Feed Components and Timing to Improve the Feed Conversion Ratio for Sustainable Aquaculture Using Starch
Abstract
:1. Introduction
2. Results
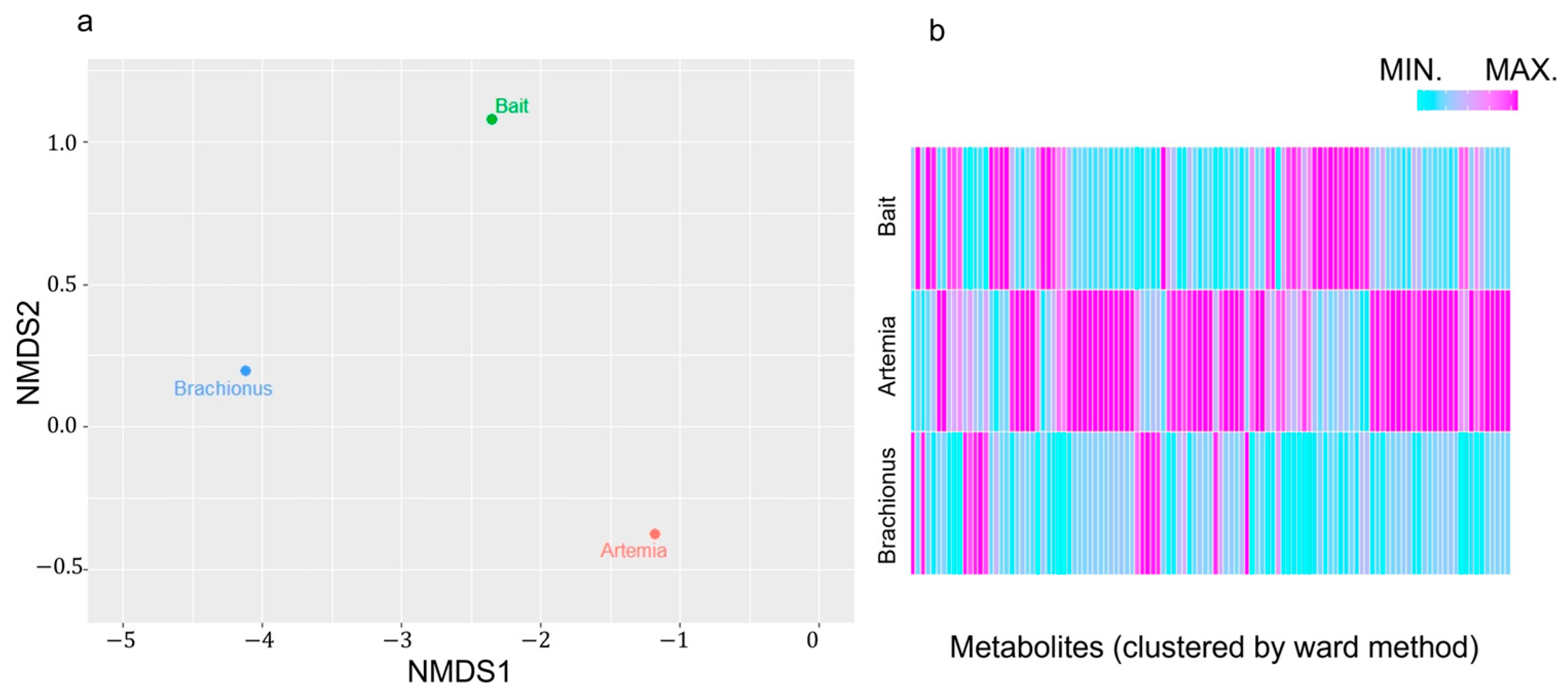
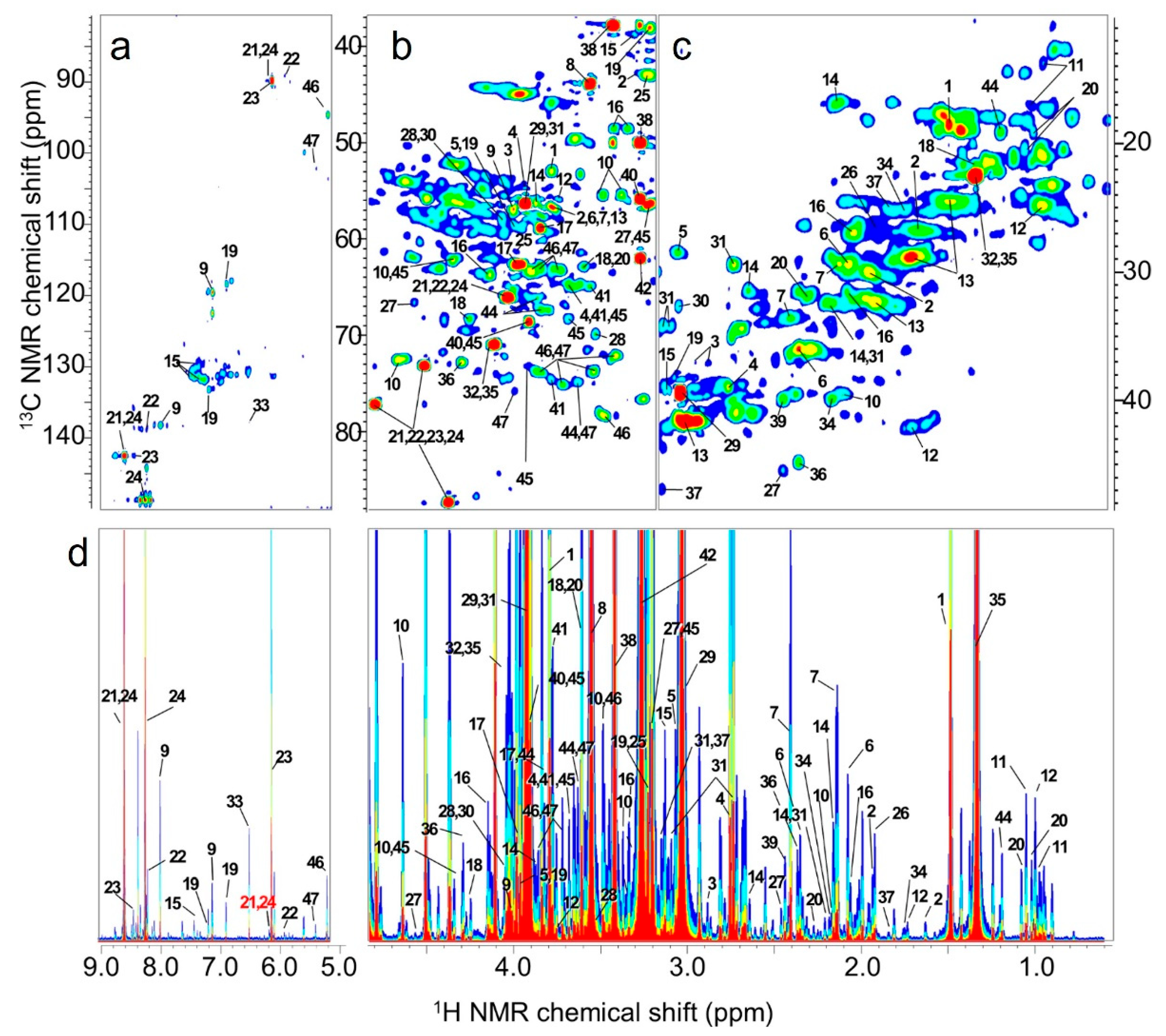
2.1. Differences in Feed Components Based on NMR Spectroscopy Data
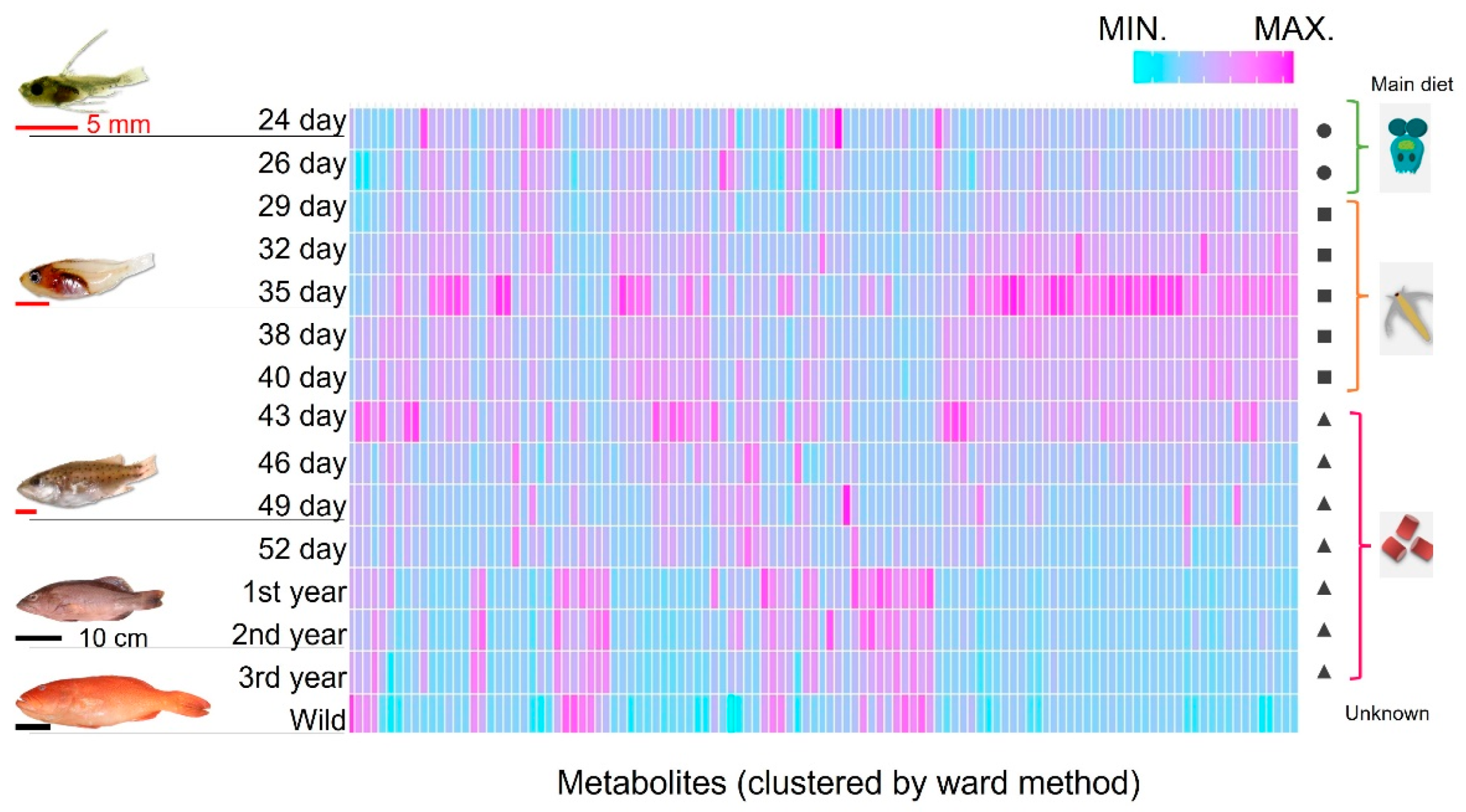
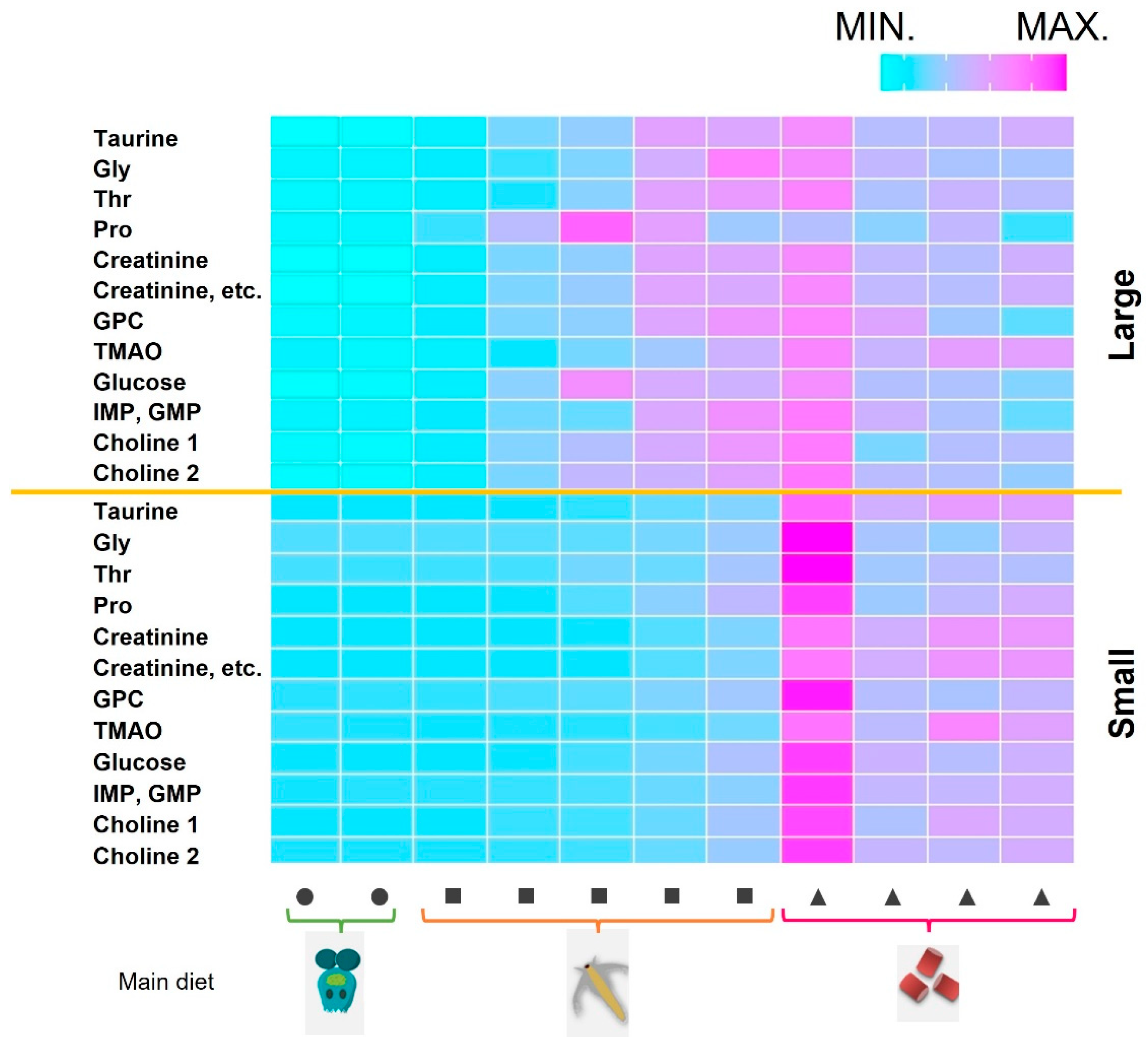
2.2. Changes in Muscle Components during Growth Stages and Comparing Farmed and Wild-Captured Adult Fish
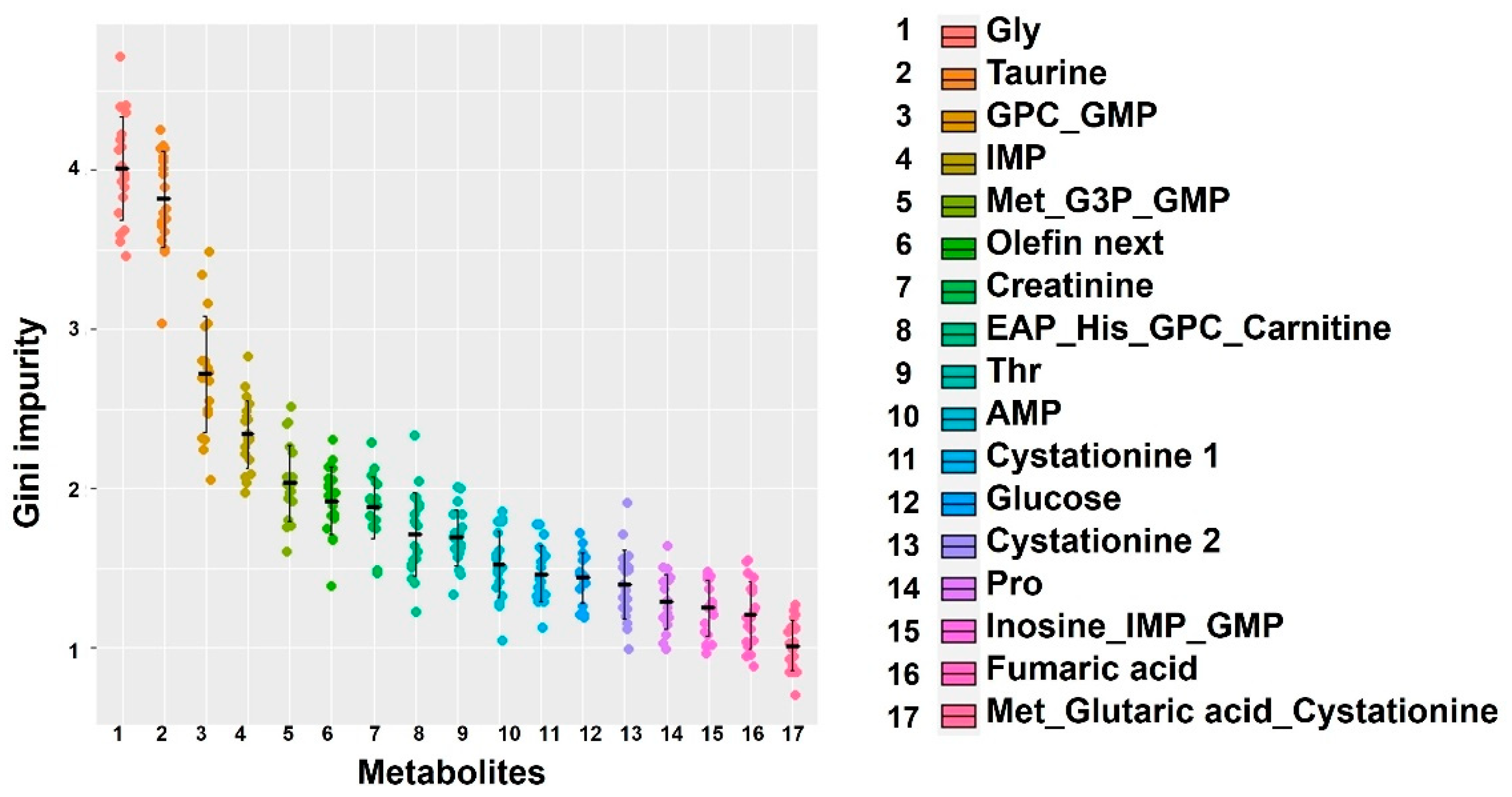
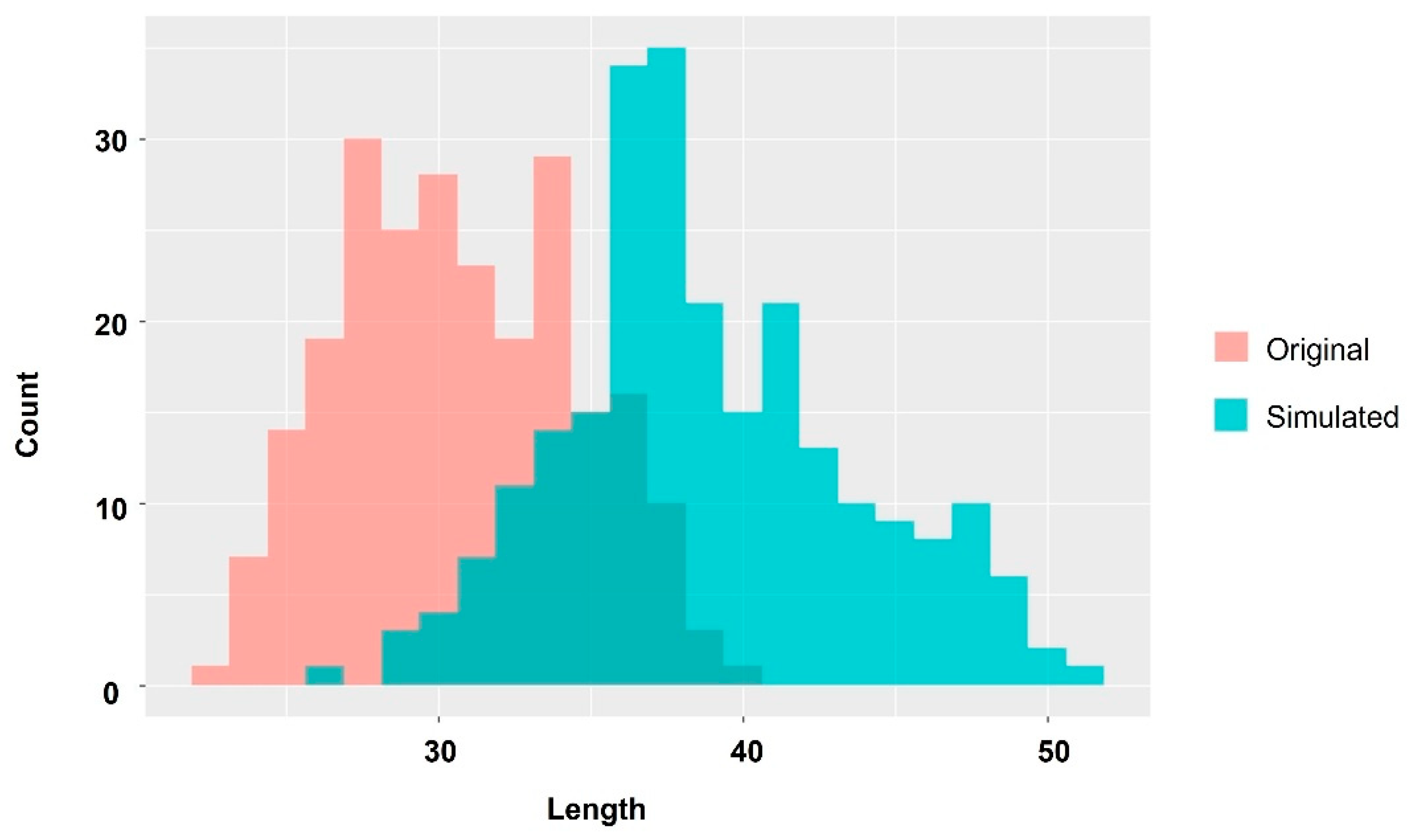
2.3. Extraction of Important Factors Determining Body Size in the Early Stages of Farming Using Machine Learning Methods
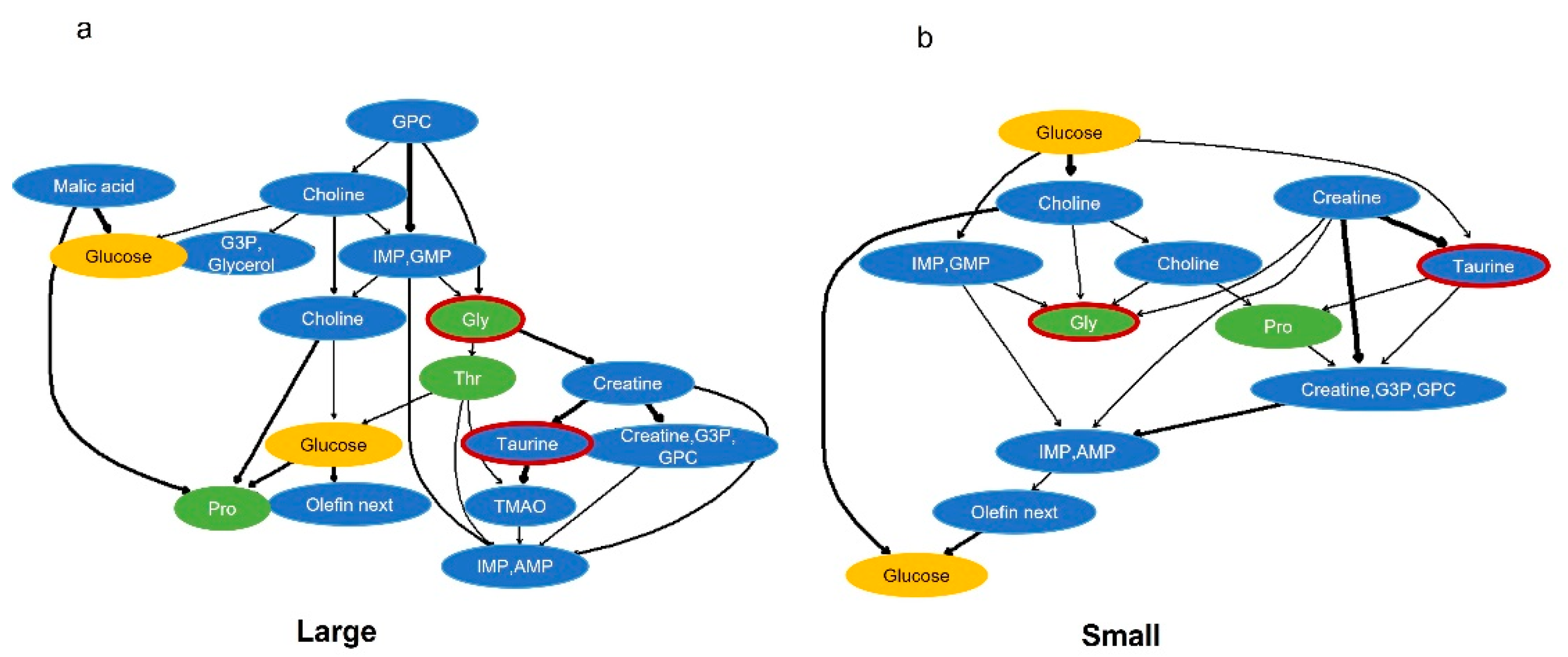
2.4. Probabilistic Causal Inference Using Components That Explain Important Factors of Body Size Classification by Bayesian Networks
2.5. Analysis of Time-Series Changes in Important Factors for Body Size
2.6. Monitoring Starch Metabolism with 13C Labeling
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Fish Samples
4.3. NMR
4.4. Annotation and Normalization of NMR Spectra
4.5. Analytics and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stead, S.M. Using systems thinking and open innovation to strengthen aquaculture policy for the United Nations Sustainable Development Goals. J. Fish Biol. 2019, 94, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A. Thought of alternate aquafeed: Conundrum in aquaculture sustainability? Proc. Proc. Zool. Soc. 2021, 74, 1–18. [Google Scholar] [CrossRef]
- Lynch, A.J.; Elliott, V.; Phang, S.C.; Claussen, J.E.; Harrison, I.; Murchie, K.J.; Steel, E.A.; Stokes, G.L. Inland fish and fisheries integral to achieving the Sustainable Development Goals. Nat. Sustain. 2020, 3, 579–587. [Google Scholar] [CrossRef]
- Naylor, R.L.; Hardy, R.W.; Buschmann, A.H.; Bush, S.R.; Cao, L.; Klinger, D.H.; Little, D.C.; Lubchenco, J.; Shumway, S.E.; Troell, M.A. A 20-year retrospective review of global aquaculture. Nature 2021, 591, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Orioles, M.; Galeotti, M.; Saccà, E.; Bulfoni, M.; Corazzin, M.; Bianchi, S.; Torge, D.; Macchiarelli, G.; Magi, G.E.; Schmidt, J.G. Effect of temperature on transfer of Midichloria-like organism and development of red mark syndrome in rainbow trout (Oncorhynchus mykiss). Aquaculture 2022, 560, 738577. [Google Scholar] [CrossRef]
- Watson, R.A.; Nowara, G.B.; Hartmann, K.; Green, B.S.; Tracey, S.R.; Carter, C.G. Marine foods sourced from farther as their use of global ocean primary production increases. Nat. Commun. 2015, 6, 7365. [Google Scholar] [CrossRef] [PubMed]
- Kok, B.; Malcorps, W.; Tlusty, M.F.; Eltholth, M.M.; Auchterlonie, N.A.; Little, D.C.; Harmsen, R.; Newton, R.W.; Davies, S.J. Fish as feed: Using economic allocation to quantify the Fish. Aquaculture 2020, 528, 735474. [Google Scholar] [CrossRef]
- Ma, M.; Hu, Q. Microalgae as feed sources and feed additives for sustainable aquaculture: Prospects and challenges. Rev. Aquacult. 2024, 16, 818–835. [Google Scholar] [CrossRef]
- Samoilys, M.A.; Squire, L.C. Preliminary observations on the spawning behavior of coral trout, Plectropomus leopardus (Pisces: Serranidae), on the Great Barrier Reef. Bull. Mar. Sci. 1994, 54, 332–342. [Google Scholar]
- Tucker, J.W., Jr. Grouper aquaculture. South Reg. Aquacult. Cent. Publ. 1999, 721, 1–11. [Google Scholar]
- Sadovy de Mitcheson, Y.; Craig, M.T.; Bertoncini, A.A.; Carpenter, K.E.; Cheung, W.W.L.; Choat, J.H.; Cornish, A.S.; Fennessy, S.T.; Ferreira, B.P.; Heemstra, P.C.; et al. Fishing groupers towards extinction: A global assessment of threats and extinction risks in a billion dollar fishery. Fish Fish. 2013, 14, 119–136. [Google Scholar] [CrossRef]
- Kamalam, B.S.; Panserat, S. Carbohydrates in fish nutrition. Int. Aquafeed 2016, 20–23. [Google Scholar] [CrossRef]
- Miyamoto, H.; Shigeta, K.; Suda, W.; Ichihashi, Y.; Nihei, N.; Matsuura, M.; Tsuboi, A.; Tominaga, N.; Aono, M.; Sato, M.; et al. An agroecological structure model of compost-soil-plant interactions for sustainable organic farming. ISME Commun. 2023, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Van Zanten, H.H.E.; Simon, W.; Van Selm, B.; Wacker, J.; Maindl, T.I.; Frehner, A.; Hijbeek, R.; Van Ittersum, M.K.; Herrero, M. Circularity in Europe strengthens the sustainability of the global food system. Nat. Food 2023, 4, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Polakof, S.; Panserat, S.; Soengas, J.L.; Moon, T.W. Glucose metabolism in fish: A review. J. Comp. Physiol. B 2012, 182, 1015–1045. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.P.; Halver, J.E. Protein and amino acid requirements of fishes. Annu. Rev. Nutr. 1986, 6, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Craig, S.R.; Helfrich, L.A.; Kuhn, D.; Schwarz, M.H. Understanding Fish Nutrition, Feeds, and Feeding; Virginia Cooperative Extension: Leesburg, VA, USA, 2017. [Google Scholar]
- Roscher, A.; Kruger, N.J.; Ratcliffe, R.G. Strategies for metabolic flux analysis in plants using isotope labelling. J. Biotechnol. 2000, 77, 81–102. [Google Scholar] [CrossRef] [PubMed]
- Petropoulou, K.; Salt, L.J.; Edwards, C.H.; Warren, F.J.; Garcia-Perez, I.; Chambers, E.S.; Alshaalan, R.; Khatib, M.; Perez-Moral, N.; Cross, K.L.; et al. A natural mutation in Pisum sativum L. (pea) alters starch assembly and improves glucose homeostasis in humans. Nat. Food 2020, 1, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Sugase, K.; Dyson, H.J.; Wright, P.E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 2007, 447, 1021–1025. [Google Scholar] [CrossRef]
- Ito, K.; Miyamoto, H.; Matsuura, M.; Ishii, C.; Tsuboi, A.; Tsuji, N.; Nakaguma, T.; Nakanishi, Y.; Kato, T.; Suda, W.; et al. Noninvasive fecal metabolic profiling for the evaluation of characteristics of thermostable lactic acid bacteria, Weizmannia coagulans SANK70258, for broiler chickens. J. Biosci. Bioeng. 2022, 134, 105–115. [Google Scholar] [CrossRef]
- Shima, H.; Sato, Y.; Sakata, K.; Asakura, T.; Kikuchi, J. Identifying a correlation among qualitative non-numeric parameters in natural fish microbe dataset using machine learning. Appl. Sci. 2022, 12, 5927. [Google Scholar] [CrossRef]
- Miyamoto, H.; Asano, F.; Ishizawa, K.; Suda, W.; Miyamoto, H.; Tsuji, N.; Matsuura, M.; Tsuboi, A.; Ishii, C.; Nakaguma, T.; et al. A potential network structure of symbiotic bacteria involved in carbon and nitrogen metabolism of wood-utilizing insect larvae. Sci. Total Environ. 2022, 836, 155520. [Google Scholar] [CrossRef]
- Shima, H.; Masuda, S.; Date, Y.; Shino, A.; Tsuboi, Y.; Kajikawa, M.; Inoue, Y.; Kanamoto, T.; Kikuchi, J. Exploring the impact of food on the gut ecosystem based on the combination of machine learning and network visualization. Nutrients 2017, 9, 1307. [Google Scholar] [CrossRef] [PubMed]
- Shima, H.; Murata, I.; Feifei, W.; Sakata, K.; Yokoyama, D.; Kikuchi, J. Identification of salmoniformes aquaculture conditions to increase creatine and anserine levels using multiomics dataset and nonnumerical information. Front. Microbiol. 2022, 13, 991819. [Google Scholar] [CrossRef]
- Reichstein, M.; Camps-Valls, G.; Stevens, B.; Jung, M.; Denzler, J.; Carvalhais, N.; Prabhat, f. Deep learning and process understanding for data-driven earth system science. Nature 2019, 566, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Kohonen, P.; Parkkinen, J.A.; Willighagen, E.L.; Ceder, R.; Wennerberg, K.; Kaski, S.; Grafström, R.C. A transcriptomics data-driven gene space accurately predicts liver cytopathology and drug-induced liver injury. Nat. Commun. 2017, 8, 15932. [Google Scholar] [CrossRef]
- St. John, J. Ontogenetic changes in the diet of the coral reef grouper Plectropomus leopardus (Serranidae):patterns in taxa, size and habitat of prey. Mar. Ecol. Prog. Ser. 1999, 180, 233–246. [Google Scholar] [CrossRef]
- Seibel, B.A.; Walsh, P.J. Trimethylamine oxide accumulation in marine animals: Relationship to acylglycerol storagej. J. Exp. Biol. 2002, 205, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Summers, G.; Wibisono, R.D.; Hedderley, D.I.; Fletcher, G.C. Trimethylamine oxide content and spoilage potential of New Zealand commercial fish species. N. Z. J. Mar. Freshw. Res. 2017, 51, 393–405. [Google Scholar] [CrossRef]
- Yin, X.; Gibbons, H.; Rundle, M.; Frost, G.; McNulty, B.A.; Nugent, A.P.; Walton, J.; Flynn, A.; Brennan, L. The relationship between fish intake and urinary trimethylamine-N-oxide. Mol. Nutr. Food Res. 2020, 64, e1900799. [Google Scholar] [CrossRef]
- Li, P.; Wu, G. Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids 2018, 50, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Salze, G.P.; Davis, D.A. Taurine: A critical nutrient for future fish feeds. Aquaculture 2015, 437, 215–229. [Google Scholar] [CrossRef]
- Rostika, R.; Iskandar, T.; Herawati, T.; Bangkit, I.; Banthani, G.; Dewanti, L.P. Growth of Sunu grouper (Plectropomus leopardus) Larvae That Given Rotivera (Bachionus rotundiformus) Enriched with taurine and glutamine. Proc. J. Phys. Conf. Ser. 2019, 1374, 012019. [Google Scholar] [CrossRef]
- Shen, G.; Wang, S.; Dong, J.; Feng, J.; Xu, J.; Xia, F.; Wang, X.; Ye, J. Metabolic effect of dietary taurine supplementation on grouper (Epinephelus coioides): A 1H-NMR-Based Metabolomics Study. Molecules 2019, 24, 2253. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Yin, B.; Liu, H.; Tan, B.; Dong, X.; Chi, S.; Yang, Q.; Zhang, S. Effects of taurine supplementation in a high-carbohydrate diet on growth performance, plasma biochemical, digestive and glucose metabolism enzymes in hybrid grouper (♀ Epinephelus fuscoguttatus × ♂ E. lanceolatus). Aquacult. Rep. 2021, 21, 100820. [Google Scholar] [CrossRef]
- Day, R.D.; German, D.P.; Manjakasy, J.M.; Farr, I.; Hansen, M.J.; Tibbetts, I.R. Enzymatic digestion in stomachless fishes: How a simple gut accommodates both herbivory and carnivory. J. Comp. Physiol. B 2011, 181, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Krogdahl, A.; Hemre, G.I.; Mommsen, T.P. Carbohydrates in fish nutrition: Digestion and absorption in postlarval stages. Aquacult. Nutr. 2005, 11, 103–122. [Google Scholar] [CrossRef]
- Fonville, J.M.; Maher, A.D.; Coen, M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Evaluation of full-resolution J-resolved 1H NMR projections of biofluids for metabonomics information retrieval and biomarker identification. Anal. Chem. 2010, 82, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, J.; Tsuboi, Y.; Komatsu, K.; Gomi, M.; Chikayama, E.; Date, Y. SpinCouple: Development of a web tool for analyzing metabolite mixtures via two-dimensional J-resolved NMR database. Anal. Chem. 2016, 88, 659–665. [Google Scholar] [CrossRef]
- Misawa, T.; Komatsu, T.; Date, Y.; Kikuchi, J. SENSI: Signal enhancement by spectral integration for the analysis of metabolic mixtures. Chem. Commun. 2016, 52, 2964–2967. [Google Scholar] [CrossRef]
- Chikayama, E.; Sekiyama, Y.; Okamoto, M.; Nakanishi, Y.; Tsuboi, Y.; Akiyama, K.; Saito, K.; Shinozaki, K.; Kikuchi, J. Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal. Chem. 2010, 82, 1653–1658. [Google Scholar] [CrossRef] [PubMed]
- Date, Y.; Kikuchi, J. Application of a deep neural network to metabolomics studies and its performance in determining important variables. Anal. Chem. 2018, 90, 1805–1810. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Sakata, K.; Date, Y.; Kikuchi, J. Regional feature extraction of various fishes based on chemical and microbial variable selection using machine learning. Anal. Methods 2018, 10, 2160–2168. [Google Scholar] [CrossRef]
- Komatsu, T.; Ohishi, R.; Shino, A.; Akashi, K.; Kikuchi, J. Multi-Spectroscopic Analysis of Seed Quality and 13C-Stable-Iotopologue Monitoring in Initial Growth Metabolism of Jatropha curcas L. Metabolites 2014, 4, 1018–1033. [Google Scholar] [CrossRef] [PubMed]
- Lewis, I.A.; Schommer, S.C.; Markley, J.L. rNMR: Open source software for identifying and quantifying metabolites in NMR spectra. Magn. Reson. Chem. 2009, 47 (Suppl. S1), S123–S126. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.G.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]
- Tikunov, Y.; Lommen, A.; de Vos, C.H.R.; Verhoeven, H.A.; Bino, R.J.; Hall, R.D.; Bovy, A.G. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 2005, 139, 1125–1137. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shima, H.; Asakura, T.; Sakata, K.; Koiso, M.; Kikuchi, J. Feed Components and Timing to Improve the Feed Conversion Ratio for Sustainable Aquaculture Using Starch. Int. J. Mol. Sci. 2024, 25, 7921. https://doi.org/10.3390/ijms25147921
Shima H, Asakura T, Sakata K, Koiso M, Kikuchi J. Feed Components and Timing to Improve the Feed Conversion Ratio for Sustainable Aquaculture Using Starch. International Journal of Molecular Sciences. 2024; 25(14):7921. https://doi.org/10.3390/ijms25147921
Chicago/Turabian StyleShima, Hideaki, Taiga Asakura, Kenji Sakata, Masahiko Koiso, and Jun Kikuchi. 2024. "Feed Components and Timing to Improve the Feed Conversion Ratio for Sustainable Aquaculture Using Starch" International Journal of Molecular Sciences 25, no. 14: 7921. https://doi.org/10.3390/ijms25147921




