G-Banding and Molecular Cytogenetics Detect Novel Translocations and Cryptic Aberrations in Human Immortal Endothelial Cells
Abstract
:1. Introduction
2. Results
2.1. G-Banding Revealed a Complex Karyogram for EA.hy926 Cells
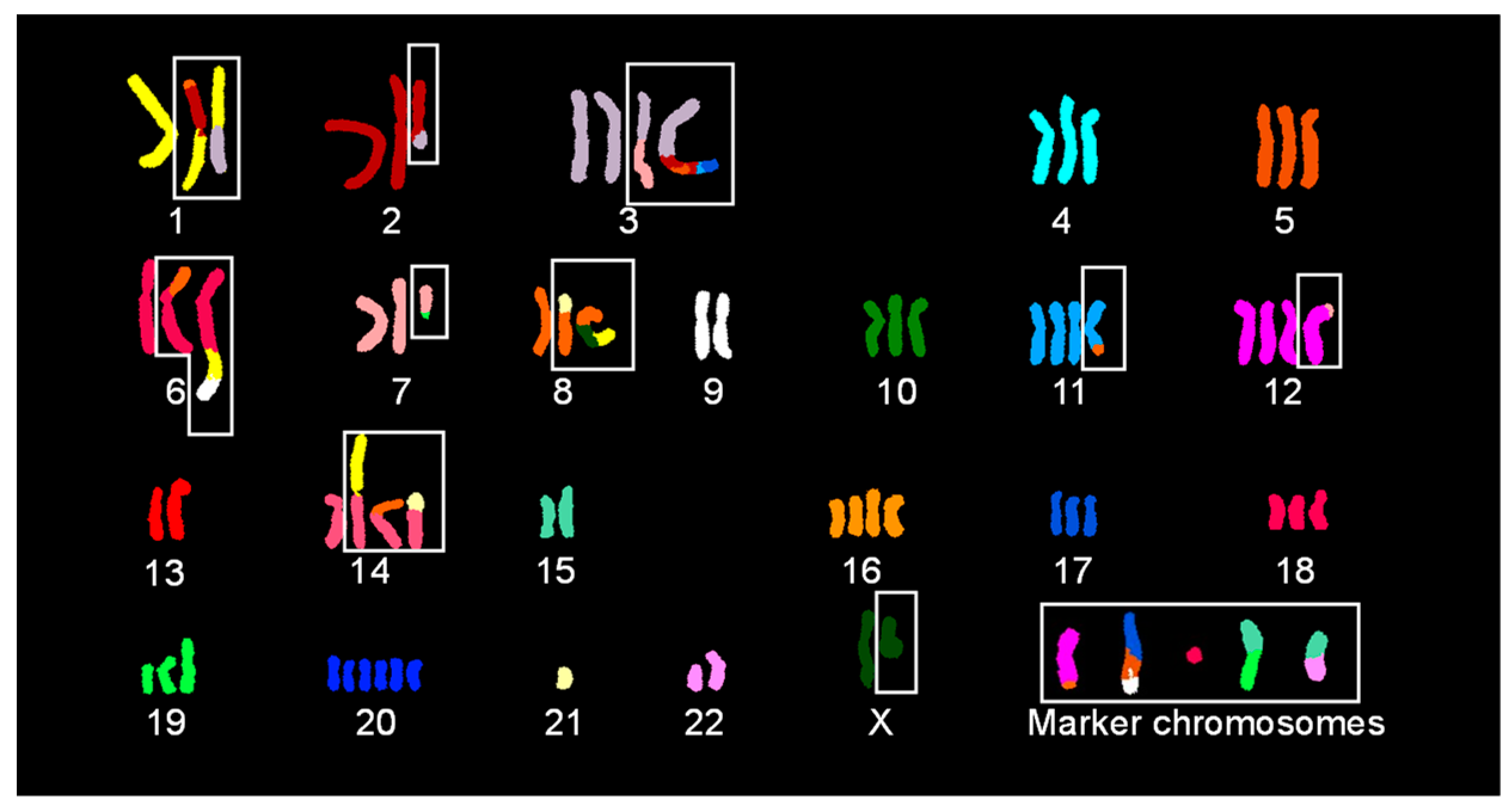
2.2. SKY Identified Simple and Complex Translocations and Revealed Unique Clones in EA.hy926 Cells
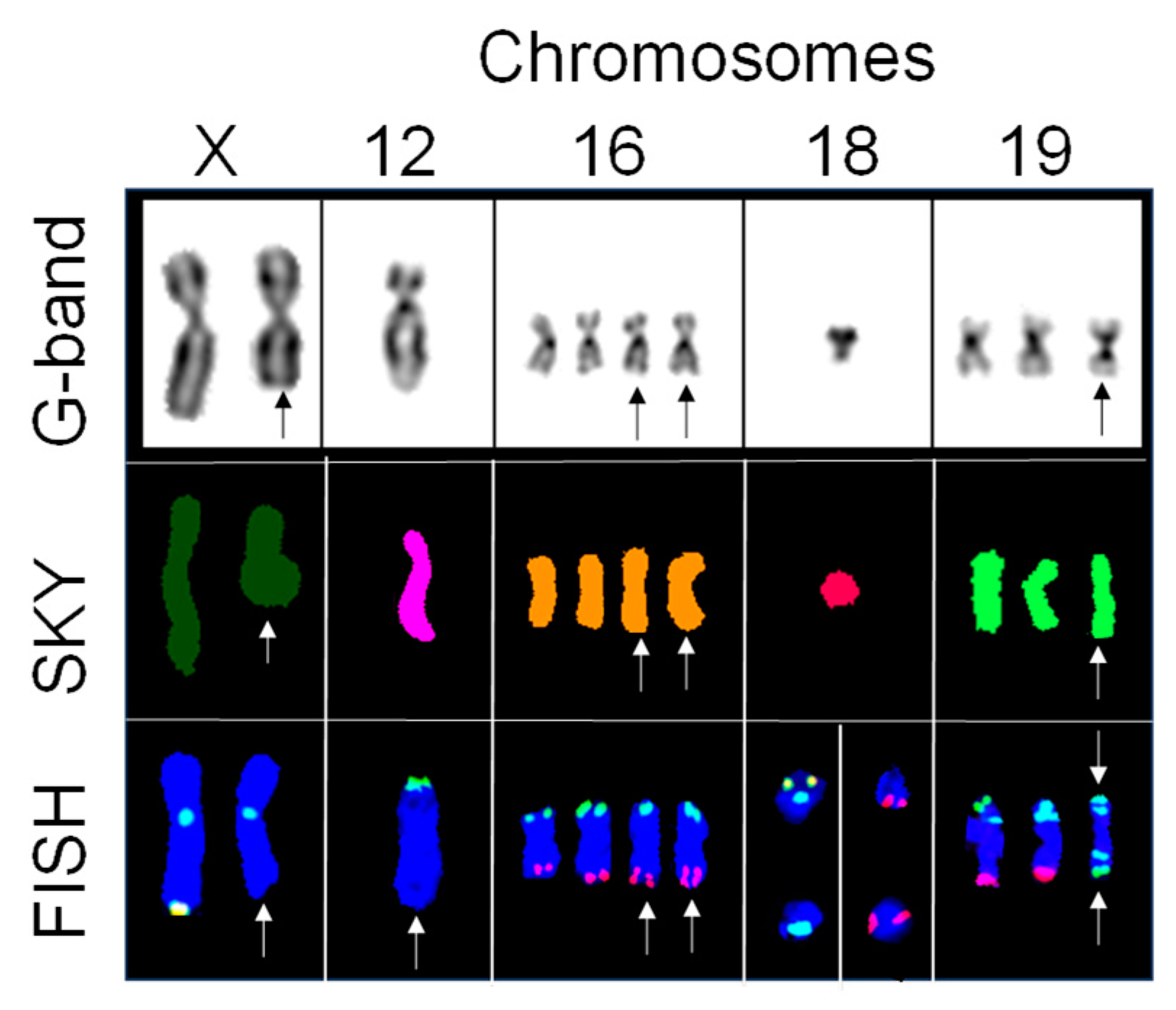
2.3. Subtelomere FISH Identified Cryptic Aberrations in Chromosomes with No Change in Spectral Emission
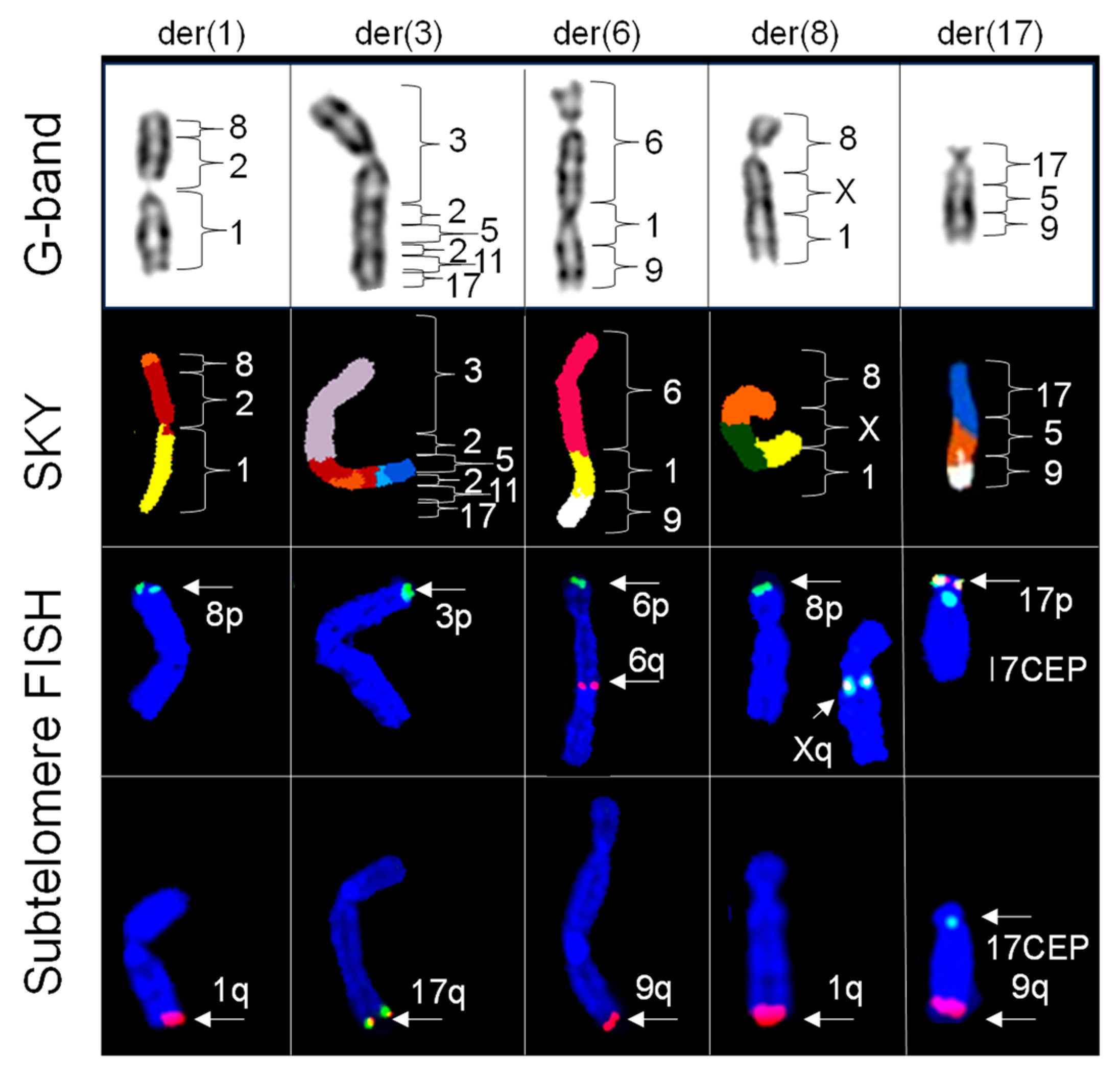
2.4. Subtelomere FISH Probes Defined the Origins of Telomeric Regions of Chromosomes Involved in Simple Translocations
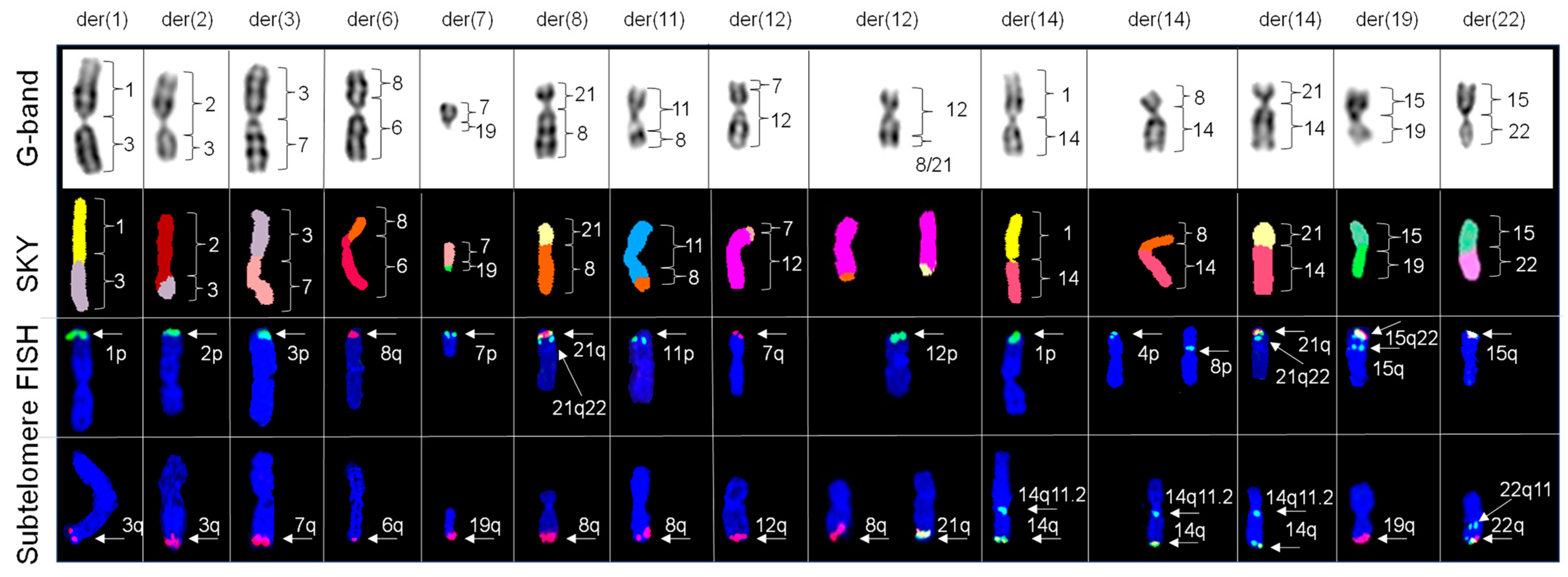
2.5. Subelomere FISH Defined the Origins of the Telomeric Regions of Chromosomes Involved in Complex Translocations
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Metaphase Chromosome Preparation
4.3. Conventional G-Banding
4.4. Spectral Karyotyping (SKY)
4.5. FISH Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deanfield, J.E.; Halcox, J.P.; Rabelink, T.J. Endothelial function and dysfunction: Testing and clinical relevance. Circulation 2007, 115, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Higashi, Y. Beneficial Effects of Supplementation on Endothelial Function: What Mechanisms? J. Atheroscler. Thromb. 2022, 29, 1268–1270. [Google Scholar] [CrossRef]
- Lerman, A.; Zeiher, A.M. Endothelial function: Cardiac events. Circulation 2005, 111, 363–368. [Google Scholar] [CrossRef]
- Rabiet, M.J.; Plantier, J.L.; Rival, Y.; Genoux, Y.; Lampugnani, M.G.; Dejana, E. Thrombin-induced increase in endothelial permeability is associated with changes in cell-to-cell junction organization. Arterioscler. Thromb. Vasc. Biol. 1996, 16, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.; Pan, S.; Beningo, K.; Hupe, D. A permanent human cell line (EA.hy926) preserves the characteristics of endothelin converting enzyme from primary human umbilical vein endothelial cells. Life Sci. 1995, 56, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Pei-Yuan, Z.; Yu-Wei, L.; Xiang-Nan, Z.; Song, T.; Rong, Z.; Xiao-Xiao, H.; Sheng-Shuai, S.; Kun, W.; Cheng-Yun, L. Overexpression of Axl reverses endothelial cells dysfunction in high glucose and hypoxia. J. Cell. Biochem. 2019, 120, 11831–11841. [Google Scholar] [CrossRef] [PubMed]
- Abdelgawad, I.Y.; Agostinucci, K.; Ismail, S.G.; Grant, M.K.O.; Zordoky, B.N. EA.hy926 Cells and HUVECs Share Similar Senescence Phenotypes but Respond Differently to the Senolytic Drug ABT-263. Cells 2022, 11i, 1992. [Google Scholar] [CrossRef]
- Lupu, C.; Lupu, F.; Dennehy, U.; Kakkar, V.V.; Scully, M.F. Thrombin induces the redistribution and acute release of tissue factor pathway inhibitor from specific granules within human endothelial cells in culture. Arterioscler. Thromb. Vasc. Biol. 1995, 15, 2055–2062. [Google Scholar] [CrossRef]
- Binz, R.L.; Pathak, R. Molecular Cytogenetics Reveals Mosaicism in Human Umbilical Vein Endothelial Cells. Genes 2022, 13, 1012. [Google Scholar] [CrossRef] [PubMed]
- Uruski, P.; Mikuła-Pietrasik, J.; Drzewiecki, M.; Budkiewicz, S.; Gładki, M.; Kurmanalina, G.; Tykarski, A.; Książek, K. Diverse functional responses to high glucose by primary and permanent hybrid endothelial cells in vitro. J. Mol. Cell. Cardiol. 2021, 156, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Edgell, C.J.; McDonald, C.C.; Graham, J.B. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. USA 1983, 80, 3734–3737. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.-J.; Wang, J.-H.; Su, W.-T.; Wang, X.-C.; Yang, F.-T.; Nie, W.-H. Characterization of two human lung adenocarcinoma cell lines by reciprocal chromosome painting. Dongwuxue Yanjiu 2010, 31, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Pfau, S.J.; Amon, A. Chromosomal instability and aneuploidy in cancer: From yeast to man. EMBO Rep. 2012, 13, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Compton, D.A. Chromosomal instability and cancer: A complex relationship with therapeutic potential. J. Clin. Investig. 2012, 122, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.J.; Ren, Y.Q.; Wang, G.P.; Song, Q.; Li, M.; Jiang, S.S.; Ning, T.; Guan, Y.S.; Yang, J.L.; Luo, F. Biological behaviors and proteomics analysis of hybrid cell line EAhy926 and its parent cell line A549. J. Exp. Clin. Cancer Res. 2009, 28, 16. [Google Scholar] [CrossRef]
- Binz, R.L.; Burns, K.; Pathak, R. Protocol for preparation and staining of chromosomes isolated from mouse and human tissues for conventional and molecular cytogenetic analysis. STAR Protoc. 2024, 5, 102897. [Google Scholar] [CrossRef] [PubMed]
- Binz, R.L.; Tian, E.; Sadhukhan, R.; Zhou, D.; Hauer-Jensen, M.; Pathak, R. Identification of novel breakpoints for locus- and region-specific translocations in 293 cells by molecular cytogenetics before and after irradiation. Sci. Rep. 2019, 9, 10554. [Google Scholar] [CrossRef] [PubMed]
- Binz, R.L.; Sadhukhan, R.; Miousse, I.R.; Garg, S.; Koturbash, I.; Zhou, D.; Hauer-Jensen, M.; Pathak, R. Dietary Methionine Deficiency Enhances Genetic Instability in Murine Immune Cells. Int. J. Mol. Sci. 2021, 22, 2378. [Google Scholar] [CrossRef] [PubMed]
- McGowan-Jordan, J.; Ros, J. Hastings; Sarah Moore. In ISCN 2020: An International System for Human Cytogenomic Nomenclature (2020); S. Karger AG: Basel, Switzerland, 2020; ISBN 978-3-318-06706-4. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Binz, R.L.; Pathak, R. G-Banding and Molecular Cytogenetics Detect Novel Translocations and Cryptic Aberrations in Human Immortal Endothelial Cells. Int. J. Mol. Sci. 2024, 25, 7941. https://doi.org/10.3390/ijms25147941
Binz RL, Pathak R. G-Banding and Molecular Cytogenetics Detect Novel Translocations and Cryptic Aberrations in Human Immortal Endothelial Cells. International Journal of Molecular Sciences. 2024; 25(14):7941. https://doi.org/10.3390/ijms25147941
Chicago/Turabian StyleBinz, Regina Lichti, and Rupak Pathak. 2024. "G-Banding and Molecular Cytogenetics Detect Novel Translocations and Cryptic Aberrations in Human Immortal Endothelial Cells" International Journal of Molecular Sciences 25, no. 14: 7941. https://doi.org/10.3390/ijms25147941





