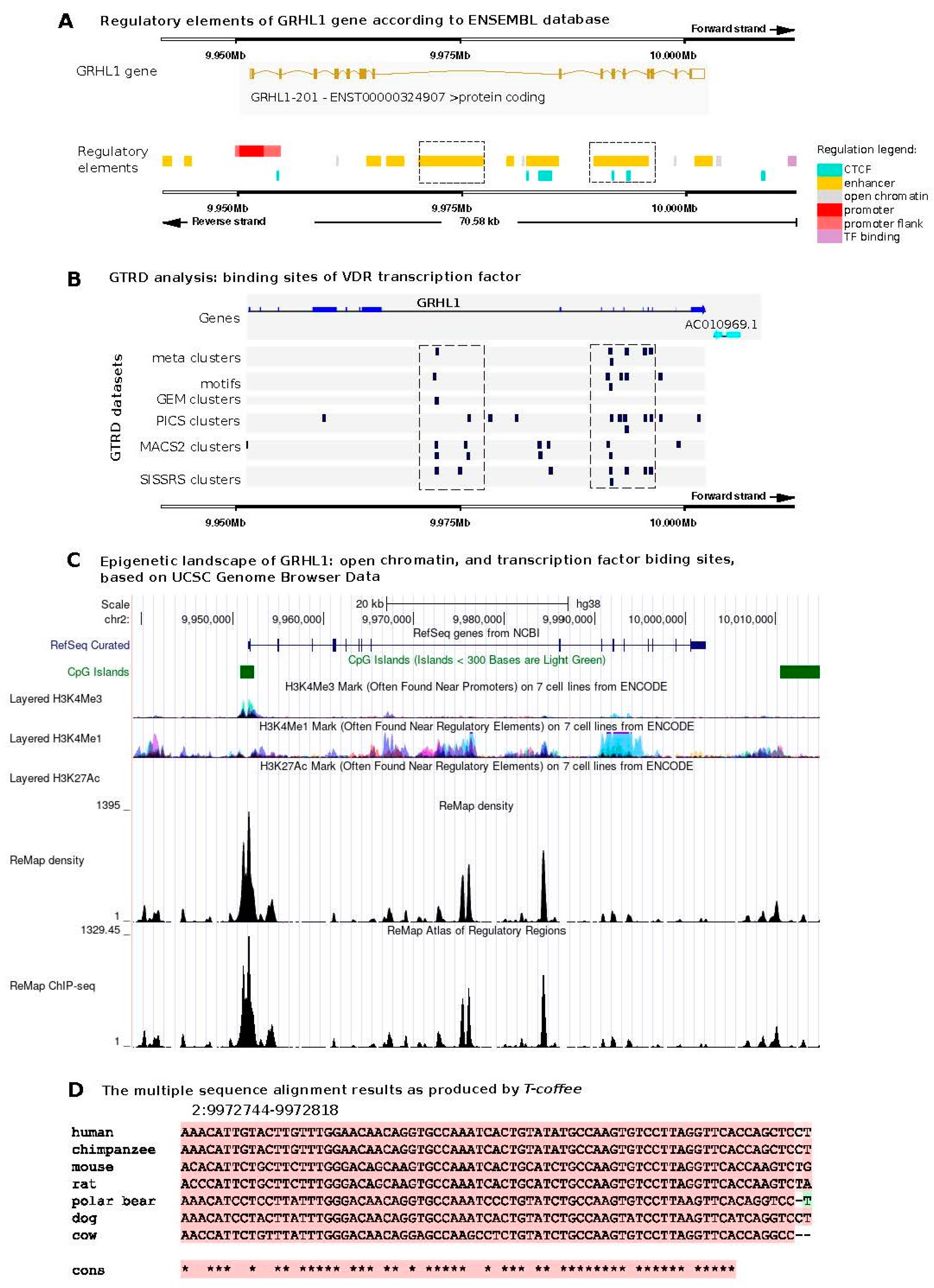
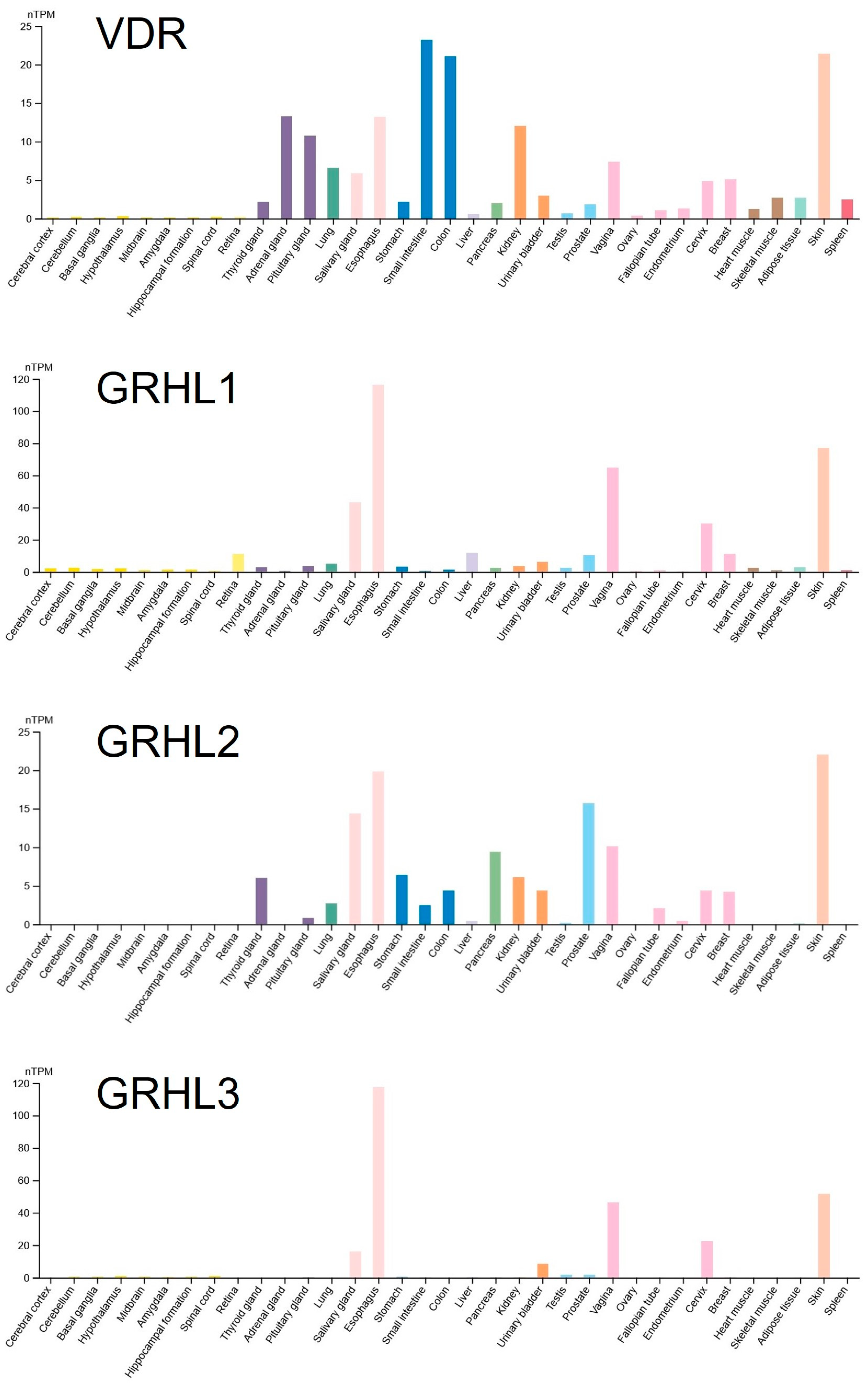
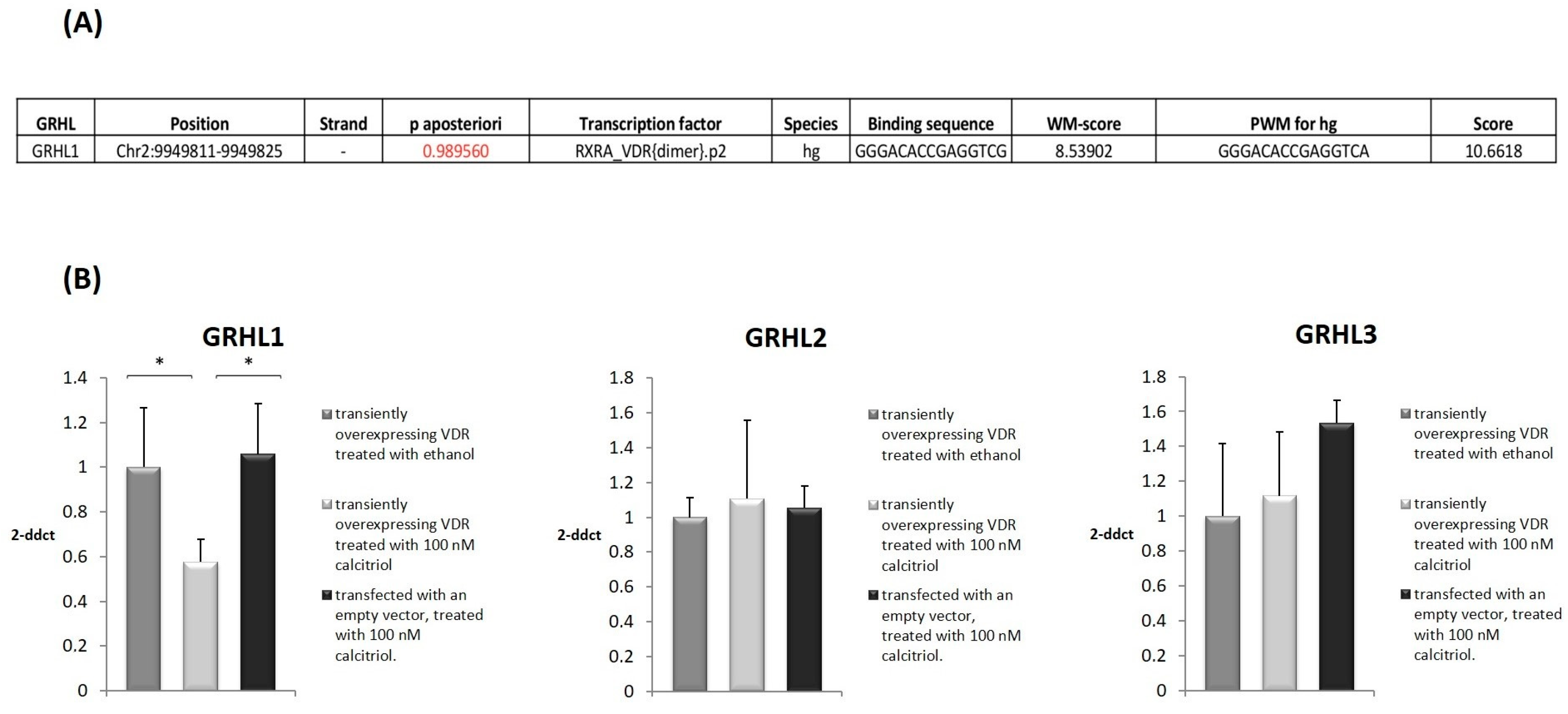
2.1. Bioinformatic Analyses of Potential Binding Sites for VDR in the Regulatory Regions of the GRHL1 Gene
2.2. VDR and Calcitriol Regulate the Expression of the GRHL1 Gene
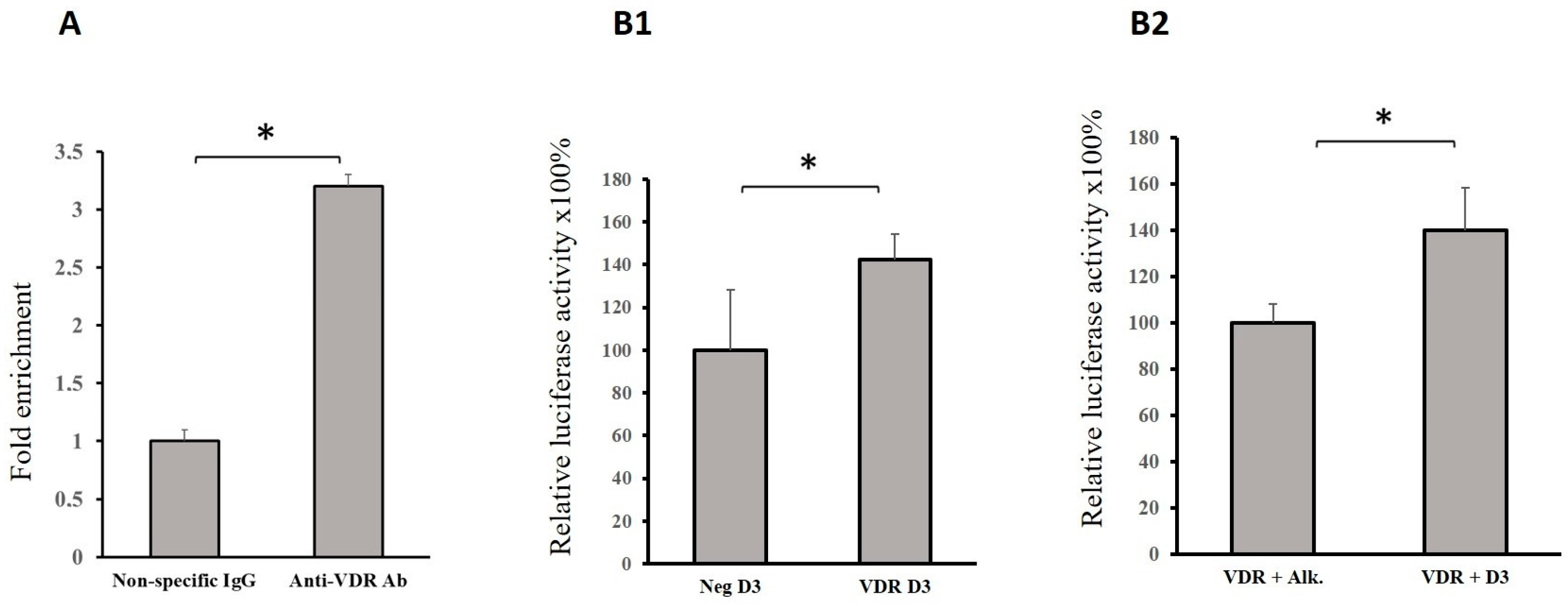
2.3. VDR Binds to a Regulatory Region of the GRHL1 Gene
3. Discussion
4. Materials and Methods
4.1. In Silico Prediction of VDR Binding Sites in the Regulatory Elements of the GRHL1 Gene
4.2. Cell Culture
4.3. Plasmids
4.4. Total RNA Extraction, Reverse Transcription, and qRT-PCR Assays
4.5. Chromatin Immunoprecipitation Assays (ChIP)
4.6. Reporter Gene Assays
4.7. Calcitriol Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pitt, M.J. Rickets and osteomalacia are still around. Radiol. Clin. N. Am. 1991, 29, 97–118. [Google Scholar] [CrossRef] [PubMed]
- Sahota, O. Osteoporosis and the role of vitamin D and calcium-vitamin D deficiency, vitamin D insufficiency and vitamin D sufficiency. Age Ageing 2000, 29, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Wharton, B.; Bishop, N. Rickets. Lancet 2003, 362, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Kechichian, E.; Ezzedine, K. Vitamin D and the Skin: An Update for Dermatologists. Am. J. Clin. Dermatol. 2018, 19, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Bikle, D.D.; Chang, S.; Crumrine, D.; Elalieh, H.; Man, M.Q.; Choi, E.H.; Dardenne, O.; Xie, Z.; Arnaud, R.S.; Feingold, K.; et al. 25 Hydroxyvitamin D 1 α-Hydroxylase Is Required for Optimal Epidermal Differentiation and Permeability Barrier Homeostasis. J. Investig. Dermatol. 2004, 122, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Nezbedova, P.; Brtko, J. 1alpha,25-dihydroxyvitamin D3 inducible transcription factor and its role in the vitamin D action. Endocr. Regul. 2004, 38, 29–38. [Google Scholar] [PubMed]
- Bikle, D.D.; Oda, Y.; Tu, C.L.; Jiang, Y. Novel mechanisms for the vitamin D receptor (VDR) in the skin and in skin cancer. J. Steroid Biochem. Mol. Biol. 2015, 148, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Sihlbom, C.; Chalkley, R.J.; Huang, L.; Rachez, C.; Chang, C.P.; Burlingame, A.L.; Freedman, L.P.; Bikle, D.D. Two distinct coactivators, DRIP/mediator and SRC/p160, are differentially involved in vitamin D receptor transactivation during keratinocyte differentiation. Mol. Endocrinol. 2003, 17, 2329–2339. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Palmer, D.J. Vitamin D and the Development of Atopic Eczema. J. Clin. Med. 2015, 4, 1036–1050. [Google Scholar] [CrossRef]
- Lim, S.K.; Ha, J.M.; Lee, Y.H.; Lee, Y.; Seo, Y.J.; Kim, C.D.; Lee, J.H.; Im, M. Comparison of Vitamin D Levels in Patients with and without Acne: A Case-Control Study Combined with a Randomized Controlled Trial. PLoS ONE 2016, 11, e0161162. [Google Scholar] [CrossRef]
- Carlberg, C. Vitamin D and Its Target Genes. Nutrients 2022, 14, 1354. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; van den Berg, P.R.; Long, M.D.; Vreugdenhil, A.; Grieshober, L.; Ochs-Balcom, H.M.; Wang, J.; Delcambre, S.; Heikkinen, S.; Carlberg, C.; et al. Integration of VDR genome wide binding and GWAS genetic variation data reveals co-occurrence of VDR and NF-kappaB binding that is linked to immune phenotypes. BMC Genom. 2017, 18, 132. [Google Scholar] [CrossRef] [PubMed]
- von Essen, M.R.; Geisler, C. VDR, the Vitamin D Receptor. In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer: New York, NY, USA, 2016; pp. 1–8. [Google Scholar]
- Dworkin, S.; Darido, C.; Georgy, S.R.; Wilanowski, T.; Srivastava, S.; Ellett, F.; Pase, L.; Han, Y.; Meng, A.; Heath, J.K.; et al. Midbrain-hindbrain boundary patterning and morphogenesis are regulated by diverse grainy head-like 2-dependent pathways. Development 2012, 139, 525–536. [Google Scholar] [CrossRef]
- Deng, Z.; Cangkrama, M.; Butt, T.; Jane, S.M.; Carpinelli, M.R. Grainyhead-like transcription factors: Guardians of the skin barrier. Vet. Dermatol. 2021, 32, 553-e152. [Google Scholar] [CrossRef] [PubMed]
- Cangkrama, M.; Darido, C.; Georgy, S.R.; Partridge, D.; Auden, A.; Srivastava, S.; Wilanowski, T.; Jane, S.M. Two Ancient Gene Families Are Critical for Maintenance of the Mammalian Skin Barrier in Postnatal Life. J. Investg. Dermatol. 2016, 136, 1438–1448. [Google Scholar] [CrossRef]
- Wilanowski, T.; Caddy, J.; Ting, S.B.; Hislop, N.R.; Cerruti, L.; Auden, A.; Zhao, L.L.; Asquith, S.; Ellis, S.; Sinclair, R.; et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 2008, 27, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Bikle, D.D. Vitamin D and the skin: Physiology and pathophysiology. Rev. Endocr. Metab. Disord. 2012, 13, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Uchida, Y.; Moradian, S.; Crumrine, D.; Elias, P.M.; Bikle, D.D. Vitamin D receptor and coactivators SRC2 and 3 regulate epidermis-specific sphingolipid production and permeability barrier formation. J. Investg. Dermatol. 2009, 129, 1367–1378. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Y.G. Vitamin D Receptor Influences Intestinal Barriers in Health and Disease. Cells 2022, 11, 1129. [Google Scholar] [CrossRef]
- Chen, C.H.; Sakai, Y.; Demay, M.B. Targeting expression of the human vitamin D receptor to the keratinocytes of vitamin D receptor null mice prevents alopecia. Endocrinology 2001, 142, 5386–5389. [Google Scholar] [CrossRef] [PubMed]
- Joko, Y.; Yamamoto, Y.; Kato, S.; Takemoto, T.; Abe, M.; Matsumoto, T.; Fukumoto, S.; Sawatsubashi, S. VDR is an essential regulator of hair follicle regression through the progression of cell death. Life Sci. Alliance 2023, 6, e202302014. [Google Scholar] [CrossRef] [PubMed]
- Kise, S.; Iijima, A.; Nagao, C.; Okada, T.; Mano, H.; Nishikawa, M.; Ikushiro, S.; Kanemoto, Y.; Kato, S.; Nakanishi, T.; et al. Functional analysis of vitamin D receptor (VDR) using adenovirus vector. J. Steroid Biochem. Mol. Biol. 2023, 230, 106275. [Google Scholar] [CrossRef] [PubMed]
- Rishikaysh, P.; Dev, K.; Diaz, D.; Qureshi, W.M.; Filip, S.; Mokry, J. Signaling involved in hair follicle morphogenesis and development. Int. J. Mol. Sci. 2014, 15, 1647–1670. [Google Scholar] [CrossRef] [PubMed]
- Narhi, K.; Jarvinen, E.; Birchmeier, W.; Taketo, M.M.; Mikkola, M.L.; Thesleff, I. Sustained epithelial beta-catenin activity induces precocious hair development but disrupts hair follicle down-growth and hair shaft formation. Development 2008, 135, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Mlacki, M.; Darido, C.; Jane, S.M.; Wilanowski, T. Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS ONE 2014, 9, e89247. [Google Scholar] [CrossRef] [PubMed]
- Kikulska, A.; Rausch, T.; Krzywinska, E.; Pawlak, M.; Wilczynski, B.; Benes, V.; Rutkowski, P.; Wilanowski, T. Coordinated expression and genetic polymorphisms in Grainyhead-like genes in human non-melanoma skin cancers. BMC Cancer 2018, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Reichrath, J.; Saternus, R.; Vogt, T. Endocrine actions of vitamin D in skin: Relevance for photocarcinogenesis of non-melanoma skin cancer, and beyond. Mol. Cell. Endocrinol. 2017, 453, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Seretis, K.; Bounas, N.; Sioka, C. The Association of Vitamin D with Non-Melanoma Skin Cancer Risk: An Umbrella Review of Systematic Reviews and Meta-Analyses. Medicina 2023, 59, 2130. [Google Scholar] [CrossRef]
- Caini, S.; Gnagnarella, P.; Stanganelli, I.; Bellerba, F.; Cocorocchio, E.; Queirolo, P.; Bendinelli, B.; Saieva, C.; Raimondi, S.; Gandini, S. Vitamin D and the Risk of Non-Melanoma Skin Cancer: A Systematic Literature Review and Meta-Analysis on Behalf of the Italian Melanoma Intergroup. Cancers 2021, 13, 4815. [Google Scholar] [CrossRef]
- Liu, M.; Freedman, L.P. Transcriptional synergism between the vitamin D3 receptor and other nonreceptor transcription factors. Mol. Endocrinol. 1994, 8, 1593–1604. [Google Scholar] [PubMed]
- Zenata, O.; Vrzal, R. Fine tuning of vitamin D receptor (VDR) activity by post-transcriptional and post-translational modifications. Oncotarget 2017, 8, 35390–35402. [Google Scholar] [CrossRef] [PubMed]
- Arriagada, G.; Paredes, R.; Olate, J.; van Wijnen, A.; Lian, J.B.; Stein, G.S.; Stein, J.L.; Onate, S.; Montecino, M. Phosphorylation at serine 208 of the 1α, 25-dihydroxy Vitamin D3 receptor modulates the interaction with transcriptional coactivators. J. Steroid Biochem. Mol. Biol. 2007, 103, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Kotarba, G.; Taracha-Wisniewska, A.; Miller, M.; Dabrowski, M.; Wilanowski, T. Transcription factors Kruppel-like factor 4 and paired box 5 regulate the expression of the Grainyhead-like genes. PLoS ONE 2021, 16, e0257977. [Google Scholar] [CrossRef] [PubMed]
- Phatak, M.; Kulkarni, S.; Miles, L.B.; Anjum, N.; Dworkin, S.; Sonawane, M. Grhl3 promotes retention of epidermal cells under endocytic stress to maintain epidermal architecture in zebrafish. PLoS Genet. 2021, 17, e1009823. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Guo, S.; Tu, Z.; Di, L.; Zha, X.; Zhou, H.; Zhang, X. Grhl3 induces human epithelial tumor cell migration and invasion via downregulation of E-cadherin. Acta Biochim. Biophys. Sin. 2016, 48, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, M.J.; Cox, R.S., III; Levine, J.H.; Murray, R.M.; Elowitz, M.B. Regulatory activity revealed by dynamic correlations in gene expression noise. Nat. Genet. 2008, 40, 1493–1498. [Google Scholar] [CrossRef] [PubMed]
- Mathiyalagan, N.; Miles, L.B.; Anderson, P.J.; Wilanowski, T.; Grills, B.L.; McDonald, S.J.; Keightley, M.C.; Charzynska, A.; Dabrowski, M.; Dworkin, S. Meta-Analysis of Grainyhead-Like Dependent Transcriptional Networks: A Roadmap for Identifying Novel Conserved Genetic Pathways. Genes 2019, 10, 876. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.J. Vitamin D Receptor Signaling Regulates Craniofacial Cartilage Development in Zebrafish. J. Dev. Biol. 2019, 7, 13. [Google Scholar] [CrossRef]
- Dworkin, S.; Simkin, J.; Darido, C.; Partridge, D.D.; Georgy, S.R.; Caddy, J.; Wilanowski, T.; Lieschke, G.J.; Doggett, K.; Heath, J.K.; et al. Grainyhead-like 3 regulation of endothelin-1 in the pharyngeal endoderm is critical for growth and development of the craniofacial skeleton. Mech. Dev. 2014, 133, 77–90. [Google Scholar] [CrossRef]
- de Vries, M.E.; Carpinelli, M.R.; Fuller, J.N.; Sutton, Y.; Partridge, D.D.; Auden, A.; Anderson, P.J.; Jane, S.M.; Dworkin, S. Grainyhead-like 2 interacts with noggin to regulate tissue fusion in mouse. Development 2024, 151, dev202420. [Google Scholar] [CrossRef] [PubMed]
- Gasperoni, J.G.; Fuller, J.N.; Darido, C.; Wilanowski, T.; Dworkin, S. Grainyhead-like (Grhl) Target Genes in Development and Cancer. Int. J. Mol. Sci. 2022, 23, 2735. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.; Kikulska, A.; Wrzesinski, T.; Rausch, T.; Kwias, Z.; Wilczynski, B.; Benes, V.; Wesoly, J.; Wilanowski, T. Potential protective role of Grainyhead-like genes in the development of clear cell renal cell carcinoma. Mol. Carcinog. 2017, 56, 2414–2423. [Google Scholar] [CrossRef] [PubMed]
- Walkowska, A.; Pawlak, M.; Jane, S.M.; Kompanowska-Jezierska, E.; Wilanowski, T. Effects of high and low sodium diet on blood pressure and heart rate in mice lacking the functional grainyhead-like 1 gene. Physiol. Res. 2017, 66, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.; Walkowska, A.; Mlacki, M.; Pistolic, J.; Wrzesinski, T.; Benes, V.; Jane, S.M.; Wesoly, J.; Kompanowska-Jezierska, E.; Wilanowski, T. Consequences of the loss of the Grainyhead-like 1 gene for renal gene expression, regulation of blood pressure and heart rate in a mouse model. Acta Biochim. Pol. 2015, 62, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Blanco, J.C.; Wang, I.M.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W.; Jurutka, P.W.; Haussler, M.R.; Ozato, K. Transcription factor TFIIB and the vitamin D receptor cooperatively activate ligand-dependent transcription. Proc. Natl. Acad. Sci. USA 1995, 92, 1535–1539. [Google Scholar] [CrossRef]
- Herdick, M.; Bury, Y.; Quack, M.; Uskokovic, M.R.; Polly, P.; Carlberg, C. Response element and coactivator-mediated conformational change of the vitamin D(3) receptor permits sensitive interaction with agonists. Mol. Pharmacol. 2000, 57, 1206–1217. [Google Scholar] [PubMed]
- Tagami, T.; Lutz, W.H.; Kumar, R.; Jameson, J.L. The Interaction of the Vitamin D Receptor with Nuclear Receptor Corepressors and Coactivators. Biochem. Biophys. Res. Commun. 1998, 253, 358–363. [Google Scholar] [CrossRef]
- Takeshita, A.; Ozawa, Y.; Chin, W.W. Nuclear receptor coactivators facilitate vitamin D receptor homodimer action on direct repeat hormone response elements. Endocrinology 2000, 141, 1281–1284. [Google Scholar] [CrossRef]
- Takeyama, K.; Masuhiro, Y.; Fuse, H.; Endoh, H.; Murayama, A.; Kitanaka, S.; Suzawa, M.; Yanagisawa, J.; Kato, S. Selective interaction of vitamin D receptor with transcriptional coactivators by a vitamin D analog. Mol. Cell. Biol. 1999, 19, 1049–1055. [Google Scholar] [CrossRef]
- Pike, J.W.; Meyer, M.B. The vitamin D receptor: New paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3). Endocrinol. Metab. Clin. N. Am. 2010, 39, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Polly, P.; Herdick, M.; Moehren, U.; Baniahmad, A.; Heinzel, T.; Carlberg, C. VDR-Alien: A novel, DNA-selective vitamin D(3) receptor-corepressor partnership. FASEB J. 2000, 14, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Rachez, C.; Suldan, Z.; Ward, J.; Chang, C.P.; Burakov, D.; Erdjument-Bromage, H.; Tempst, P.; Freedman, L.P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998, 12, 1787–1800. [Google Scholar] [CrossRef] [PubMed]
- Warwick, T.; Schulz, M.H.; Günther, S.; Gilsbach, R.; Neme, A.; Carlberg, C.; Brandes, R.P.; Seuter, S. A hierarchical regulatory network analysis of the vitamin D induced transcriptome reveals novel regulators and complete VDR dependency in monocytes. Sci. Rep. 2021, 11, 6518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Baudino, T.A.; Dowd, D.R.; Tokumaru, H.; Wang, W.; MacDonald, P.N. Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J. Biol. Chem. 2001, 276, 40614–40620. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.C.; Whitfield, G.K.; Oza, A.K.; Dang, H.T.; Price, J.N.; Galligan, M.A.; Jurutka, P.W.; Thompson, P.D.; Haussler, C.A.; Haussler, M.R. Characterization of unique DNA-binding and transcriptional-activation functions in the carboxyl-terminal extension of the zinc finger region in the human vitamin D receptor. Biochemistry 1999, 38, 16347–16358. [Google Scholar] [CrossRef] [PubMed]
- Haussler, M.R.; Jurutka, P.W.; Mizwicki, M.; Norman, A.W. Vitamin D receptor (VDR)-mediated actions of 1alpha,25(OH)2vitamin D3: Genomic and non-genomic mechanisms. Best Pr. Res. Clin. Endocrinol. Metab. 2011, 25, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.T.; Tuckey, R.C.; Jetten, A.M.; Holick, M.F. Recent Advances in Vitamin D Biology: Something New under the Sun. J. Investg. Dermatol. 2023, 143, 2340–2342. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.T.; Tuckey, R.C.; Jenkinson, C.; Li, W.; Jetten, A.M. Alternative pathways for vitamin D metabolism. In Feldman and Pike’s Vitamin D; Elsevier: Amsterdam, The Netherlands, 2024; pp. 85–109. [Google Scholar]
- Slominski, A.T.; Brozyna, A.A.; Zmijewski, M.A.; Janjetovic, Z.; Kim, T.K.; Slominski, R.M.; Tuckey, R.C.; Mason, R.S.; Jetten, A.M.; Guroji, P.; et al. The Role of Classical and Novel Forms of Vitamin D in the Pathogenesis and Progression of Nonmelanoma Skin Cancers. Adv. Exp. Med. Biol. 2020, 1268, 257–283. [Google Scholar]
- Kim, T.K.; Slominski, R.M.; Pyza, E.; Kleszczynski, K.; Tuckey, R.C.; Reiter, R.J.; Holick, M.F.; Slominski, A.T. Evolutionary formation of melatonin and vitamin D in early life forms: Insects take centre stage. Biol. Rev. 2024. [Google Scholar] [CrossRef]
- Arnold, P.; Erb, I.; Pachkov, M.; Molina, N.; van Nimwegen, E. MotEvo: Integrated Bayesian probabilistic methods for inferring regulatory sites and motifs on multiple alignments of DNA sequences. Bioinformatics 2011, 28, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Kolmykov, S.; Yevshin, I.; Kulyashov, M.; Sharipov, R.; Kondrakhin, Y.; Makeev, V.J.; Kulakovskiy, I.V.; Kel, A.; Kolpakov, F. GTRD: An integrated view of transcription regulation. Nucleic Acids Res. 2021, 49, D104–D111. [Google Scholar] [CrossRef] [PubMed]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Binding Site | Primer | Sequence (5′→3′) |
|---|---|---|
| VDR in GRHL1 promoter | Forward | GGGCACAGAGGAGGGACT |
| Reverse | GAGACAGAAGACGGGGACAC |
| Transcription Factor | Organism | Tissue/Organ/Cell Type | Details of Interaction | Source |
|---|---|---|---|---|
| Liver | ||||
| RXR | Human, mouse, rat | Liver | Forms heterodimers with VDR to regulate gene transcription. | [47] |
| TFIIB | Human, mouse | Liver | Interacts directly with VDR to facilitate transcription initiation. | [47] |
| p300/CBP | Human, mouse, rat | Liver | Acts as a histone acetyltransferase, enhancing transcription by relaxing DNA. | [48] |
| HNF-4α | Human, mouse, rat | Liver | Interacts with VDR to regulate metabolic processes in liver cells. | [48] |
| Kidney | ||||
| RXR | Human, mouse, rat | Kidney | Forms heterodimers with VDR to regulate gene transcription. | [47] |
| TFIIB | Human, mouse | Kidney | Interacts directly with VDR to facilitate transcription initiation. | [47] |
| p300/CBP | Human, mouse, rat | Kidney | Acts as a histone acetyltransferase, enhancing transcription by relaxing DNA. | [48] |
| HNF-4α | Human, mouse, rat | Kidney | Interacts with VDR to regulate metabolic processes in kidney cells. | [48] |
| Intestine | ||||
| RXR | Human, mouse, rat | Intestine | Forms heterodimers with VDR to regulate gene transcription. | [47] |
| TFIIB | Human, mouse | Intestine | Interacts directly with VDR to facilitate transcription initiation. | [47] |
| p300/CBP | Human, rat, mouse | Intestine | Acts as a histone acetyltransferase, enhancing transcription by relaxing DNA. | [48] |
| HNF-4α | Human, mouse, rat | Intestine | Interacts with VDR to regulate metabolic processes in intestinal cells. | [48] |
| Bone | ||||
| RXR | Human, mouse, rat | Osteoblasts, osteoclasts | Forms heterodimers with VDR to regulate gene transcription. | [47] |
| NCoR | Human, mouse | Osteoblasts, osteoclasts | Acts as a corepressor that interacts with VDR to suppress transcription. | [49] |
| SMRT | Human, mouse | Osteoblasts, osteoclasts | Another corepressor that interacts with VDR to regulate gene expression. | [49] |
| SRC-1 | Human, mouse | Osteoblasts, osteoclasts | Coactivator that enhances VDR-mediated transcription. | [50] |
| Muscle Cells | ||||
| MyoD | Human, mouse | Myoblasts | Interacts with VDR to influence muscle cell differentiation and growth. | [49] |
| Skin Cells | ||||
| TIF2 | Human, mouse | Keratinocytes | Coactivator that interacts with VDR to enhance transcription in skin cells. | [51] |
| Nervous System | ||||
| NF-1 | Human, mouse | Neurons, glial cells | Interacts with VDR to regulate genes involved in nervous system development and function. | [32] |
| FOXO3 | Human, mouse | Neurons, glial cells | Interacts with VDR to regulate genes involved in stress resistance, metabolism, and neuronal function. | [32] |
| Cancer Cells | ||||
| c-Myc | Human, mouse | Various cancer cells (colon, breast, prostate) | Oncogene that interacts with VDR, influencing cell proliferation and cancer progression. | [32] |
| HOXB13 | Human, mouse | Prostate cancer cells | Interacts with VDR in prostate cancer cells to regulate gene expression related to cancer progression. | [52] |
| Immune System | ||||
| NF-κB | Human, mouse, rat | Immune cells (T cells, B cells) | Modulates immune responses and inflammation through interaction with VDR. | [32] |
| AP-1 | Human, mouse, rat | Immune cells (T cells, B cells) | Regulates gene expression in response to a variety of stimuli, interacting with VDR to modulate immune functions. | [32] |
| Alien | Human, mouse | Immune cells (T cells, B cells) | Acts as a corepressor that interacts with VDR to modulate transcriptional repression. | [53] |
| BCL6 | Human, mouse | B cells, T cells | Regulates differentiation and function of T cells and B cells, interacting with VDR. | [32] |
| STAT3 | Human, mouse | Various immune cells (T cells, NK cells, macrophages) | Involved in the signaling pathways that mediate immune responses and interacts with VDR. | [32] |
| GATA3 | Human, mouse | T cells, Th2 cells | Interacts with VDR to regulate immune responses, particularly in T-helper 2 (Th2) cells. | [32] |
| NFAT | Human, mouse | Immune cells (T cells, B cells) | Interacts with VDR to modulate the expression of immune-related genes. | [32] |
| IRF1 | Human, mouse | Immune cells (T cells, B cells) | Interacts with VDR to regulate the transcription of interferon-responsive genes. | [32] |
| c-Fos | Human, mouse | Immune cells (T cells, B cells) | Component of the AP-1 transcription factor, interacts with VDR to modulate immune responses. | [32] |
| Development | ||||
| Oct4 | Human, mouse | Embryonic stem cells | Interacts with VDR to regulate gene expression during development. | [54] |
| Wnt Signaling | ||||
| TCF/LEF | Human, mouse, rat | Various cells (liver, kidney, intestine, bone) | Mediates gene expression changes in response to Wnt signaling, interacts with VDR. | [48] |
| TCF7L2 | Human | Various tissues (liver, kidney, intestine, bone) | Interacts with VDR in the Wnt signaling pathway to regulate gene expression. | [55] |
| Transcription Regulation | ||||
| GRIP1 | Human, mouse | Various cells (liver, kidney, intestine, bone) | Coactivator that interacts with VDR to enhance transcriptional activation. | [54] |
| RAC3 | Human, mouse | Various cells (liver, kidney, intestine, bone) | Coactivator that works with VDR to enhance transcription. | [54] |
| SKIP | Human, mouse | Various cells (liver, kidney, intestine, bone) | Involved in the assembly of the transcriptional complex with VDR. | [56] |
| VDRE Binding Proteins | Human | Various cells (liver, kidney, intestine, bone) | Proteins that bind to Vitamin D Response Elements (VDREs) to regulate gene expression in coordination with VDR. | [57] |
| General | ||||
| TRAM-1 | Human, mouse, rat | Various tissues (liver, kidney, intestine, bone) | Coactivator that enhances VDR-mediated transcription. | [50] |
| TIF1 | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Interacts with VDR to modulate transcription. | [51] |
| TRAP220 | Human, mouse, rat | Various tissues (liver, kidney, intestine, bone) | Part of the mediator complex, linking transcriptional regulators to the RNA polymerase II initiation complex. | [54] |
| SUG1 | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Component of the 26S proteasome, involved in non-proteolytic roles like nuclear receptor-mediated transcription. | [54] |
| BAF60a | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Part of the SWI/SNF chromatin remodeling complex, facilitates transcriptional activation with VDR. | [54] |
| NCoA1 | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Enhances the transcriptional activities of steroid hormone receptors like VDR. | [51] |
| c-Jun | Human, mouse, rat | Various tissues (liver, kidney, intestine, bone) | Component of the AP-1 transcription factor, interacts with VDR to respond to stress signals and inflammation. | [32] |
| CREB | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Interacts with VDR to mediate cAMP response element-binding protein transcriptional activities. | [48] |
| COUP-TF | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Interacts with VDR to regulate gene expression across various tissues. | [52] |
| USF1 | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Interacts with VDR to regulate gene expression across various tissues. | [52] |
| GFI1 | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Interacts with VDR to regulate gene expression across various tissues. | [52] |
| SUG2 | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Component of the 26S proteasome, involved in nuclear receptor-mediated transcription with VDR. | [54] |
| VDR-AP (alternative pocket) | Human, mouse | Various tissues (liver, kidney, intestine, bone) | Represents a different binding conformation within VDR that might interact uniquely with ligands or coregulators. | [58] |
| Direction | Sequences of Oligonucleotides (5′→3′) |
|---|---|
| Forward | actGAGCTCcaaacaacaggctgcatgga |
| Reverse | actgCTCGAGcgagagtctgttttggacgt |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taracha-Wisniewska, A.; Parks, E.G.C.; Miller, M.; Lipinska-Zubrycka, L.; Dworkin, S.; Wilanowski, T. Vitamin D Receptor Regulates the Expression of the Grainyhead-Like 1 Gene. Int. J. Mol. Sci. 2024, 25, 7913. https://doi.org/10.3390/ijms25147913
Taracha-Wisniewska A, Parks EGC, Miller M, Lipinska-Zubrycka L, Dworkin S, Wilanowski T. Vitamin D Receptor Regulates the Expression of the Grainyhead-Like 1 Gene. International Journal of Molecular Sciences. 2024; 25(14):7913. https://doi.org/10.3390/ijms25147913
Chicago/Turabian StyleTaracha-Wisniewska, Agnieszka, Emma G. C. Parks, Michal Miller, Lidia Lipinska-Zubrycka, Sebastian Dworkin, and Tomasz Wilanowski. 2024. "Vitamin D Receptor Regulates the Expression of the Grainyhead-Like 1 Gene" International Journal of Molecular Sciences 25, no. 14: 7913. https://doi.org/10.3390/ijms25147913
APA StyleTaracha-Wisniewska, A., Parks, E. G. C., Miller, M., Lipinska-Zubrycka, L., Dworkin, S., & Wilanowski, T. (2024). Vitamin D Receptor Regulates the Expression of the Grainyhead-Like 1 Gene. International Journal of Molecular Sciences, 25(14), 7913. https://doi.org/10.3390/ijms25147913









