Discovery of the Natural Bibenzyl Compound Erianin in Dendrobium Inhibiting the Growth and EMT of Gastric Cancer through Downregulating the LKB1-SIK2/3-PARD3 Pathway
Abstract
:1. Introduction
2. Results
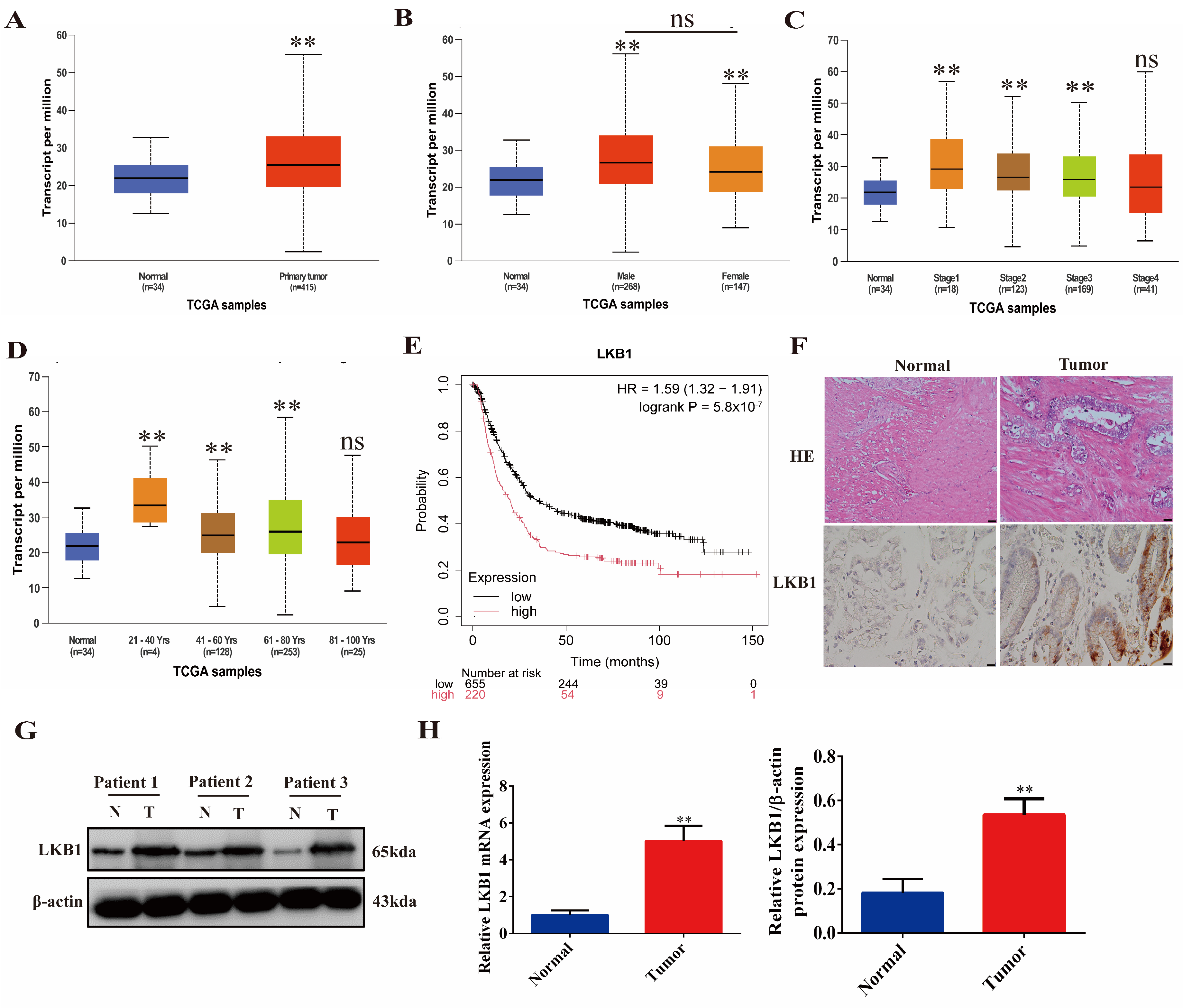
2.1. LKB1 Is Highly Expressed in Gastric Cancer and Associated with Poor Patient Prognosis
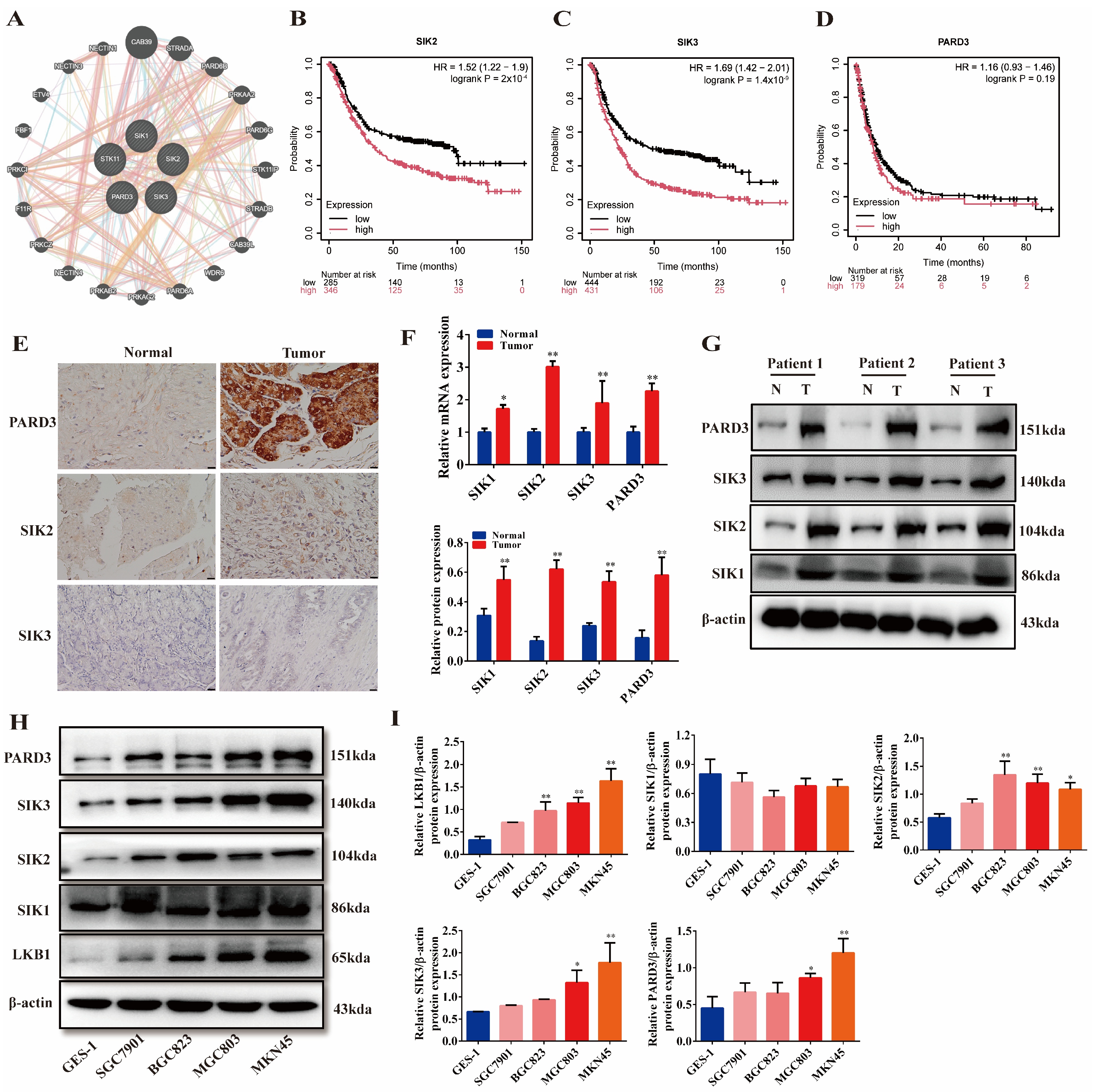
2.2. LKB1 Is Closely Related to SIKs and PARD3, and They Are Highly Expressed in Gastric Cancer
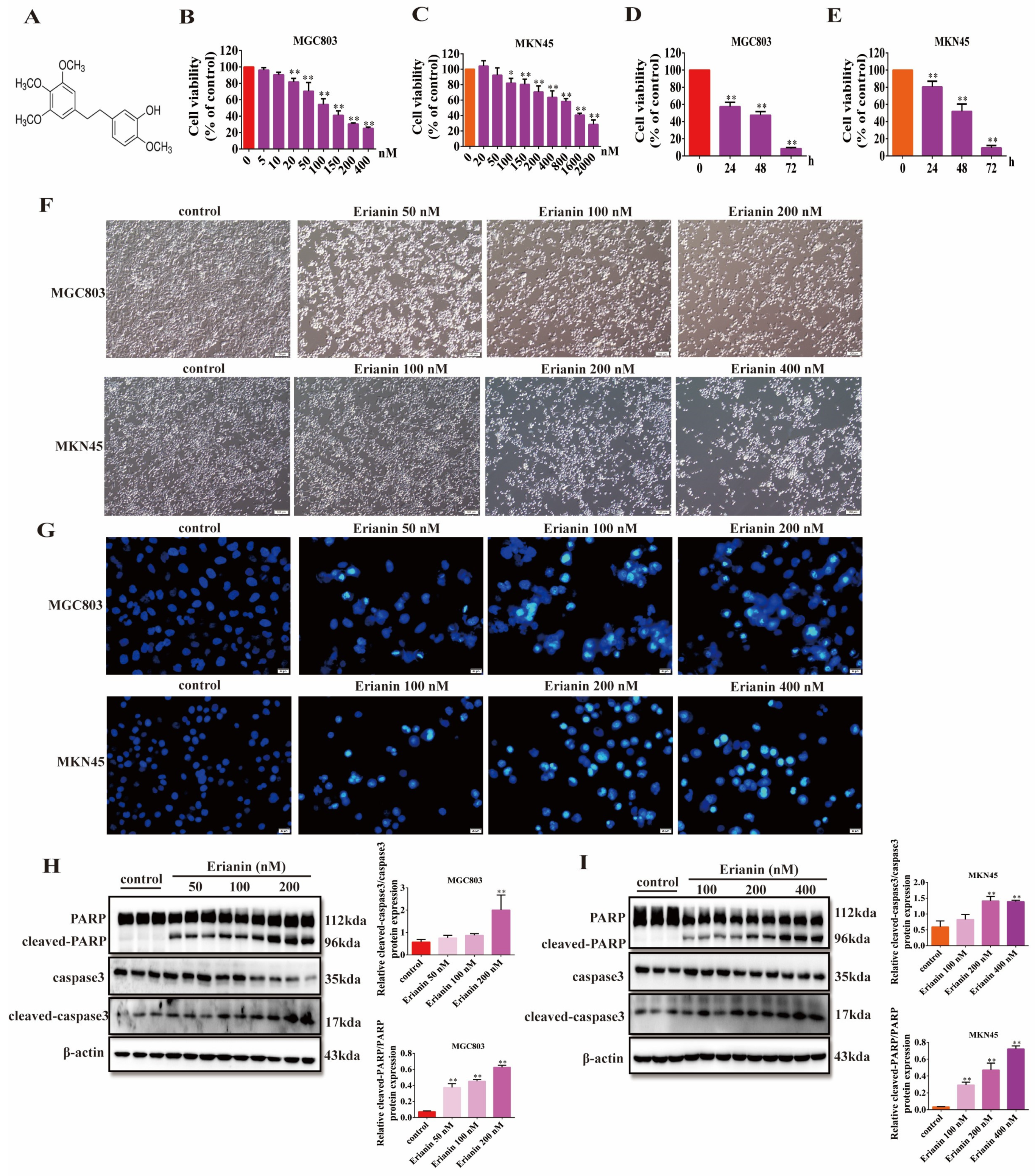
2.3. Erianin Inhibits GC Cell Viability and Induces Apoptosis
2.4. Erianin Inhibits the Expression of LKB1, SIK2/3 and PARD3 in GC Cells
2.5. Erianin Inhibits GC Cell Migration, Invasion, and EMT Properties
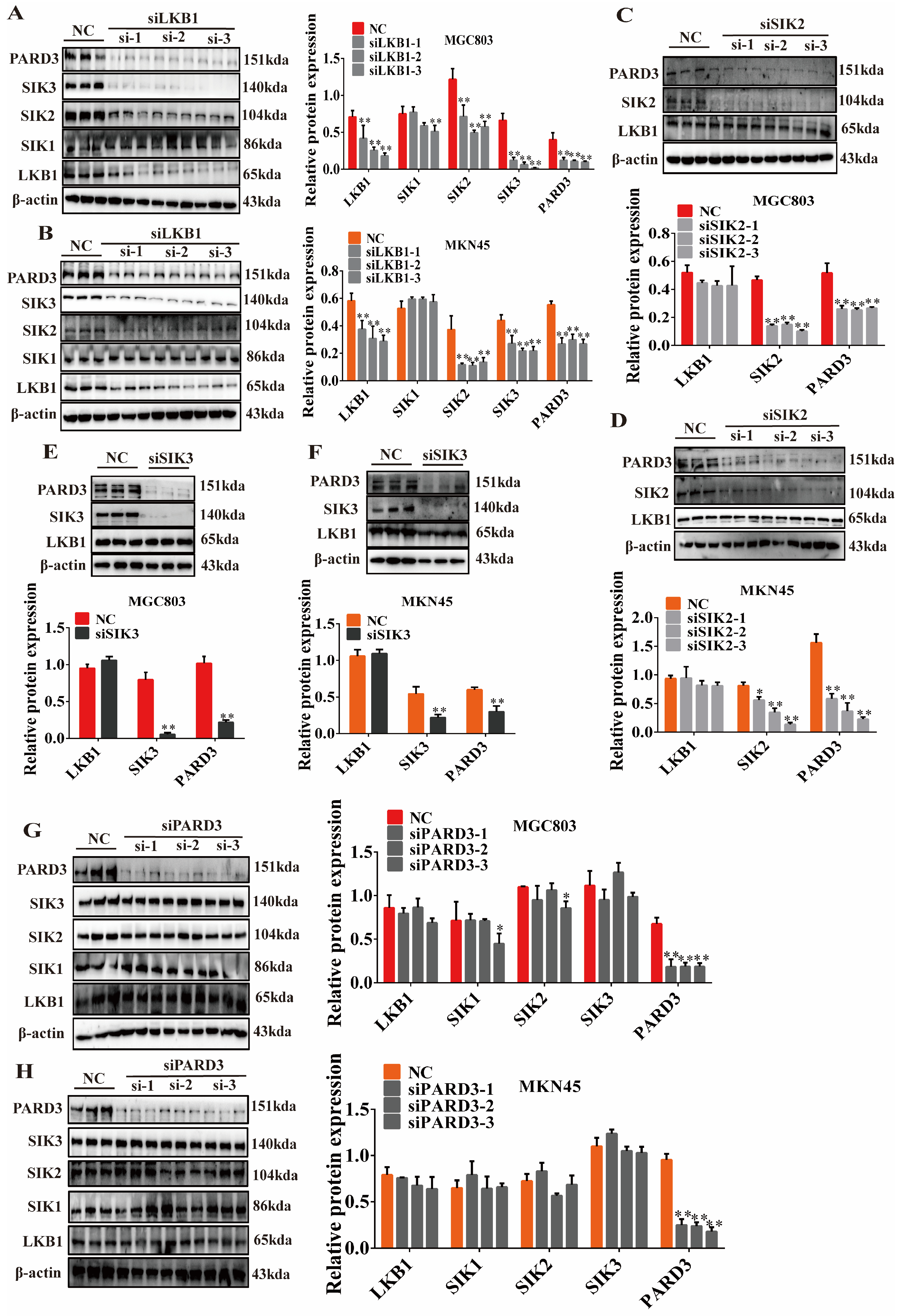
2.6. Regulatory Relationship of LKB1-SIK2/3-PARD3
2.7. Erianin Inhibits Tumor Growth in CDX Mice
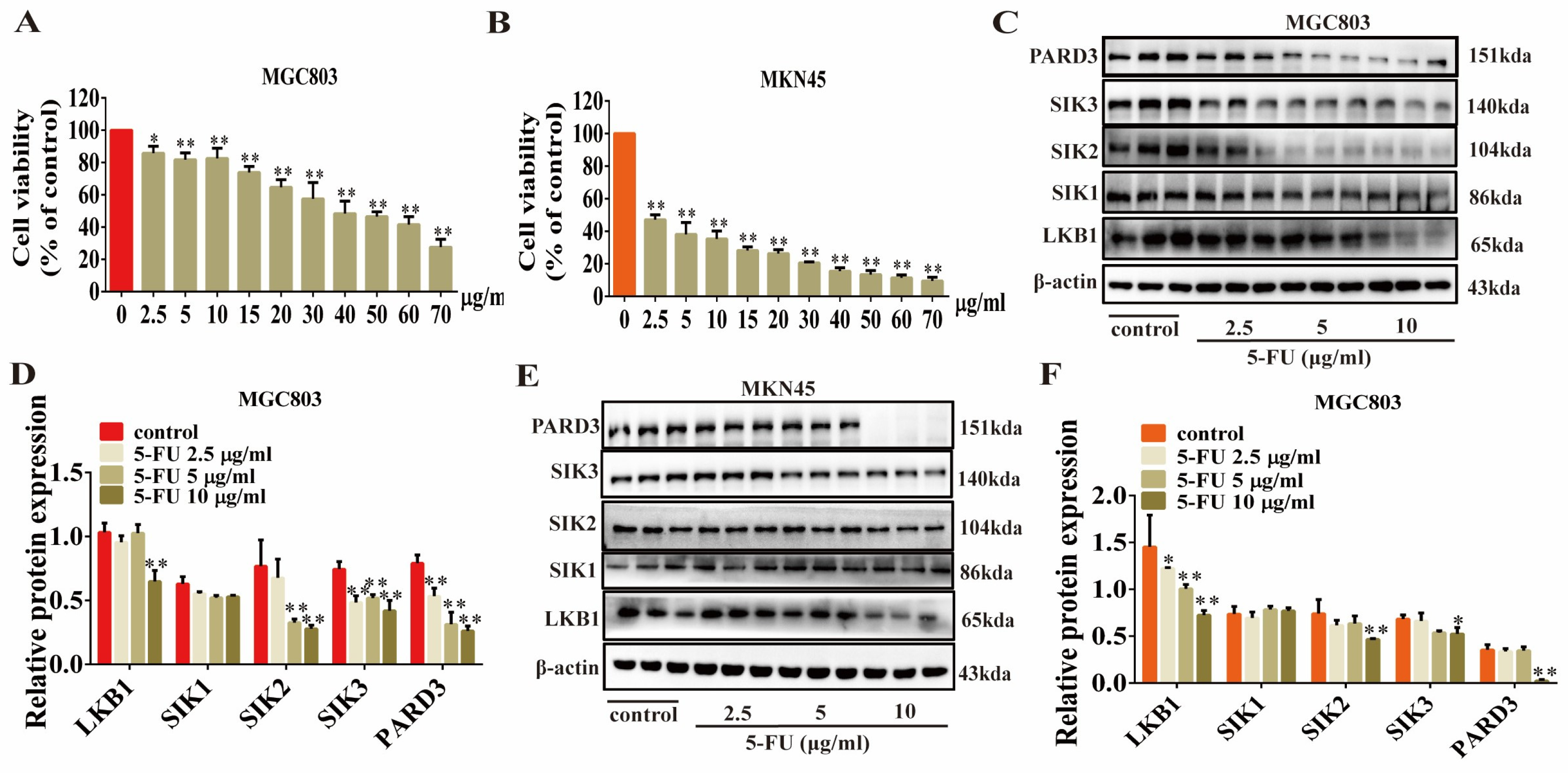
2.8. 5-FU Inhibits the Expression of LKB1, SIK2/3, and PARD3 in GC Cells
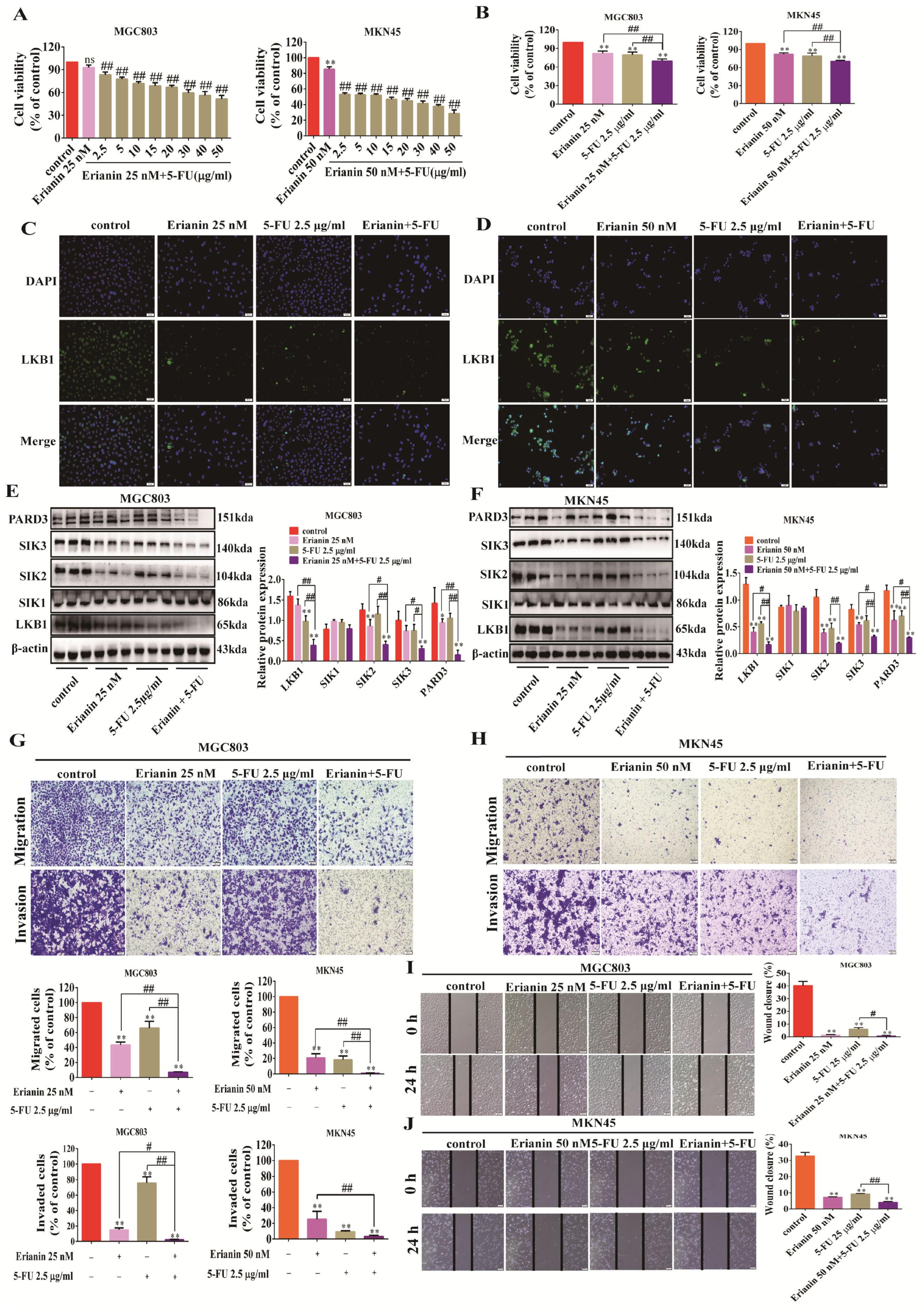
2.9. 5-FU Combined with Erianin Jointly Inhibits the Expression of LKB1-SIK2/3-PARD3 Axis
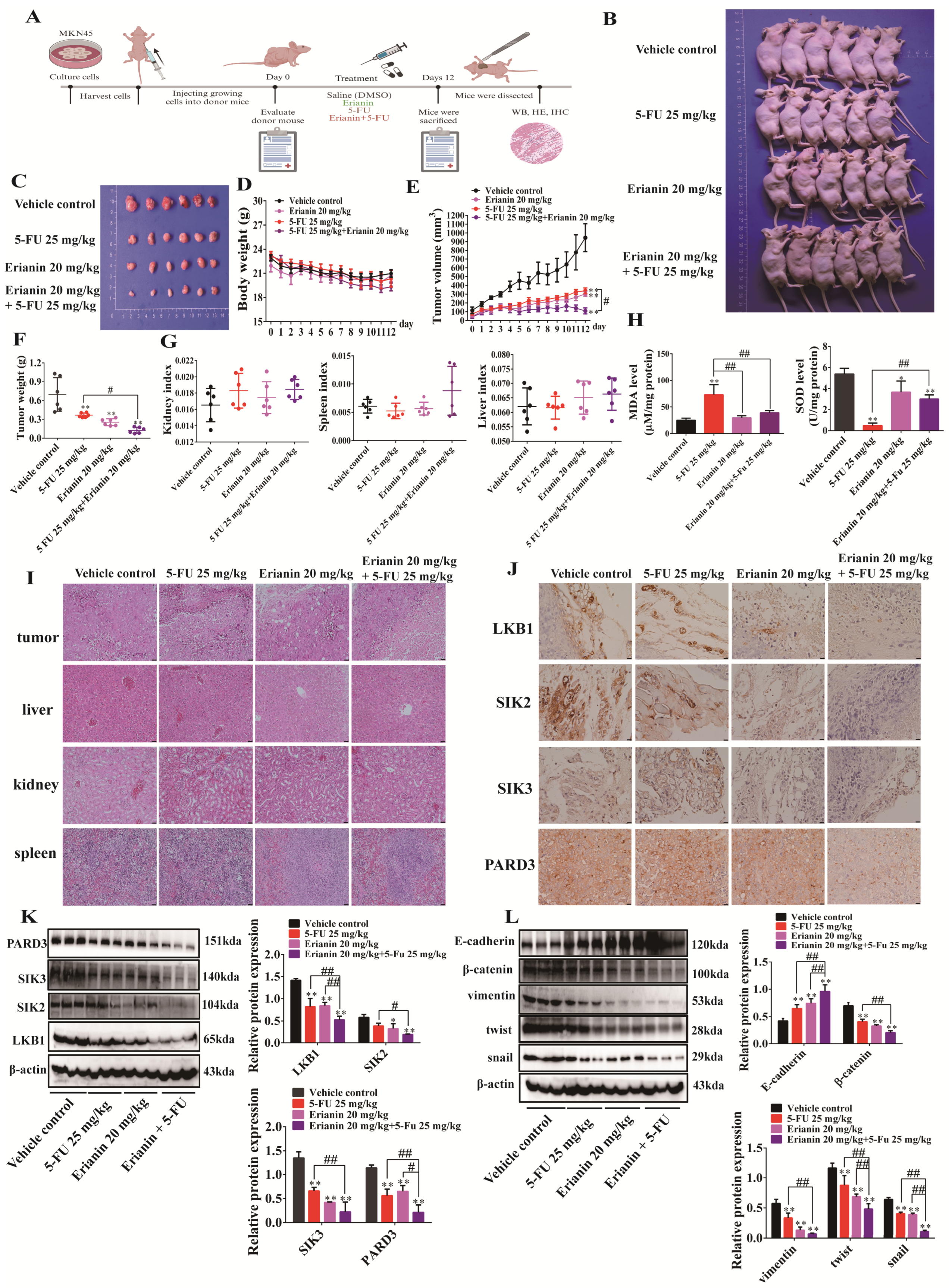
2.10. Erianin Combined with 5-FU Inhibits Tumor Growth in CDX Mice
3. Discussion
4. Materials and Methods
4.1. Drugs and Reagents
4.2. Clinical Patients
4.3. Cell Culture
4.4. Small Interfering RNA (siRNA) Transfection
4.5. Molecular Docking
4.6. Gene Expression and Survival Curves
4.7. Gene Correlation Analysis
4.8. Cell Viability Assay
4.9. Real-Time Quantitative PCR
4.10. Western Blot
4.11. Hoechst 33342 Staining
4.12. Wound-Healing Assay
4.13. Migration and Invasion Assays
4.14. Immunofluorescence
4.15. Animal Experiment
4.16. Immunohistochemistry (IHC)
4.17. Hematoxylin and Eosin (H&E) Staining
4.18. Superoxide Dismutase (SOD) and Malondialdehyde (MDA) Assay
4.19. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Health Commission of the People’s Republic of China N. National guidelines for diagnosis and treatment of thyroid cancer 2022 in China (English version). Chin. J. Cancer Res. 2022, 34, 131–150. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Gastroenterol. Rev. 2019, 14, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Ladeiras-Lopes, R.; Pereira, A.K.; Nogueira, A.; Pinheiro-Torres, T.; Pinto, I.; Santos-Pereira, R.; Lunet, N. Smoking and gastric cancer: Systematic review and meta-analysis of cohort studies. Cancer Causes Control 2008, 19, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, L.; Rushton, L. Stomach cancer and occupational exposure to asbestos: A meta-analysis of occupational cohort studies. Br. J. Cancer 2015, 112, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Jha, H.C. Status of Epstein-Barr Virus Coinfection with Helicobacter pylori in Gastric Cancer. J. Oncol. 2017, 2017, 3456264. [Google Scholar] [CrossRef] [PubMed]
- Johnston, F.M.; Beckman, M. Updates on Management of Gastric Cancer. Curr. Oncol. Rep. 2019, 21, 67. [Google Scholar] [CrossRef]
- Yang, W.J.; Zhao, H.P.; Yu, Y.; Wang, J.H.; Guo, L.; Liu, J.Y.; Pu, J.; Lv, J. Updates on global epidemiology, risk and prognostic factors of gastric cancer. World J. Gastroenterol. 2023, 29, 2452–2468. [Google Scholar] [CrossRef] [PubMed]
- Satake, H.; Kondo, M.; Mizumoto, M.; Kotake, T.; Okita, Y.; Ogata, T.; Hatachi, Y.; Yasui, H.; Miki, A.; Imai, Y.; et al. Phase I Study of Neoadjuvant Chemotherapy with Capecitabine and Oxaliplatin for Locally Advanced Gastric Cancer. Anticancer Res. 2017, 37, 3703–3710. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Park, J.Y.; Camargo, M.C.; Lunet, N.; Forman, D.; Soerjomataram, I. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035. Gut 2020, 69, 823–829. [Google Scholar] [CrossRef]
- Ito, Y.; Miyashiro, I.; Ishikawa, T.; Akazawa, K.; Fukui, K.; Katai, H.; Nunobe, S.; Oda, I.; Isobe, Y.; Tsujitani, S.; et al. Determinant Factors on Differences in Survival for Gastric Cancer Between the United States and Japan Using Nationwide Databases. J. Epidemiol. 2021, 31, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Liu, J.; Xu, Z.; Wang, D.; Zhu, Q.; Chen, Q.; Xu, W. Research progress on the pharmacological mechanism, in vivo metabolism and structural modification of Erianin. Biomed. Pharmacother. 2024, 173, 116295. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q.; Wei, F.; Liu, N. Progressive study of effects of erianin on anticancer activity. Onco Targets Ther. 2019, 12, 5457–5465. [Google Scholar] [CrossRef]
- Yang, A.; Sun, Z.; Liu, R.; Liu, X.; Zhang, Y.; Zhou, Y.; Qiu, Y.; Zhang, X. Transferrin-Conjugated Erianin-Loaded Liposomes Suppress the Growth of Liver Cancer by Modulating Oxidative Stress. Front. Oncol. 2021, 11, 727605. [Google Scholar] [CrossRef]
- Chen, P.; Wu, Q.; Feng, J.; Yan, L.; Sun, Y.; Liu, S.; Xiang, Y.; Zhang, M.; Pan, T.; Chen, X.; et al. Erianin, a novel dibenzyl compound in Dendrobium extract, inhibits lung cancer cell growth and migration via calcium/calmodulin-dependent ferroptosis. Signal Transduct. Target. Ther. 2020, 5, 51. [Google Scholar] [CrossRef]
- Xiang, Y.; Chen, X.; Wang, W.; Zhai, L.; Sun, X.; Feng, J.; Duan, T.; Zhang, M.; Pan, T.; Yan, L.; et al. Natural Product Erianin Inhibits Bladder Cancer Cell Growth by Inducing Ferroptosis via NRF2 Inactivation. Front. Pharmacol. 2021, 12, 775506. [Google Scholar] [CrossRef]
- Wang, Y.; Chu, F.; Lin, J.; Li, Y.; Johnson, N.; Zhang, J.; Gai, C.; Su, Z.; Cheng, H.; Wang, L.; et al. Erianin, the main active ingredient of Dendrobium chrysotoxum Lindl, inhibits precancerous lesions of gastric cancer (PLGC) through suppression of the HRAS-PI3K-AKT signaling pathway as revealed by network pharmacology and in vitro experimental verification. J. Ethnopharmacol. 2021, 279, 114399. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Patel, A.S.; Kim, E.; Li, H.; Chen, Y.; Li, S.; Liu, S.; Dilly, J.; Kapner, K.S.; Zhang, N.; et al. Adeno-to-squamous transition drives resistance to KRAS inhibition in LKB1 mutant lung cancer. Cancer Cell 2024, 42, 413–428 e417. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Hebert, K.; McConnell, E.; Cullen, N.; Cheng, T.; Awoyode, S.; Martin, E.; Chen, W.; Wu, T.; Alahari, S.K.; et al. LKB1 signaling and patient survival outcomes in hepatocellular carcinoma. Pharmacol. Res. 2023, 192, 106757. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, H.; Wang, X.; Zhao, W.; Chen, J.J. Expression and transcriptional profiling of the LKB1 tumor suppressor in cervical cancer cells. Gynecol. Oncol. 2014, 134, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.P.; Jamieson, N.B.; Karim, S.A.; Athineos, D.; Ridgway, R.A.; Nixon, C.; McKay, C.J.; Carter, R.; Brunton, V.G.; Frame, M.C.; et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology 2010, 139, 586–597.e6. [Google Scholar] [CrossRef] [PubMed]
- Proteau, S.; Krossa, I.; Husser, C.; Gueguinou, M.; Sella, F.; Bille, K.; Irondelle, M.; Dalmasso, M.; Barouillet, T.; Cheli, Y.; et al. LKB1-SIK2 loss drives uveal melanoma proliferation and hypersensitivity to SLC8A1 and ROS inhibition. EMBO Mol. Med. 2023, 15, e17719. [Google Scholar] [CrossRef]
- Morikawa, A.; Williams, T.Y.; Dirix, L.; Colpaert, C.; Goodman, M.; Lyles, R.H.; Zhong, D.; Zhou, W. Allelic imbalances of chromosomes 8p and 18q and their roles in distant relapse of early stage, node-negative breast cancer. Breast Cancer Res. 2005, 7, R1051–R1057. [Google Scholar] [CrossRef]
- Sanchez-Cespedes, M.P.P.; Esteller, M.; Nomoto, S.; Trink, B.; Engles, J.M.; Westra, W.H.; Herman, J.G.; Sidransky, D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002, 62, 3659–3662. [Google Scholar]
- Long, L.L.M.S.; Guo, Z.Q.; Zhang, Y.P.; Fan, Z.; Liu, L.J.; Liu, L.; Han, D.D.; Leng, M.X.; Wang, J.; Guo, X.J.; et al. PARP Inhibition Induces Synthetic Lethality and Adaptive Immunity in LKB1-Mutant Lung Cancer. Cancer Res. 2023, 83, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Ramsey, M.R.; Hayes, D.N.; Fan, C.; McNamara, K.; Kozlowski, P.; Torrice, C.; Wu, M.C.; Shimamura, T.; Perera, S.A.; et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007, 448, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Liu, F.; Liu, X.; Khuri, F.R.; Marcus, A.I.; Li, M.; Zhou, W. Re-expression of LKB1 in LKB1-mutant EKVX cells leads to resistance to paclitaxel through the up-regulation of MDR1 expression. Lung Cancer 2015, 88, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Park, W.S.; Moon, Y.W.; Yang, Y.M.; Kim, Y.S.; Kim, Y.D.; Fuller, B.G.; Vortmeyer, A.O.; Fogt, F.; Lubensky, I.A.; Zhuang, Z. Mutations of the STK11 gene in sporadic gastric carcinoma. Int. J. Oncol. 1998, 13, 601–604. [Google Scholar] [CrossRef]
- Feng, S.; Wei, F.; Shi, H.; Chen, S.; Wang, B.; Huang, D.; Luo, L. Roles of salt-inducible kinases in cancer (Review). Int. J. Oncol. 2023, 63, 118. [Google Scholar] [CrossRef]
- Tesch, R.; Rak, M.; Raab, M.; Berger, L.M.; Kronenberger, T.; Joerger, A.C.; Berger, B.T.; Abdi, I.; Hanke, T.; Poso, A.; et al. Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors. J. Med. Chem. 2021, 64, 8142–8160. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lin, F.; Xu, C.; Xu, Y. LINC00662 facilitates osteosarcoma progression via sponging miR-103a-3p and regulating SIK2 expression. J. Tissue Eng. Regen. Med. 2021, 15, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, E.M.; Montell, D.J. Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development 2004, 131, 5243–5251. [Google Scholar] [CrossRef] [PubMed]
- Jan, Y.J.; Ko, B.S.; Liu, T.A.; Wu, Y.M.; Liang, S.M.; Chen, S.C.; Wang, J.; Liou, J.Y. Expression of partitioning defective 3 (Par-3) for predicting extrahepatic metastasis and survival with hepatocellular carcinoma. Int. J. Mol. Sci. 2013, 14, 1684–1697. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Nagasaka, K.; Kawana, K.; Taguchi, A.; Uehara, Y.; Yoshida, M.; Sato, M.; Nishida, H.; Fujimoto, A.; Inoue, T.; et al. Expression of Par3 polarity protein correlates with poor prognosis in ovarian cancer. BMC Cancer 2016, 16, 897. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.J.; Xue, W.; Peng, J.; Wang, Y.; Wei, L.; Yang, Z.; Zhu, H.H.; Fang, Y.X.; Gao, W.Q. Elevated expression of Par3 promotes prostate cancer metastasis by forming a Par3/aPKC/KIBRA complex and inactivating the hippo pathway. J. Exp. Clin. Cancer Res. 2017, 36, 139. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jin, J. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging 2019, 11, 4876–4889. [Google Scholar] [CrossRef]
- Vanlandewijck, M.; Dadras, M.S.; Lomnytska, M.; Mahzabin, T.; Lee Miller, M.; Busch, C.; Brunak, S.; Heldin, C.H.; Moustakas, A. The protein kinase SIK downregulates the polarity protein Par3. Oncotarget 2017, 9, 5716–5735. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Jha, H.C.; Sun, Z.; Upadhyay, S.K.; El-Naccache, D.W.; Singh, R.K.; Sahu, S.K.; Robertson, E.S. KSHV-Mediated Regulation of Par3 and SNAIL Contributes to B-Cell Proliferation. PLoS Pathog. 2016, 12, e1005801. [Google Scholar] [CrossRef]
- Zen, K.; Yasui, K.; Gen, Y.; Dohi, O.; Wakabayashi, N.; Mitsufuji, S.; Itoh, Y.; Zen, Y.; Nakanuma, Y.; Taniwaki, M.; et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene 2009, 28, 2910–2918. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, S.M.; Mohapatra, G.; Rivera, M.N.; Winokur, D.; Greninger, P.; Nitta, M.; Sadow, P.M.; Sooriyakumar, G.; Brannigan, B.W.; Ulman, M.J.; et al. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 2010, 70, 2158–2164. [Google Scholar] [CrossRef]
- McCaffrey, L.M.; Montalbano, J.; Mihai, C.; Macara, I.G. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell 2012, 22, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Liu, P.; Wang, Z.C.; Zou, L.; Santiago, S.; Garbitt, V.; Gjoerup, O.V.; Iglehart, J.D.; Miron, A.; Richardson, A.L.; et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci. Signal. 2009, 2, ra35. [Google Scholar] [CrossRef]
- Lin, X.; Takemori, H.; Katoh, Y.; Doi, J.; Horike, N.; Makino, A.; Nonaka, Y.; Okamoto, M. Salt-inducible kinase is involved in the ACTH/cAMP-dependent protein kinase signaling in Y1 mouse adrenocortical tumor cells. Mol. Endocrinol. 2001, 15, 1264–1276. [Google Scholar] [CrossRef] [PubMed]
- Bricambert, J.; Miranda, J.; Benhamed, F.; Girard, J.; Postic, C.; Dentin, R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Investig. 2010, 120, 4316–4331. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, L.; Kothandan, G.; Manoharan, R. Berberine and Emodin abrogates breast cancer growth and facilitates apoptosis through inactivation of SIK3-induced mTOR and Akt signaling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165897. [Google Scholar] [CrossRef]
- Du, W.Q.; Zheng, J.N.; Pei, D.S. The diverse oncogenic and tumor suppressor roles of salt-inducible kinase (SIK) in cancer. Expert. Opin. Ther. Targets 2016, 20, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; Sakamoto, K.; Bayascas, J.R. LKB1-dependent signaling pathways. Annu. Rev. Biochem. 2006, 75, 137–163. [Google Scholar] [CrossRef] [PubMed]
- Molina, E.; Hong, L.; Chefetz, I. AMPKalpha-like proteins as LKB1 downstream targets in cell physiology and cancer. J. Mol. Med. 2021, 99, 651–662. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, Q.; Sun, Q.; Xu, Z.-X.; Zhou, H.; Wang, Y. LKB1 deficiency-induced metabolic reprogramming in tumorigenesis and non-neoplastic diseases. Mol. Metab. 2021, 44, 101131. [Google Scholar] [CrossRef] [PubMed]
- Bouchekioua-Bouzaghou, K.; Poulard, C.; Rambaud, J.; Lavergne, E.; Hussein, N.; Billaud, M.; Bachelot, T.; Chabaud, S.; Mader, S.; Dayan, G.; et al. LKB1 when associated with methylatedERα is a marker of bad prognosis in breast cancer. Int. J. Cancer 2014, 135, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Liao, Z.; Liang, H.; Chen, X.; Zhang, B.; Chu, L. Upregulation of liver kinase B1 predicts poor prognosis in hepatocellular carcinoma. Int. J. Oncol. 2018, 53, 1913–1926. [Google Scholar] [CrossRef] [PubMed]
- Dahmani, R.; Just, P.A.; Delay, A.; Canal, F.; Finzi, L.; Prip-Buus, C.; Lambert, M.; Sujobert, P.; Buchet-Poyau, K.; Miller, E.; et al. A novel LKB1 isoform enhances AMPK metabolic activity and displays oncogenic properties. Oncogene 2015, 34, 2337–2346. [Google Scholar] [CrossRef]
- Kim, H.; Kim, D.; Ku, S.H.; Kim, K.; Kim, S.H.; Kwon, I.C. MicroRNA-mediated non-viral direct conversion of embryonic fibroblasts to cardiomyocytes: Comparison of commercial and synthetic non-viral vectors. J. Biomater. Sci. Polym. Ed. 2017, 28, 1070–1085. [Google Scholar] [CrossRef]
- Shu, G.; Sun, H.; Zhang, T.; Zhu, A.; Lei, X.; Wang, C.; Song, A.; Deng, X. Theaflavine inhibits hepatic stellate cell activation by modulating the PKA/LKB1/AMPK/GSK3beta cascade and subsequently enhancing Nrf2 signaling. Eur. J. Pharmacol. 2023, 956, 175964. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: Improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Gyorffy, B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br. J. Pharmacol. 2024, 181, 362–374. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Liu, C.; Wang, X.; Wang, X.; Zhang, Y.; Min, W.; Yu, P.; Miao, J.; Shen, W.; Chen, S.; Zhou, S.; et al. A new LKB1 activator, piericidin analogue S14, retards renal fibrosis through promoting autophagy and mitochondrial homeostasis in renal tubular epithelial cells. Theranostics 2022, 12, 7158–7179. [Google Scholar] [CrossRef]
- Gao, Y.L.H.; Wang, P.; Wang, J.; Yao, X. SIK1 suppresses colorectal cancer metastasis and chemoresistance via the TGF-β signaling pathway. J. Cancer 2023, 14, 2455–2467. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Qi, L.; Wang, L. Overexpression of circ_CELSR1 facilitates paclitaxel resistance of ovarian cancer by regulating miR-149-5p/SIK2 axis. Anticancer Drugs 2021, 32, 496–507. [Google Scholar] [CrossRef]
- Cheng, T.C.; Sayseng, J.O.; Tu, S.H.; Juan, T.C.; Fang, C.L.; Liao, Y.C.; Chu, C.Y.; Chang, H.W.; Yen, Y.; Chen, L.C.; et al. Curcumin-induced antitumor effects on triple-negative breast cancer patient-derived xenograft tumor mice through inhibiting salt-induced kinase-3 protein. J. Food Drug Anal. 2021, 29, 622–637. [Google Scholar] [CrossRef]
- Li, J.; Xu, H.; Wang, Q.; Fu, P.; Huang, T.; Anas, O.; Zhao, H.; Xiong, N. Pard3 suppresses glioma invasion by regulating RhoA through atypical protein kinase C/NF-kappaB signaling. Cancer Med. 2019, 8, 2288–2302. [Google Scholar] [CrossRef]
- Chen, H.Y.; Lin, P.H.; Shih, Y.H.; Wang, K.L.; Hong, Y.H.; Shieh, T.M.; Huang, T.C.; Hsia, S.M. Natural Antioxidant Resveratrol Suppresses Uterine Fibroid Cell Growth and Extracellular Matrix Formation In Vitro and In Vivo. Antioxidants 2019, 8, 99. [Google Scholar] [CrossRef]
- Zhang, G.; Li, Z.; Dong, J.; Zhou, W.; Zhang, Z.; Que, Z.; Zhu, X.; Xu, Y.; Cao, N.; Zhao, A. Acacetin inhibits invasion, migration and TGF-beta1-induced EMT of gastric cancer cells through the PI3K/Akt/Snail pathway. BMC Complement. Med. Ther. 2022, 22, 10. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ma, Y.; Chen, S.; Guo, S.; Hu, J.; Yue, T.; Zhang, J.; Zhu, J.; Wang, P.; Chen, G.; et al. SPARC enhances 5-FU chemosensitivity in gastric cancer by modulating epithelial-mesenchymal transition and apoptosis. Biochem. Biophys. Res. Commun. 2021, 558, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.W.Y.; Li, X.; Yang, A.; Li, Z.; Wang, D. The anti-carcinogenesis properties of erianin in the modulation of oxidative stress-mediated apoptosis and immune response in liver cancer. Aging 2019, 11, 10284–10300. [Google Scholar] [CrossRef]
- Rui, T.; Wang, H.; Li, Q.; Cheng, Y.; Gao, Y.; Fang, X.; Ma, X.; Chen, G.; Gao, C.; Gu, Z.; et al. Deletion of ferritin H in neurons counteracts the protective effect of melatonin against traumatic brain injury-induced ferroptosis. J. Pineal Res. 2021, 70, e12704. [Google Scholar] [CrossRef]
- Weng, Q.; Sun, H.; Fang, C.; Xia, F.; Liao, H.; Lee, J.; Wang, J.; Xie, A.; Ren, J.; Guo, X.; et al. Catalytic activity tunable ceria nanoparticles prevent chemotherapy-induced acute kidney injury without interference with chemotherapeutics. Nat. Commun. 2021, 12, 1436. [Google Scholar] [CrossRef] [PubMed]



| siRNA Primer | Sequence (5′-3′) |
|---|---|
| siLKB1-1-F | GGGCCAAGCUCAUCGGCAATT |
| siLKB1-1-R | UUGCCGAUGAGCUUGGCCCTT |
| siLKB1-2-F | UGUAUAUGGUGAUGGAGUATT |
| siLKB1-2-R | UACUCCAUCACCAUAUACATT |
| siLKB1-3-F | GGGACAACAUCUACAAGUUTT |
| siLKB1-3-R | AACUUGUAGAUGUUGUCCCTT |
| siSIK2-1-F | GGUAUGUCCCGGUGAAUUATT |
| siSIK2-1-R | UAAUUCACCAGGACAUACCTT |
| siSIK2-2-F | UUGCAGAACAAGAGCUAUATT |
| siSIK2-2-R | UAUAGCUCUUGUUCUGCAATT |
| siSIK2-3-F | GGCUAGAACCAAAGGAAUUTT |
| siSIK2-3-R | AAUUCCUUUGGUUCUAGCCTT |
| siSIK3-F | GUAGAAUGGCAGAAAAGGATT |
| siSIK3-R | UCCUUUCUGCCAUUCUACTT |
| siPARD3-1-F | GAGAUAAGGAGAAGGAUAATT |
| siPARD3-1-R | UUAUCCUUCUCCUUAUCUCTT |
| siPARD3-2-F | GGGCAAAUCCCAAGAGGAATT |
| siPARD3-2-R | UUCCUCUUGGGAUUUGCCCTT |
| siPARD3-3-F | ACAUGGAGAUGGAGGAAUATT |
| siPARD3-3-R | UAUUCCUCCAUCUCCAUGUTT |
| NC-F | UUCUCCGAACGUGUCACGUTT |
| NC-R | ACGUGACACGUUCGGAGAATT |
| Gene Primer | Sequence (5′-3′) |
|---|---|
| LKB1-F | TGTCGGTGGGTATGGACAC |
| LKB1-R | CCTTGCCGTAAGAGCCTTCC |
| SIK1-F | GAGTCACCAAAACGCAGGTTG |
| SIK1-R | AGTGACGATGTAAAGCATGTCC |
| SIK2-F | TGAGCAGGTTCTTCGACTGAsT |
| SIK2-R | AGATCGCATCAGTCTCACGTT |
| SIK3-F | TCAGCAGCAACCTGAGAACT |
| SIK3-R | ACAAGGGACGGTGCCCATAG |
| PARD3-F | CAGACAGAACTACTAACTTCGCC |
| PARD3-R | ATGC CTCGGATGAAGAGTCCT |
| β-actin-F | CTCACCATGGATGATGATATCGC |
| β-actin-R | AGGAATCCTTCTGACCCATGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Liu, Q.; Liu, L.; Wang, D.; Liu, J.; Zhu, Q.; Xu, Z.; Chen, Q.; Xu, W. Discovery of the Natural Bibenzyl Compound Erianin in Dendrobium Inhibiting the Growth and EMT of Gastric Cancer through Downregulating the LKB1-SIK2/3-PARD3 Pathway. Int. J. Mol. Sci. 2024, 25, 7973. https://doi.org/10.3390/ijms25147973
Wei X, Liu Q, Liu L, Wang D, Liu J, Zhu Q, Xu Z, Chen Q, Xu W. Discovery of the Natural Bibenzyl Compound Erianin in Dendrobium Inhibiting the Growth and EMT of Gastric Cancer through Downregulating the LKB1-SIK2/3-PARD3 Pathway. International Journal of Molecular Sciences. 2024; 25(14):7973. https://doi.org/10.3390/ijms25147973
Chicago/Turabian StyleWei, Xin, Qunshan Liu, Liu Liu, Dan Wang, Jiajia Liu, Qizhi Zhu, Ziming Xu, Qi Chen, and Weiping Xu. 2024. "Discovery of the Natural Bibenzyl Compound Erianin in Dendrobium Inhibiting the Growth and EMT of Gastric Cancer through Downregulating the LKB1-SIK2/3-PARD3 Pathway" International Journal of Molecular Sciences 25, no. 14: 7973. https://doi.org/10.3390/ijms25147973
APA StyleWei, X., Liu, Q., Liu, L., Wang, D., Liu, J., Zhu, Q., Xu, Z., Chen, Q., & Xu, W. (2024). Discovery of the Natural Bibenzyl Compound Erianin in Dendrobium Inhibiting the Growth and EMT of Gastric Cancer through Downregulating the LKB1-SIK2/3-PARD3 Pathway. International Journal of Molecular Sciences, 25(14), 7973. https://doi.org/10.3390/ijms25147973




