Long-Term In Vitro Culture Alters Gene Expression Pattern of Genes Involved in Ontological Groups Representing Cellular Processes
Abstract
1. Introduction
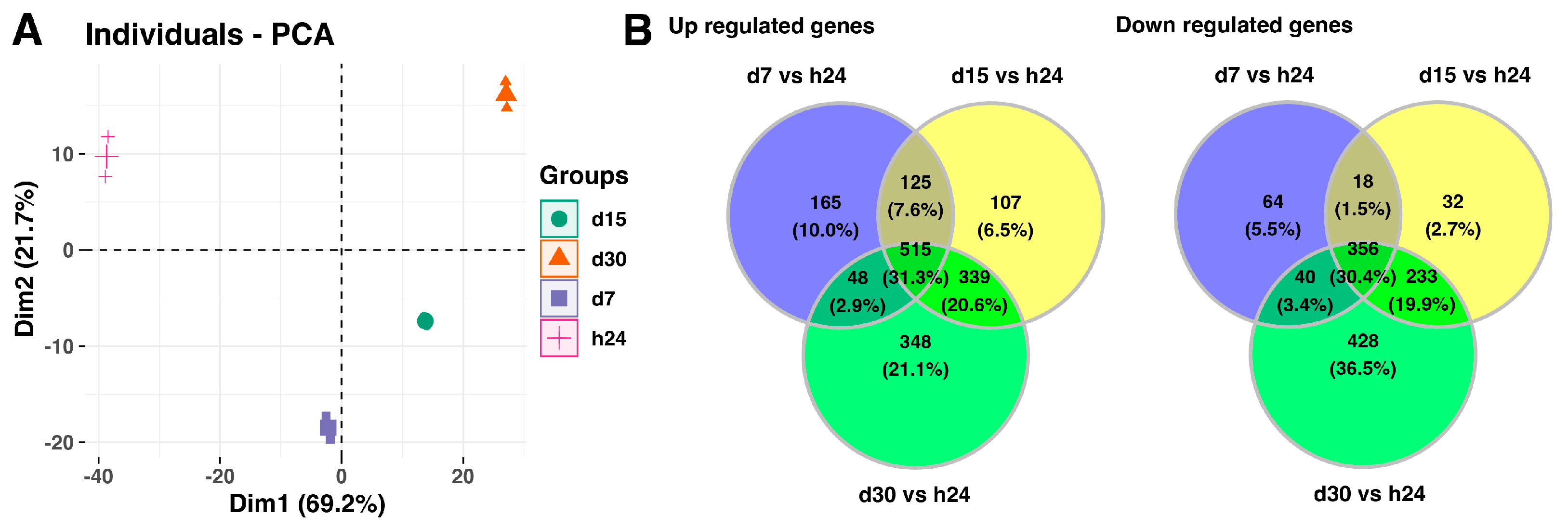
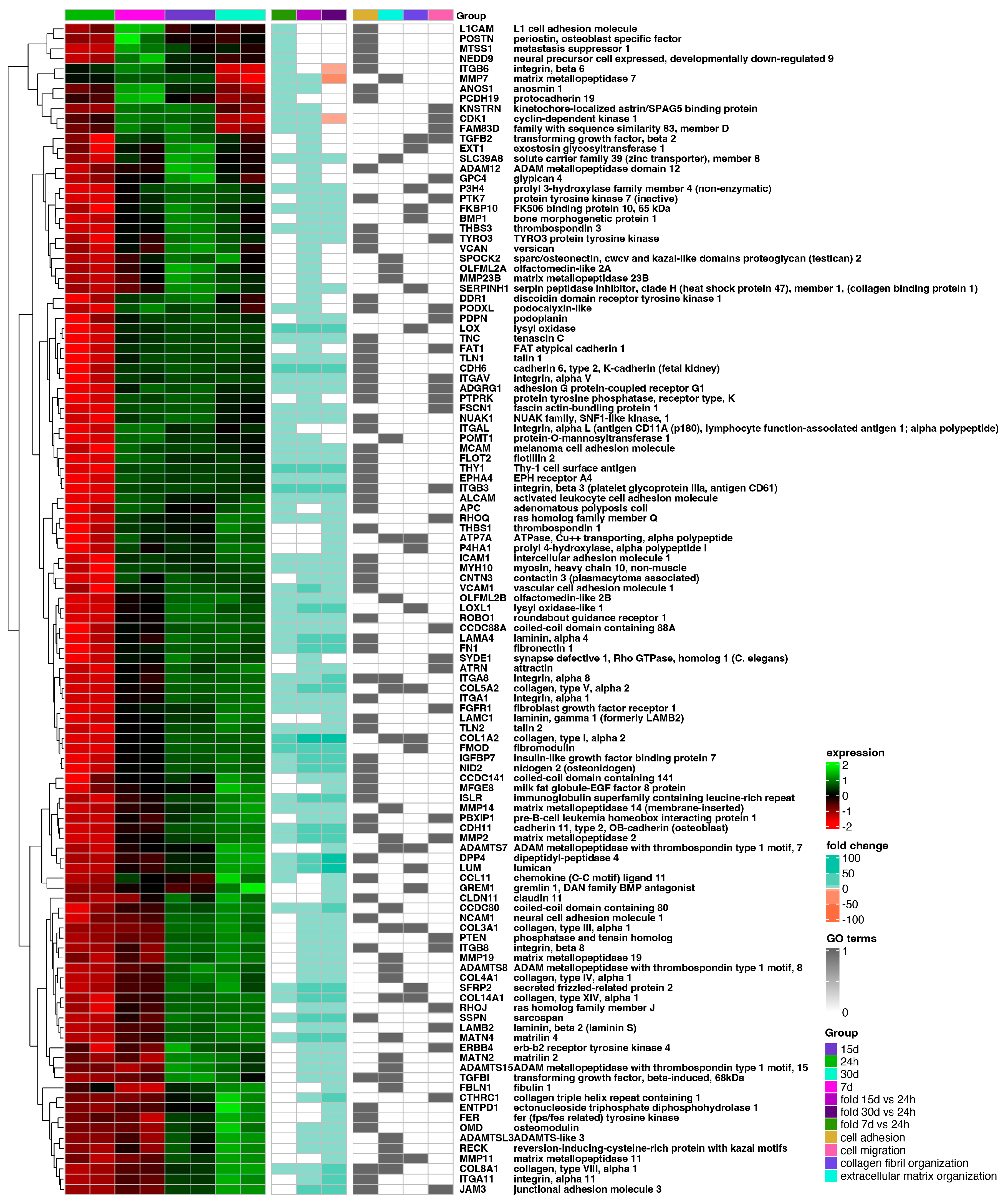
2. Results
3. Discussion
4. Materials and Methods
4.1. Tissue Collection
4.2. Primary Long-Term Cell Culture of Porcine Oviductal Epithelial Cells (POECs)
4.3. RNA Isolation from Porcine Oviductal Epithelial Cells (POECs)
4.4. Microarray Expression Study
4.5. Microarray Data Analysis
4.6. mRNA Co-Expression Analysis—Clustering of mRNA Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Avilés, M.; Coy, P.; Rizos, D. The oviduct: A key organ for the success of early reproductive events. Anim. Front. 2015, 5, 25–31. [Google Scholar] [CrossRef]
- Rybska, M.; Knap, S.; Jankowski, M.; Jeseta, M.; Bukowska, D.; Antosik, P.; Nowicki, M.; Zabel, M.; Kempisty, B.; Jaśkowski, J.M. Characteristic of factors influencing the proper course of folliculogenesis in mammals. Med. J. Cell Biol. 2018, 6, 33–38. [Google Scholar] [CrossRef]
- Kölle, S.; Hughes, B.; Steele, H. Early embryo-maternal communication in the oviduct: A review. Mol. Reprod. Dev. 2020, 87, 650–662. [Google Scholar] [CrossRef]
- Kölle, S.; Reese, S.; Kummer, W. New aspects of gamete transport, fertilization, and embryonic development in the oviduct gained by means of live cell imaging. Theriogenology 2010, 73, 786–795. [Google Scholar] [CrossRef]
- Li, S.; Winuthayanon, W. Oviduct: Roles in fertilization and early embryo development. J. Endocrinol. 2017, 232, R1–R26. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Shin, J.; Jang, J.; Hwang, S.; Kim, J.; Kong, J.; Yang, H. 17Beta-Estradiol Regulates NUCB2/ Nesfatin-1 Expression in MouseOviduct. Dev. Reprod. 2020, 24, 43. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Palma-Vera, S.E.; Kempisty, B.; Rucinski, M.; Vernunft, A.; Schoen, J. In Vitro Mimicking of Estrous Cycle Stages: Dissecting the Impact of Estradiol and Progesterone on Oviduct Epithelium. Endocrinology 2018, 159, 3421–3432. [Google Scholar] [CrossRef]
- Barton, B.E.; Herrera, G.G.; Anamthathmakula, P.; Rock, J.K.; Willie, A.M.; Harris, E.A.; Takemaru, K.I.; Winuthayanon, W. Roles of steroid hormones in oviductal function. Reproduction 2020, 159, R125. [Google Scholar] [CrossRef]
- Wu, E.; Vastenhouw, N.L. From mother to embryo: A molecular perspective on zygotic genome activation. Curr. Top. Dev. Biol. 2020, 140, 209–254. [Google Scholar] [CrossRef]
- Duranthon, V.; Watson, A.J.; Lonergan, P. Preimplantation embryo programming: Transcription, epigenetics, and culture environment. Reproduction 2008, 135, 141–150. [Google Scholar] [CrossRef]
- Leese, H.J.; Hugentobler, S.A.; Gray, S.M.; Morris, D.G.; Sturmey, R.G.; Whitear, S.L.; Sreenan, J.M. Female reproductive tract fluids: Composition, mechanism of formation and potential role in the developmental origins of health and disease. Reprod. Fertil. Dev. 2008, 20, 1–8. [Google Scholar] [CrossRef]
- Bauersachs, S.; Wolf, E. Transcriptome analyses of bovine, porcine and equine endometrium during the pre-implantation phase. Anim. Reprod. Sci. 2012, 134, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Kulus, M.; Kranc, W.; Wojtanowicz-markiewicz, K.; Celichowski, P.; Światły-Błaszkiewicz, A.; Matuszewska, E.; Sujka-kordowska, P.; Konwerska, A.; Zdun, M.; Bryl, R.; et al. New Gene Markers Expressed in Porcine Oviductal Epithelial Cells Cultured Primary In Vitro Are Involved in Ontological Groups Representing Physiological Processes of Porcine Oocytes. Int. J. Mol. Sci. 2021, 22, 2082. [Google Scholar] [CrossRef]
- Hasan, M.M.; Viil, J.; Lättekivi, F.; Ord, J.; Reshi, Q.U.A.; Jääger, K.; Velthut-Meikas, A.; Andronowska, A.; Jaakma, Ü.; Salumets, A.; et al. Bovine Follicular Fluid and Extracellular Vesicles Derived from Follicular Fluid Alter the Bovine Oviductal Epithelial Cells Transcriptome. Int. J. Mol. Sci. 2020, 21, 5365. [Google Scholar] [CrossRef] [PubMed]
- Danesh Mesgaran, S.; Sharbati, J.; Einspanier, R.; Gabler, C. mRNA expression pattern of selected candidate genes differs in bovine oviductal epithelial cells in vitro compared with the in vivo state and during cell culture passages. Reprod. Biol. Endocrinol. 2016, 14, 44. [Google Scholar] [CrossRef]
- Kulus, M.; Józkowiak, M.; Kulus, J.; Popis, M.; Borowiec, B.; Stefańska, K.; Celichowski, P.; Nawrocki, M.J.; Bukowska, D.; Brüssow, K.P.; et al. “Cell cycle process”, “cell division” and “cell proliferation” belong to ontology groups highly regulated during long–term culture of porcine oviductal epithelial cells. Med. J. Cell Biol. 2019, 7, 15–24. [Google Scholar] [CrossRef]
- Kulus, M.; Kulus, J.; Popis, M.; Borowiec, B.; Stefańska, K.; Celichowski, P.; Nawrocki, M.J.; Brüssow, K.P.; Kempisty, B.; Jeseta, M.; et al. “Cell cycle” and ‘cell death’-Related genes are differentially expressed during long—Term in vitro real-time cultivation of porcine oviductal epithelial cells. Med. J. Cell Biol. 2019, 7, 90–99. [Google Scholar] [CrossRef]
- Chen, S.; Einspanier, R.; Schoen, J. Long-term culture of primary porcine oviduct epithelial cells: Validation of a comprehensive in vitro model for reproductive science. Theriogenology 2013, 80, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Chamier-Gliszczyńska, A.; Brazert, M.; Sujka-Kordowska, P.; Popis, M.; Ozegowska, K.; Stefańska, K.; Kocherova, I.; Celichowski, P.; Kulus, M.; Bukowska, D.; et al. Genes involved in angiogenesis and circulatory system development are differentially expressed in porcine epithelial oviductal cells during long-term primary in vitro culture—A transcriptomic study. Med. J. Cell Biol. 2018, 6, 163–173. [Google Scholar] [CrossRef]
- Stefańska, K.; Kocherova, I.; Knap, S.; Kulus, M.; Celichowski, P.; Jeseta, M. The genes regulating maintenance of cellular protein location are differentially expressed in porcine epithelial oviductal cells during longterm in vitro cultivation. Med. J. Cell Biol. 2019, 7, 77–85. [Google Scholar] [CrossRef]
- Stefańska, K.; Knap, S.; Kulus, M.; Kocherova, I.; Celichowski, P.; Jeseta, M.; Machatkova, M.; Bukowska, D.; Antosik, P. Differential expression pattern of genes involved in oxygen metabolism in epithelial oviductal cells during primary in vitro culture. Med. J. Cell Biol. 2019, 7, 66–76. [Google Scholar] [CrossRef]
- Bendarska-Czerwińska, A.; Zmarzły, N.; Morawiec, E.; Panfil, A.; Bryś, K.; Czarniecka, J.; Ostenda, A.; Dziobek, K.; Sagan, D.; Boroń, D.; et al. Endocrine disorders and fertility and pregnancy: An update. Front. Endocrinol. 2023, 13, 970439. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, F.; Muto, M.G.; Lee, Y.; Elvin, J.A.; Callahan, M.J.; Feltmate, C.; Garber, J.E.; Cramer, D.W.; Crum, C.P. The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am. J. Surg. Pathol. 2006, 30, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Coffey, D.M.; Creighton, C.J.; Yu, Z.; Hawkins, S.M.; Matzuk, M.M. High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proc. Natl. Acad. Sci. USA 2012, 109, 3921–3926. [Google Scholar] [CrossRef]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 2017, 8, 1093. [Google Scholar] [CrossRef]
- Kyo, S.; Ishikawa, N.; Nakamura, K.; Nakayama, K. The fallopian tube as origin of ovarian cancer: Change of diagnostic and preventive strategies. Cancer Med. 2020, 9, 421. [Google Scholar] [CrossRef] [PubMed]
- Monde’jarmonde’jar, I.; Acunã, O.S.; Izquierdo-Rico, M.J.; Coy, P.; Avile’s, M.A. The Oviduct: Functional Genomic and Proteomic Approach. Reprod. Domest. Anim. 2012, 47, 22–29. [Google Scholar] [CrossRef]
- Maillo, V.; Lopera-Vasquez, R.; Hamdi, M.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Maternal-embryo interaction in the bovine oviduct: Evidence from in vivo and in vitro studies. Theriogenology 2016, 86, 443–450. [Google Scholar] [CrossRef]
- Maillo, V.; Gaora, P.; Forde, N.; Besenfelder, U.; Havlicek, V.; Burns, G.W.; Spencer, T.E.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Oviduct-embryo interactions in cattle: Two-way traffic or a one-way street? Biol. Reprod. 2015, 92, 144–145. [Google Scholar] [CrossRef]
- White, K.L.; Hehnke, K.; Rickords, L.F.; Southern, L.L.; Thompson, D.L.; Wood, T.C. Early Embryonic Development in Vitro by Coculture with Oviductal Epithelial Cells in Pigs. Biol. Reprod. 1989, 41, 425–430. [Google Scholar] [CrossRef]
- Graf, A.; Krebs, S.; Heininen-Brown, M.; Zakhartchenko, V.; Blum, H.; Wolf, E. Genome activation in bovine embryos: Review of the literature and new insights from RNA sequencing experiments. Anim. Reprod. Sci. 2014, 149, 46–58. [Google Scholar] [CrossRef]
- Budna, J.; Celichowski, P.; Knap, S.; Jankowski, M.; Magas, M.; Nawrocki, M.J.; Ramlau, P.; Nowicki, A.; Rojewska, M.; Chermuła, B.; et al. Fatty acids related genes expression undergo substantial changes in porcine oviductal epithelial cells during long-term primary culture. Med. J. Cell Biol. 2018, 6, 39–47. [Google Scholar] [CrossRef]
- AmiGO 2: Term Details for “Cell Adhesion” (GO:0007155). Available online: https://amigo.geneontology.org/amigo/term/GO:0007155 (accessed on 16 March 2024).
- Croxatto, H.B. Physiology of gamete and embryo transport through the Fallopian tube. Reprod. Biomed. Online 2002, 4, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Talbot, P.; Shur, B.D.; Myles, D.G. Cell adhesion and fertilization: Steps in oocyte transport, sperm-zona pellucida interactions, and sperm-egg fusion. Biol. Reprod. 2003, 68, 1–9. [Google Scholar] [CrossRef] [PubMed]
- D’Occhio, M.J.; Campanile, G.; Zicarelli, L.; Visintin, J.A.; Baruselli, P.S. Adhesion molecules in gamete transport, fertilization, early embryonic development, and implantation—Role in establishing a pregnancy in cattle: A review. Mol. Reprod. Dev. 2020, 87, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.M.; Joussen, S.; Schellenberg, A.; Lin, Q.; Zenke, M.; Wagner, W. Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell 2012, 11, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Rubin, H. Cell aging in vivo and in vitro. Mech. Ageing Dev. 1997, 98, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Kulus, M.; Kranc, W.; Sujka-Kordowska, P.; Mozdziak, P.; Jankowski, M.; Konwerska, A.; Kulus, J.; Bukowska, D.; Skowroński, M.; Piotrowska-Kempisty, H.; et al. The processes of cellular growth, aging, and programmed cell death are involved in lifespan of ovarian granulosa cells during short-term IVC—Study based on animal model. Theriogenology 2020, 148, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Chermuła, B.; Kranc, W.; Jopek, K.; Budna-Tukan, J.; Hutchings, G.; Dompe, C.; Moncrieff, L.; Janowicz, K.; Józkowiak, M.; Jeseta, M.; et al. Human Cumulus Cells in Long-Term In Vitro Culture Reflect Differential Expression Profile of Genes Responsible for Planned Cell Death and Aging—A Study of New Molecular Markers. Cells 2020, 9, 1265. [Google Scholar] [CrossRef]
- Banliat, C.; Tsikis, G.; Labas, V.; Teixeira-Gomes, A.P.; Com, E.; Lavigne, R.; Pineau, C.; Guyonnet, B.; Mermillod, P.; Saint-Dizier, M. Identification of 56 Proteins Involved in Embryo–Maternal Interactions in the Bovine Oviduct. Int. J. Mol. Sci. 2020, 21, 466. [Google Scholar] [CrossRef]
- Pillai, V.V.; Weber, D.M.; Phinney, B.S.; Selvaraj, V. Profiling of proteins secreted in the bovine oviduct reveals diverse functions of this luminal microenvironment. PLoS ONE 2017, 12, e0188105. [Google Scholar] [CrossRef] [PubMed]
- Cebrian-Serrano, A.; Salvador, I.; García-Roselló, E.; Pericuesta, E.; Pérez-Cerezales, S.; Gutierrez-Adán, A.; Coy, P.; Silvestre, M.A. Effect of the Bovine Oviductal Fluid on In Vitro Fertilization, Development and Gene Expression of In Vitro-Produced Bovine Blastocysts. Reprod. Domest. Anim. 2013, 48, 331–338. [Google Scholar] [CrossRef]
- Pradeep, M.A.; Jagadeesh, J.; De, A.K.; Kaushik, J.K.; Malakar, D.; Kumar, S.; Dang, A.K.; Das, S.K.; Mohanty, A.K. Purification, sequence characterization and effect of goat oviduct-specific glycoprotein on in vitro embryo development. Theriogenology 2011, 75, 1005–1015. [Google Scholar] [CrossRef]
- Buhi, W.C. Characterization and biological roles of oviduct-specific, oestrogen-dependent glycoprotein. Reproduction 2002, 123, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Mao, W.; Zhang, Y.; Huang, N.; Liu, B.; Gao, L.; Zhang, S.; Cao, J. The prostaglandin E2 receptor PTGER2 and prostaglandin F2α receptor PTGFR mediate oviductal glycoprotein 1 expression in bovine oviductal epithelial cells. J. Reprod. Dev. 2018, 64, 101–108. [Google Scholar] [CrossRef]
- Choudhary, S.; Kumaresan, A.; Kumar, M.; Chhillar, S.; Malik, H.; Kumar, S.; Kaushik, J.K.; Datta, T.K.; Mohanty, A.K. Effect of recombinant and native buffalo OVGP1 on sperm functions and in vitro embryo development: A comparative study. J. Anim. Sci. Biotechnol. 2017, 8, 69. [Google Scholar] [CrossRef]
- Nelson, R.N.; Chakravarthi, V.P.; Ratri, A.; Hong, X.; Gossen, J.A.; Christenson, L.K. Granulosa Cell Specific Loss of Adar in Mice Delays Ovulation, Oocyte Maturation and Leads to Infertility. Int. J. Mol. Sci. 2022, 23, 14001. [Google Scholar] [CrossRef] [PubMed]
- Moniaux, N.; Escande, F.; Batra, S.K.; Porchet, N.; Laine, A.; Aubert, J.P. Alternative splicing generates a family of putative secreted and membrane-associated MUC4 mucins. Eur. J. Biochem. 2000, 267, 4536–4544. [Google Scholar] [CrossRef]
- Yeh, J.C.; Ong, E.; Fukuda, M. Molecular cloning and expression of a novel beta-1, 6-N-acetylglucosaminyltransferase that forms core 2, core 4, and I branches. J. Biol. Chem. 1999, 274, 3215–3221. [Google Scholar] [CrossRef]
- Redzovic, A.; Laskarin, G.; Dominovic, M.; Haller, H.; Rukavina, D. Mucins help to avoid alloreactivity at the maternal fetal interface. Clin. Dev. Immunol. 2013, 2013, 542152. [Google Scholar] [CrossRef]
- Jang, J.S.; Lee, J.H.; Jung, N.C.; Choi, S.Y.; Park, S.Y.; Yoo, J.Y.; Song, J.Y.; Seo, H.G.; Lee, H.S.; Lim, D.S. Rsad2 is necessary for mouse dendritic cell maturation via the IRF7-mediated signaling pathway. Cell Death Dis. 2018, 9, 823. [Google Scholar] [CrossRef]
- Song, G.; Bazer, F.W.; Spencer, T.E. Pregnancy and interferon tau regulate RSAD2 and IFIH1 expression in the ovine uterus. Reproduction 2007, 133, 285–295. [Google Scholar] [CrossRef]
- Schmaltz-Panneau, B.; Cordova, A.; Dhorne-Pollet, S.; Hennequet-Antier, C.; Uzbekova, S.; Martinot, E.; Doret, S.; Martin, P.; Mermillod, P.; Locatelli, Y. Early bovine embryos regulate oviduct epithelial cell gene expression during in vitro co-culture. Anim. Reprod. Sci. 2014, 149, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Qi, S.H.; Shu, B.; Chen, L.; Xie, J.L.; Xu, Y.B.; Liu, X.S. Fibroblast growth factor-binding protein facilitates the growth and migration of skin-derived precursors. J. Cutan. Med. Surg. 2011, 15, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Abuharbeid, S.; Czubayko, F.; Aigner, A. The fibroblast growth factor-binding protein FGF-BP. Int. J. Biochem. Cell Biol. 2006, 38, 1463–1468. [Google Scholar] [CrossRef]
- Taetzsch, T.; Brayman, V.L.; Valdez, G. FGF binding proteins (FGFBPs): Modulators of FGF signaling in the developing, adult, and stressed nervous system. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1864 Pt B, 2983–2991. [Google Scholar] [CrossRef]
- Lee, H.O.; Choe, H.; Seo, K.; Lee, H.; Lee, J.; Kim, J. Fgfbp1 is essential for the cellular survival during zebrafish embryogenesis. Mol. Cells 2010, 29, 501–507. [Google Scholar] [CrossRef]
- Lee, J.A.; Yerbury, J.J.; Farrawell, N.; Shearer, R.F.; Constantinescu, P.; Hatters, D.M.; Schroder, W.A.; Suhrbier, A.; Wilson, M.R.; Saunders, D.N.; et al. SerpinB2 (PAI-2) Modulates Proteostasis via Binding Misfolded Proteins and Promotion of Cytoprotective Inclusion Formation. PLoS ONE 2015, 10, e0130136. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.R.; Cater, J.H.; Ranson, M. PZP and PAI-2: Structurally-diverse, functionally similar pregnancy proteins? Int. J. Biochem. Cell Biol. 2016, 79, 113–117. [Google Scholar] [CrossRef]
- Kim, J.; Jang, K.T.; Kim, K.H.; Park, J.W.; Chang, B.J.; Lee, K.H.; Lee, J.K.; Heo, J.S.; Choi, S.H.; Choi, D.W.; et al. Aberrant maspin expression is involved in early carcinogenesis of gallbladder cancer. Tumour Biol. 2010, 31, 471–476. [Google Scholar] [CrossRef]
- Sinha, K.K.; Vinay, J.; Parida, S.; Singh, S.P.; Dixit, M. Association and functional significance of genetic variants present in regulatory elements of SERPINB5 gene in gallbladder cancer. Gene 2022, 808, 145989. [Google Scholar] [CrossRef]
- Machowska, M.; Wachowicz, K.; Sopel, M.; Rzepecki, R. Nuclear location of tumor suppressor protein maspin inhibits proliferation of breast cancer cells without affecting proliferation of normal epithelial cells. BMC Cancer 2014, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Longhi, M.T.; Magalhães, M.; Reina, J.; Freitas, V.M.; Cella, N. EGFR Signaling Regulates Maspin/SerpinB5 Phosphorylation and Nuclear Localization in Mammary Epithelial Cells. PLoS ONE 2016, 11, e0159856. [Google Scholar] [CrossRef]
- Kane, R.; Godson, C.; O’Brien, C. Chordin-like 1, a bone morphogenetic protein-4 antagonist, is upregulated by hypoxia in human retinal pericytes and plays a role in regulating angiogenesis. Mol. Vis. 2008, 14, 1138. [Google Scholar] [PubMed]
- Wang, Y.W.; Wu, C.H.; Lin, T.Y.; Luo, C.W. Expression profiling of ovarian BMP antagonists reveals the potential interaction between TWSG1 and the chordin subfamily in the ovary. Mol. Cell. Endocrinol. 2021, 538, 111457. [Google Scholar] [CrossRef] [PubMed]
- Budna, J.; Rybska, M.; Ciesiółka, S.; Bryja, A.; Borys, S.; Kranc, W.; Wojtanowicz-Markiewicz, K.; Jeseta, M.; Sumelka, E.; Bukowska, D.; et al. Expression of genes associated with BMP signaling pathway in porcine oocytes before and after IVM—A microarray approach. Reprod. Biol. Endocrinol. 2017, 15, 43. [Google Scholar] [CrossRef]
- Kedem, A.; Ulanenko-Shenkar, K.; Yung, Y.; Youngster, M.; Avraham, S.; Yerushalmi, G.; Hourvitz, A. The Involvement of Lumican in Human Ovulatory Processes. Reprod. Sci. 2022, 29, 366–373. [Google Scholar] [CrossRef]
- Bonavina, G.; Taylor, H.S. Endometriosis-associated infertility: From pathophysiology to tailored treatment. Front. Endocrinol. 2022, 13, 1020827. [Google Scholar] [CrossRef] [PubMed]
- Imrich, S.; Hachmeister, M.; Gires, O. EpCAM and its potential role in tumor-initiating cells. Cell Adhes. Migr. 2012, 6, 30–38. [Google Scholar] [CrossRef]
- Keller, L.; Werner, S.; Pantel, K. Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165. [Google Scholar] [CrossRef]
- Gires, O.; Pan, M.; Schinke, H.; Canis, M.; Baeuerle, P.A. Expression and function of epithelial cell adhesion molecule EpCAM: Where are we after 40 years? Cancer Metastasis Rev. 2020, 39, 969–987. [Google Scholar] [CrossRef]
- González, B.; Denzel, S.; Mack, B.; Conrad, M.; Gires, O. EpCAM is involved in maintenance of the murine embryonic stem cell phenotype. Stem Cells 2009, 27, 1782–1791. [Google Scholar] [CrossRef]
- van der Gun, B.T.F.; Melchers, L.J.; Ruiters, M.H.J.; de Leij, L.F.M.H.; McLaughlin, P.M.J.; Rots, M.G. EpCAM in carcinogenesis: The good, the bad or the ugly. Carcinogenesis 2010, 31, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.C.; Sankpal, N.V.; Gillanders, W.E. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules 2021, 11, 956. [Google Scholar] [CrossRef] [PubMed]
- Stelcer, E.; Komarowska, H.; Jopek, K.; Żok, A.; Iżycki, D.; Malińska, A.; Szczepaniak, B.; Komekbai, Z.; Karczewski, M.; Wierzbicki, T.; et al. Biological response of adrenal carcinoma and melanoma cells to mitotane treatment. Oncol. Lett. 2022, 23, 120. [Google Scholar] [CrossRef]
- Budna, J.; Chachuła, A.; Kaźmierczak, D.; Rybska, M.; Ciesiółka, S.; Bryja, A.; Kranc, W.; Borys, S.; Zok, A.; Bukowska, D.; et al. Morphogenesis-related gene-expression profile in porcine oocytes before and after in vitro maturation. Zygote 2017, 25, 331–340. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Szyszka, M.; Paschke, L.; Tyczewska, M.; Jopek, K.; Celichowski, P.; Milecka, P.; Sultanova, G.; Stelcer, E.; Malinska, A.; Malendowicz, L.K.; et al. Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations. Int. J. Mol. Sci. 2019, 20, 5412. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.V. Genefilter: Genefilter: Methods for Filtering Genes from High-Throughput Experiments. R Package Version 1.74.1. Available online: https://bioconductor.org/packages/release/bioc/html/genefilter.html (accessed on 5 April 2023).
- Fresno, C.; Fernández, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef]
- Benjamini, Y.; Cohen, R. Weighted false discovery rate controlling procedures for clinical trials. Biostatistics 2017, 18, 91–104. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Golkar-Narenji, A.; Antosik, P.; Nolin, S.; Rucinski, M.; Jopek, K.; Zok, A.; Sobolewski, J.; Jankowski, M.; Zdun, M.; Bukowska, D.; et al. Gene Ontology Groups and Signaling Pathways Regulating the Process of Avian Satellite Cell Differentiation. Genes 2022, 13, 242. [Google Scholar] [CrossRef] [PubMed]
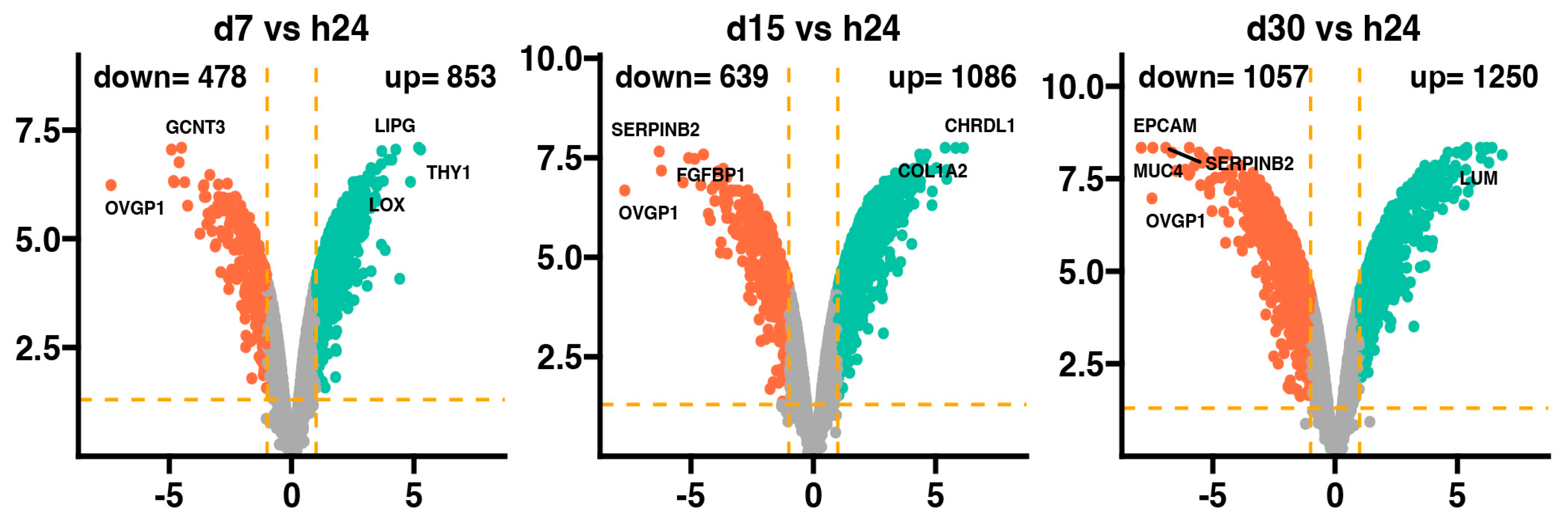



| Gene Symbol | Gene Name | Fold Change |
| THY1 | Thy−1 cell surface antigen | 38.75 |
| LIPG | lipase, endothelial | 36.55 |
| LOX | lysyl oxidase | 29.05 |
| ANKRD1 | ankyrin repeat domain 1 (cardiac muscle) | 21.40 |
| UNC45B | unc−45 homolog B (C. elegans) | 19.12 |
| LOC106508700 | protein prune homolog 2−like | 17.16 |
| COL1A2 | collagen, type I, alpha 2 | 15.57 |
| TMSB15A | thymosin beta 15a | 14.21 |
| UBE2QL1 | ubiquitin−conjugating enzyme E2Q family−like 1 | 13.45 |
| FMOD | Fibromodulin | 12.88 |
| TXNIP | thioredoxin interacting protein | −12.17 |
| SLC28A3 | solute carrier family 28 (concentrative nucleoside transporter), member 3 | −13.42 |
| OASL | 2−5−oligoadenylate synthetase−like | −19.01 |
| GPX2 | glutathione peroxidase 2 | −20.67 |
| CD274 | CD274 molecule | −22.60 |
| SERPINB2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | −24.27 |
| FGFBP1 | fibroblast growth factor binding protein 1 | −27.95 |
| RSAD2 | radical S−adenosyl methionine domain containing 2 | −28.39 |
| GCNT3 | glucosaminyl (N−acetyl) transferase 3, mucin type | −30.10 |
| OVGP1 | oviductal glycoprotein 1, 120 kDa | −166.91 |
| Gene Symbol | Gene Name | Fold Change |
| CHRDL1 | chordin−like 1 | 70.42 |
| COL1A2 | collagen, type I, alpha 2 | 56.38 |
| AGTR1 | angiotensin II receptor, type 1 | 44.11 |
| LOX | lysyl oxidase | 43.85 |
| FMOD | Fibromodulin | 41.89 |
| THY1 | Thy−1 cell surface antigen | 32.74 |
| SULF1 | sulfatase 1 | 29.09 |
| COL14A1 | collagen, type XIV, alpha 1 | 28.88 |
| FBN1 | fibrillin 1 | 27.55 |
| CDH11 | cadherin 11, type 2, OB−cadherin (osteoblast) | 24.48 |
| LOC100524999 | placenta−specific gene 8 protein | −18.51 |
| OASL | 2−5−oligoadenylate synthetase−like | −19.59 |
| CD274 | CD274 molecule | −22.51 |
| GPX2 | glutathione peroxidase 2 | −24.24 |
| MUC1 | mucin 1, cell surface associated | −28.79 |
| GCNT3 | glucosaminyl (N−acetyl) transferase 3, mucin type | −34.27 |
| ANKRD22 | ankyrin repeat domain 22 | −40.07 |
| FGFBP1 | fibroblast growth factor binding protein 1 | −74.13 |
| SERPINB2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | −78.88 |
| OVGP1 | oviductal glycoprotein 1, 120 kDa | −209.47 |
| Gene Symbol | Gene Name | Fold Change |
| LUM | Lumican | 114.07 |
| CHRDL1 | chordin−like 1 | 85.62 |
| AGTR1 | angiotensin II receptor, type 1 | 78.21 |
| PRLR | prolactin receptor | 73.13 |
| DPP4 | dipeptidyl−peptidase 4 | 71.86 |
| FBN1 | fibrillin 1 | 62.85 |
| COL1A2 | collagen, type I, alpha 2 | 61.33 |
| LOX | lysyl oxidase | 46.65 |
| LOC102164159 | myozenin−2−like | 43.49 |
| LOC106508700 | protein prune homolog 2−like | 41.92 |
| CLDN7 | claudin 7 | −61.38 |
| SPINT1 | serine peptidase inhibitor, Kunitz type 1 | −62.17 |
| APOBEC1 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 | −63.28 |
| AQP9 | aquaporin 9 | −69.25 |
| FGFBP1 | fibroblast growth factor binding protein 1 | −90.37 |
| SERPINB5 | serpin peptidase inhibitor, clade B (ovalbumin), member 5 | −101.70 |
| SERPINB2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | −121.38 |
| MUC4 | mucin 4, cell surface associated | −175.03 |
| OVGP1 | oviductal glycoprotein 1, 120 kDa | −179.30 |
| EPCAM | epithelial cell adhesion molecule | −243.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zgórecka, W.; Kranc, W.; Blatkiewicz, M.; Kamiński, K.; Farzaneh, M.; Bryja, A.; Mozdziak, P.; Antosik, P.; Zabel, M.; Podhorska-Okołów, M.; et al. Long-Term In Vitro Culture Alters Gene Expression Pattern of Genes Involved in Ontological Groups Representing Cellular Processes. Int. J. Mol. Sci. 2024, 25, 7109. https://doi.org/10.3390/ijms25137109
Zgórecka W, Kranc W, Blatkiewicz M, Kamiński K, Farzaneh M, Bryja A, Mozdziak P, Antosik P, Zabel M, Podhorska-Okołów M, et al. Long-Term In Vitro Culture Alters Gene Expression Pattern of Genes Involved in Ontological Groups Representing Cellular Processes. International Journal of Molecular Sciences. 2024; 25(13):7109. https://doi.org/10.3390/ijms25137109
Chicago/Turabian StyleZgórecka, Wiktoria, Wiesława Kranc, Małgorzata Blatkiewicz, Kacper Kamiński, Maryam Farzaneh, Artur Bryja, Paul Mozdziak, Paweł Antosik, Maciej Zabel, Marzenna Podhorska-Okołów, and et al. 2024. "Long-Term In Vitro Culture Alters Gene Expression Pattern of Genes Involved in Ontological Groups Representing Cellular Processes" International Journal of Molecular Sciences 25, no. 13: 7109. https://doi.org/10.3390/ijms25137109
APA StyleZgórecka, W., Kranc, W., Blatkiewicz, M., Kamiński, K., Farzaneh, M., Bryja, A., Mozdziak, P., Antosik, P., Zabel, M., Podhorska-Okołów, M., Dzięgiel, P., Kempisty, B., & Bukowska, D. (2024). Long-Term In Vitro Culture Alters Gene Expression Pattern of Genes Involved in Ontological Groups Representing Cellular Processes. International Journal of Molecular Sciences, 25(13), 7109. https://doi.org/10.3390/ijms25137109






