Transcriptome Analysis of Mesenchymal Progenitor Cells Revealed Molecular Insights into Metabolic Dysfunction and Inflammation in Polycystic Ovary Syndrome
Abstract
:1. Introduction
2. Results
2.1. iPSC Establishment and MPC Derivation
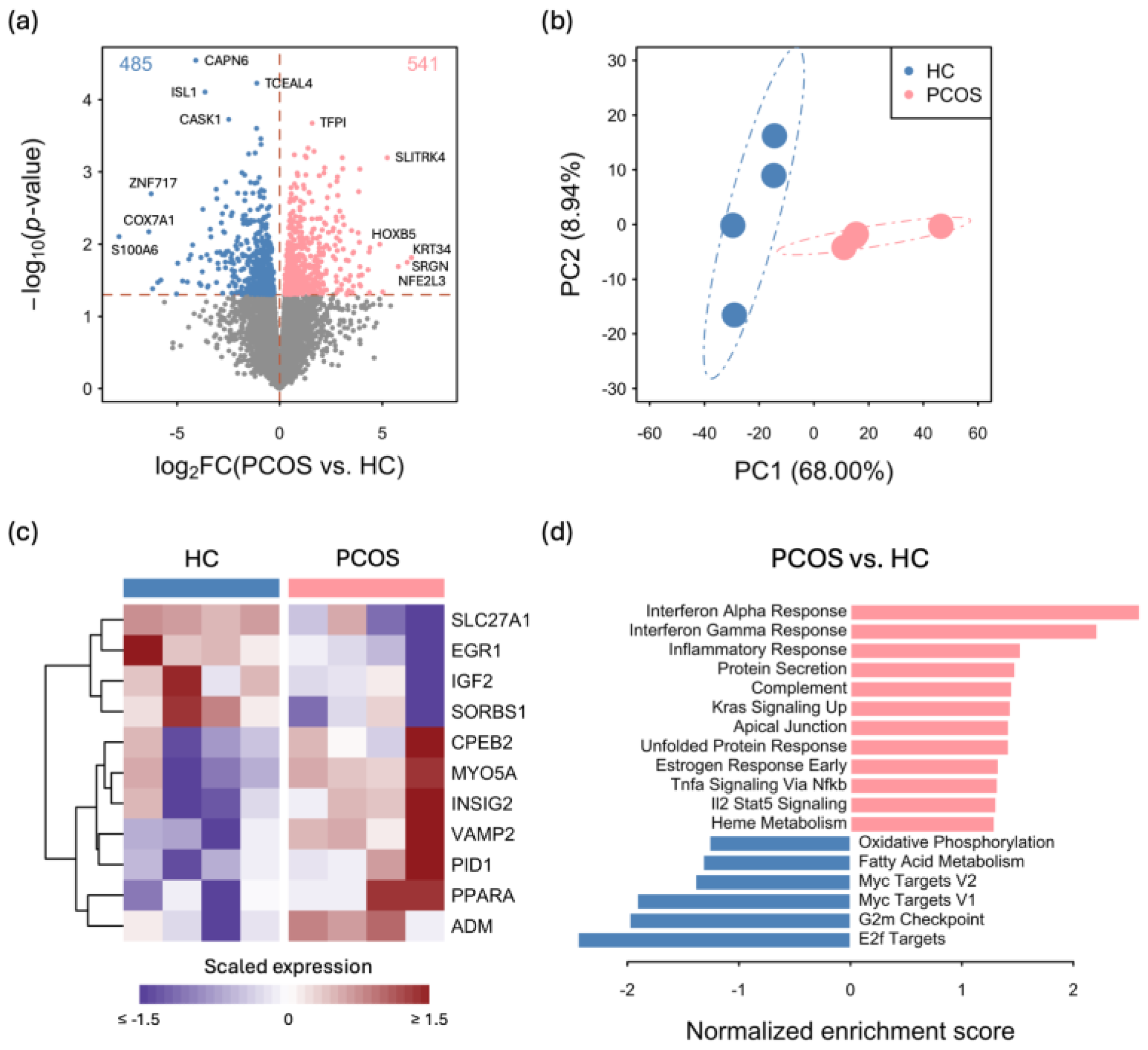
2.2. Transcriptome Differences between iPSCMs from PCOS Patients and Those from Healthy Controls
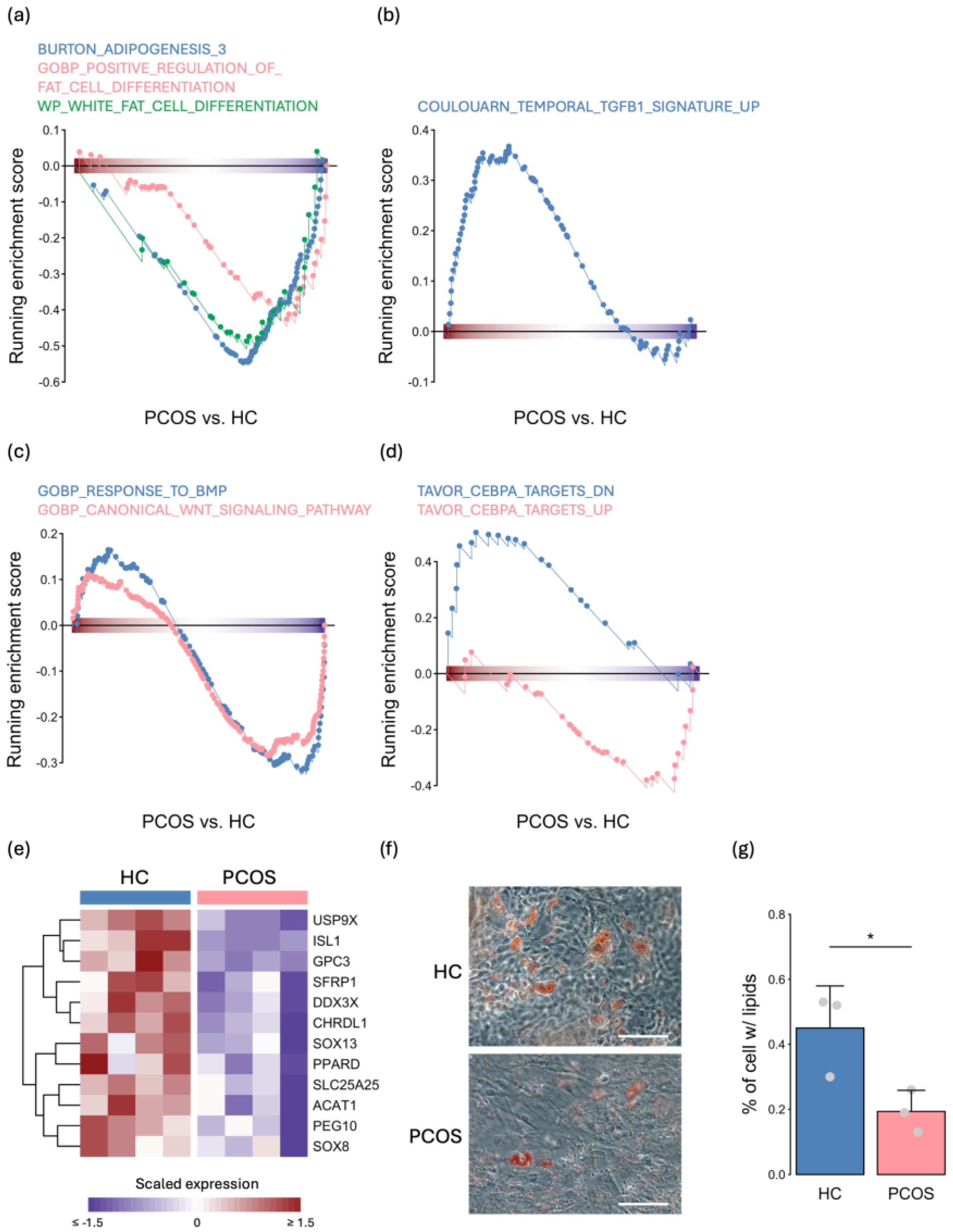
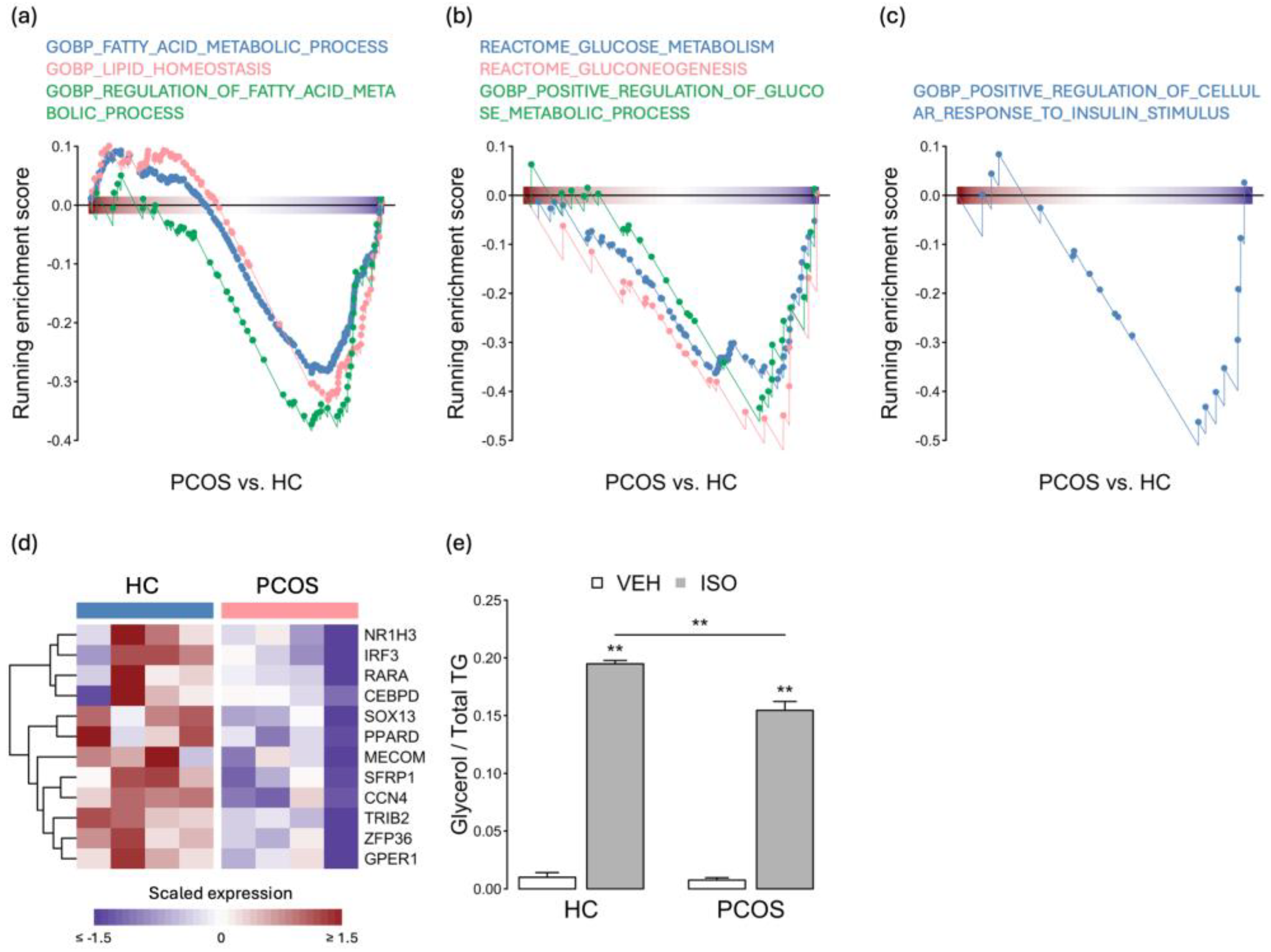
2.3. The Expression Levels of Adipogenesis- and Metabolism-Related Genes Were Decreased in PCOS iPSCMs
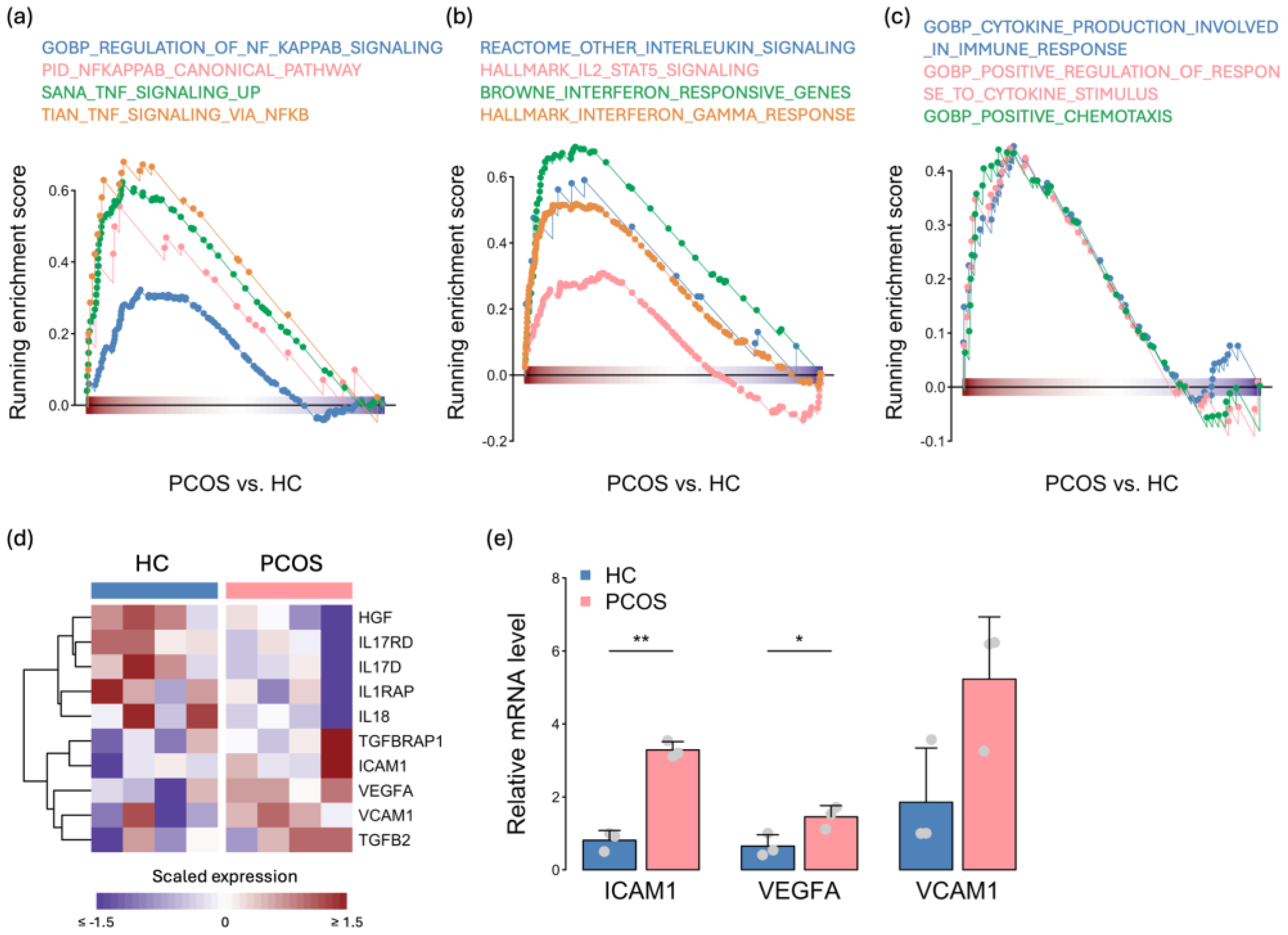
2.4. Activation of the Immune Response in PCOS iPSCMs
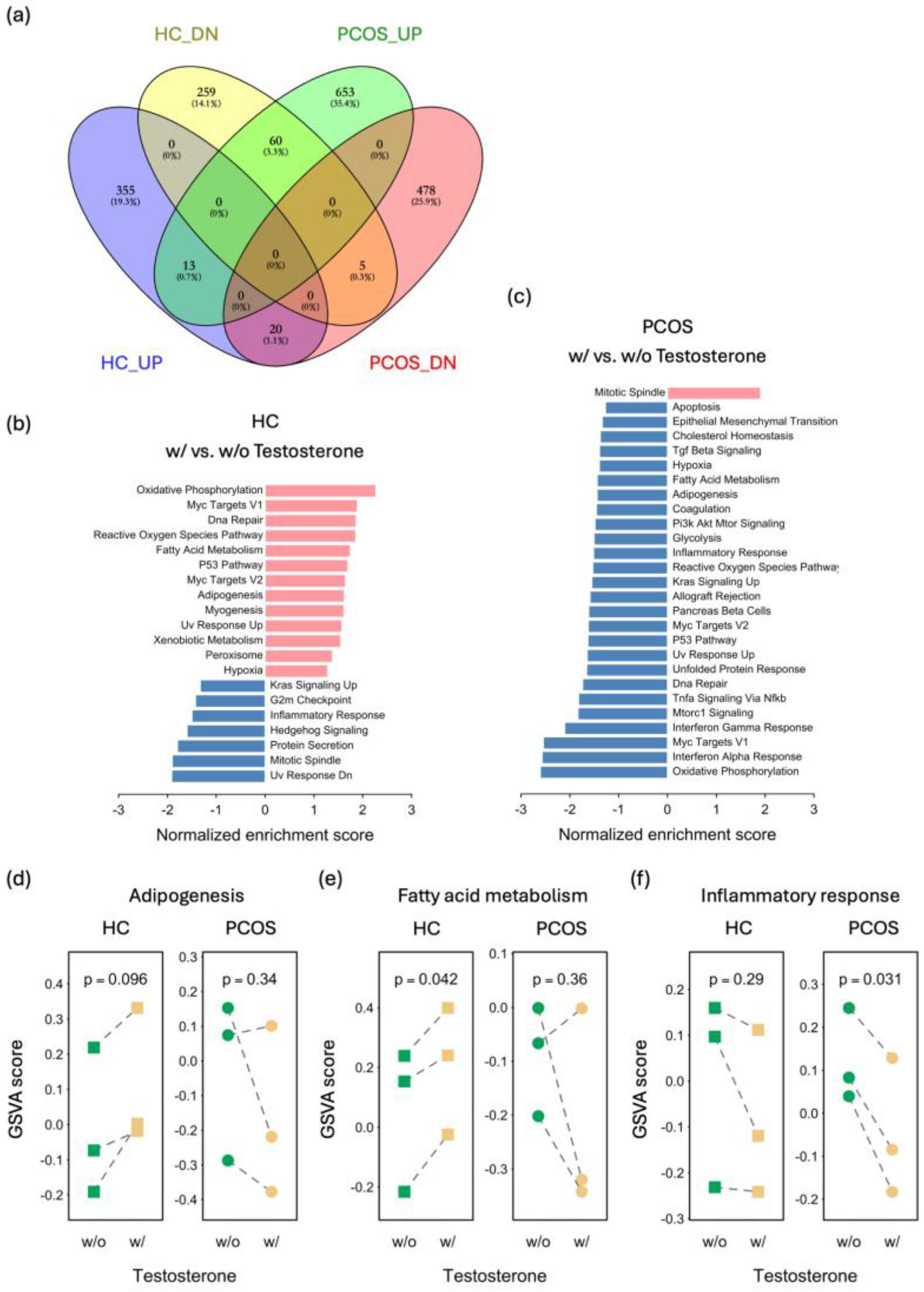
2.5. Distinct Effects of Testosterone Treatment on PCOS and HC iPSCMs
3. Discussion
4. Materials and Methods
4.1. iPSC Derivation and Maintenance
4.2. MPC Differentiation and Testosterone Treatment
4.3. Teratoma Formation
4.4. Staining for Alkaline Phosphatase Activity and Immunocytochemistry
4.5. Induction of Specific Lineages (Adipocytes, Osteoblasts, and Chondrocytes), Oil Red O Staining, Alizarin Red S Staining, and Alcian Blue Staining
4.6. RNA Sequencing and Data Analysis
4.7. Flow Cytometry Analysis
4.8. Lipolysis Assay and Triglyceride Quantification
4.9. Real-Time Quantitative PCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ehrmann, D.A. Polycystic ovary syndrome. N. Engl. J. Med. 2005, 352, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Homburg, R. Polycystic ovary syndrome. Best Pract. Res. Clin. Obstet. Gynaecol. 2008, 22, 261–274. [Google Scholar] [CrossRef]
- Rotterdam ESHRE/ASRM Sponsored PCOS consensus workshop group: Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum. Reprod. 2004, 19, 41–47. [CrossRef] [PubMed]
- Goodarzi, M.O.; Dumesic, D.A.; Chazenbalk, G.; Azziz, R. Polycystic ovary syndrome: Etiology, pathogenesis and diagnosis. Nat. Rev. Endocrinol. 2011, 7, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Vélez, L.M.; Motta, A.B. Association between polycystic ovary syndrome and metabolic syndrome. Curr. Med. Chem. 2014, 21, 3999–4012. [Google Scholar] [CrossRef] [PubMed]
- Kosova, G.; Urbanek, M. Genetics of the polycystic ovary syndrome. Mol. Cell. Endocrinol. 2013, 373, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Rababa’h, A.M.; Matani, B.R.; Yehya, A. An update of polycystic ovary syndrome: Causes and therapeutics options. Heliyon 2022, 8, e11010. [Google Scholar] [CrossRef]
- Barber, T.M.; McCarthy, M.I.; Wass, J.A.; Franks, S. Obesity and polycystic ovary syndrome. Clin. Endocrinol. 2006, 65, 137–145. [Google Scholar] [CrossRef]
- Dapas, M.; Dunaif, A. Deconstructing a Syndrome: Genomic Insights Into PCOS Causal Mechanisms and Classification. Endocr. Rev. 2022, 43, 927–965. [Google Scholar] [CrossRef] [PubMed]
- Novosadova, E.V.; Arsenyeva, E.L.; Antonov, S.A.; Vanyushina, Y.N.; Malova, T.V.; Komissarov, A.A.; Illarioshkin, S.N.; Khaspekov, L.G.; Andreeva, L.A.; Myasoedov, N.F.; et al. The Use of Human Induced Pluripotent Stem Cells for Testing Neuroprotective Activity of Pharmacological Compounds. Biochemistry 2019, 84, 1296–1305. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Corbett, J.L.; Duncan, S.A. iPSC-Derived Hepatocytes as a Platform for Disease Modeling and Drug Discovery. Front. Med. 2019, 6, 265. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shi, Y. Mesenchymal stem/stromal cells (MSCs): Origin, immune regulation, and clinical applications. Cell. Mol. Immunol. 2023, 20, 555–557. [Google Scholar] [CrossRef]
- Jiang, W.; Xu, J. Immune modulation by mesenchymal stem cells. Cell Prolif. 2020, 53, e12712. [Google Scholar] [CrossRef]
- Song, N.; Scholtemeijer, M.; Shah, K. Mesenchymal Stem Cell Immunomodulation: Mechanisms and Therapeutic Potential. Trends Pharmacol. Sci. 2020, 41, 653–664. [Google Scholar] [CrossRef]
- Weiss, A.R.R.; Dahlke, M.H. Immunomodulation by Mesenchymal Stem Cells (MSCs): Mechanisms of Action of Living, Apoptotic, and Dead MSCs. Front. Immunol. 2019, 10, 1191. [Google Scholar] [CrossRef]
- Wu, Q.; Ortegon, A.M.; Tsang, B.; Doege, H.; Feingold, K.R.; Stahl, A. FATP1 is an insulin-sensitive fatty acid transporter involved in diet-induced obesity. Mol. Cell. Biol. 2006, 26, 3455–3467. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell. Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Diamanti-Kandarakis, E.; Paterakis, T.; Alexandraki, K.; Piperi, C.; Aessopos, A.; Katsikis, I.; Katsilambros, N.; Kreatsas, G.; Panidis, D. Indices of low-grade chronic inflammation in polycystic ovary syndrome and the beneficial effect of metformin. Hum. Reprod. 2006, 21, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Victor, V.M.; Rovira-Llopis, S.; Banuls, C.; Diaz-Morales, N.; Martinez de Maranon, A.; Rios-Navarro, C.; Alvarez, A.; Gomez, M.; Rocha, M.; Hernandez-Mijares, A. Insulin Resistance in PCOS Patients Enhances Oxidative Stress and Leukocyte Adhesion: Role of Myeloperoxidase. PLoS ONE 2016, 11, e0151960. [Google Scholar] [CrossRef]
- Echiburu, B.; Perez-Bravo, F.; Galgani, J.E.; Sandoval, D.; Saldias, C.; Crisosto, N.; Maliqueo, M.; Sir-Petermann, T. Enlarged adipocytes in subcutaneous adipose tissue associated to hyperandrogenism and visceral adipose tissue volume in women with polycystic ovary syndrome. Steroids 2018, 130, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Manneras-Holm, L.; Leonhardt, H.; Kullberg, J.; Jennische, E.; Oden, A.; Holm, G.; Hellstrom, M.; Lonn, L.; Olivecrona, G.; Stener-Victorin, E.; et al. Adipose tissue has aberrant morphology and function in PCOS: Enlarged adipocytes and low serum adiponectin, but not circulating sex steroids, are strongly associated with insulin resistance. J. Clin. Endocrinol. Metab. 2011, 96, E304–E311. [Google Scholar] [CrossRef]
- Dumesic, D.A.; Akopians, A.L.; Madrigal, V.K.; Ramirez, E.; Margolis, D.J.; Sarma, M.K.; Thomas, A.M.; Grogan, T.R.; Haykal, R.; Schooler, T.A.; et al. Hyperandrogenism Accompanies Increased Intra-Abdominal Fat Storage in Normal Weight Polycystic Ovary Syndrome Women. J. Clin. Endocrinol. Metab. 2016, 101, 4178–4188. [Google Scholar] [CrossRef] [PubMed]
- Chazenbalk, G.; Chen, Y.H.; Heneidi, S.; Lee, J.M.; Pall, M.; Chen, Y.D.; Azziz, R. Abnormal expression of genes involved in inflammation, lipid metabolism, and Wnt signaling in the adipose tissue of polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E765–E770. [Google Scholar] [CrossRef]
- Keller, E.; Chazenbalk, G.D.; Aguilera, P.; Madrigal, V.; Grogan, T.; Elashoff, D.; Dumesic, D.A.; Abbott, D.H. Impaired preadipocyte differentiation into adipocytes in subcutaneous abdominal adipose of PCOS-like female rhesus monkeys. Endocrinology 2014, 155, 2696–2703. [Google Scholar] [CrossRef] [PubMed]
- Chazenbalk, G.; Singh, P.; Irge, D.; Shah, A.; Abbott, D.H.; Dumesic, D.A. Androgens inhibit adipogenesis during human adipose stem cell commitment to preadipocyte formation. Steroids 2013, 78, 920–926. [Google Scholar] [CrossRef]
- Glenske, K.; Schuler, G.; Arnhold, S.; Elashry, M.I.; Wagner, A.S.; Barbeck, M.; Neumann, E.; Muller-Ladner, U.; Schnettler, R.; Wenisch, S. Effects of testosterone and 17beta-estradiol on osteogenic and adipogenic differentiation capacity of human bone-derived mesenchymal stromal cells of postmenopausal women. Bone Rep. 2019, 11, 100226. [Google Scholar] [CrossRef]
- Gupta, V.; Bhasin, S.; Guo, W.; Singh, R.; Miki, R.; Chauhan, P.; Choong, K.; Tchkonia, T.; Lebrasseur, N.K.; Flanagan, J.N.; et al. Effects of dihydrotestosterone on differentiation and proliferation of human mesenchymal stem cells and preadipocytes. Mol. Cell. Endocrinol. 2008, 296, 32–40. [Google Scholar] [CrossRef]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell. Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef]
- Orisaka, M.; Mizutani, T.; Miyazaki, Y.; Shirafuji, A.; Tamamura, C.; Fujita, M.; Tsuyoshi, H.; Yoshida, Y. Chronic low-grade inflammation and ovarian dysfunction in women with polycystic ovarian syndrome, endometriosis, and aging. Front Endocrinol. 2023, 14, 1324429. [Google Scholar] [CrossRef]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell. Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.; Liu, Z.; Ren, Y.A.; Wun, W.S.; Zhou, W.; Kenigsberg, S.; Librach, C.; Valdes, C.; Gibbons, W.; Richards, J. Enhanced Inflammatory Transcriptome in the Granulosa Cells of Women With Polycystic Ovarian Syndrome. J. Clin. Endocrinol. Metab. 2016, 101, 3459–3468. [Google Scholar] [CrossRef]
- Piltonen, T.T.; Chen, J.; Erikson, D.W.; Spitzer, T.L.; Barragan, F.; Rabban, J.T.; Huddleston, H.; Irwin, J.C.; Giudice, L.C. Mesenchymal stem/progenitors and other endometrial cell types from women with polycystic ovary syndrome (PCOS) display inflammatory and oncogenic potential. J. Clin. Endocrinol. Metab. 2013, 98, 3765–3775. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Morreale, H.F.; Luque-Ramirez, M.; Gonzalez, F. Circulating inflammatory markers in polycystic ovary syndrome: A systematic review and metaanalysis. Fertil. Steril. 2011, 95, 1048–1058 e1041–1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ruan, Y.C.; Yang, Y.J.; Wang, K.; Liang, S.S.; Han, Y.B.; Teng, X.M.; Yang, J.Z. Follicular hyperandrogenism downregulates aromatase in luteinized granulosa cells in polycystic ovary syndrome women. Reproduction 2015, 150, 289–296. [Google Scholar] [CrossRef]
- Schulte, D.M.; Muller, N.; Neumann, K.; Oberhauser, F.; Faust, M.; Gudelhofer, H.; Brandt, B.; Krone, W.; Laudes, M. Pro-inflammatory wnt5a and anti-inflammatory sFRP5 are differentially regulated by nutritional factors in obese human subjects. PLoS ONE 2012, 7, e32437. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Huang, Y.; Yu, Y.; Li, R.; Li, M.; Liu, N.; Liu, P.; Qiao, J. Up-regulated expression of WNT5a increases inflammation and oxidative stress via PI3K/AKT/NF-kappaB signaling in the granulosa cells of PCOS patients. J. Clin. Endocrinol. Metab. 2015, 100, 201–211. [Google Scholar] [CrossRef]
- Trigunaite, A.; Dimo, J.; Jorgensen, T.N. Suppressive effects of androgens on the immune system. Cell. Immunol. 2015, 294, 87–94. [Google Scholar] [CrossRef]
- Wang, J.; Yin, T.; Liu, S. Dysregulation of immune response in PCOS organ system. Front Immunol. 2023, 14, 1169232. [Google Scholar] [CrossRef]
- Kuppusamy, P.; Kim, D.; Soundharrajan, I.; Hwang, I.; Choi, K.C. Adipose and Muscle Cell Co-Culture System: A Novel In Vitro Tool to Mimic the In Vivo Cellular Environment. Biology 2020, 10, 6. [Google Scholar] [CrossRef]
- Venter, C.; Niesler, C. A triple co-culture method to investigate the effect of macrophages and fibroblasts on myoblast proliferation and migration. Biotechniques 2018, 64, 52–58. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.-C.; Chen, P.-L.; Hsu, C.-L. Transcriptome Analysis of Mesenchymal Progenitor Cells Revealed Molecular Insights into Metabolic Dysfunction and Inflammation in Polycystic Ovary Syndrome. Int. J. Mol. Sci. 2024, 25, 7948. https://doi.org/10.3390/ijms25147948
Huang M-C, Chen P-L, Hsu C-L. Transcriptome Analysis of Mesenchymal Progenitor Cells Revealed Molecular Insights into Metabolic Dysfunction and Inflammation in Polycystic Ovary Syndrome. International Journal of Molecular Sciences. 2024; 25(14):7948. https://doi.org/10.3390/ijms25147948
Chicago/Turabian StyleHuang, Mei-Chi, Pei-Lung Chen, and Chia-Lang Hsu. 2024. "Transcriptome Analysis of Mesenchymal Progenitor Cells Revealed Molecular Insights into Metabolic Dysfunction and Inflammation in Polycystic Ovary Syndrome" International Journal of Molecular Sciences 25, no. 14: 7948. https://doi.org/10.3390/ijms25147948




