The Integrated Bioinformatic Approach Reveals the Prognostic Significance of LRP1 Expression in Ovarian Cancer
Abstract
:1. Introduction
2. Results
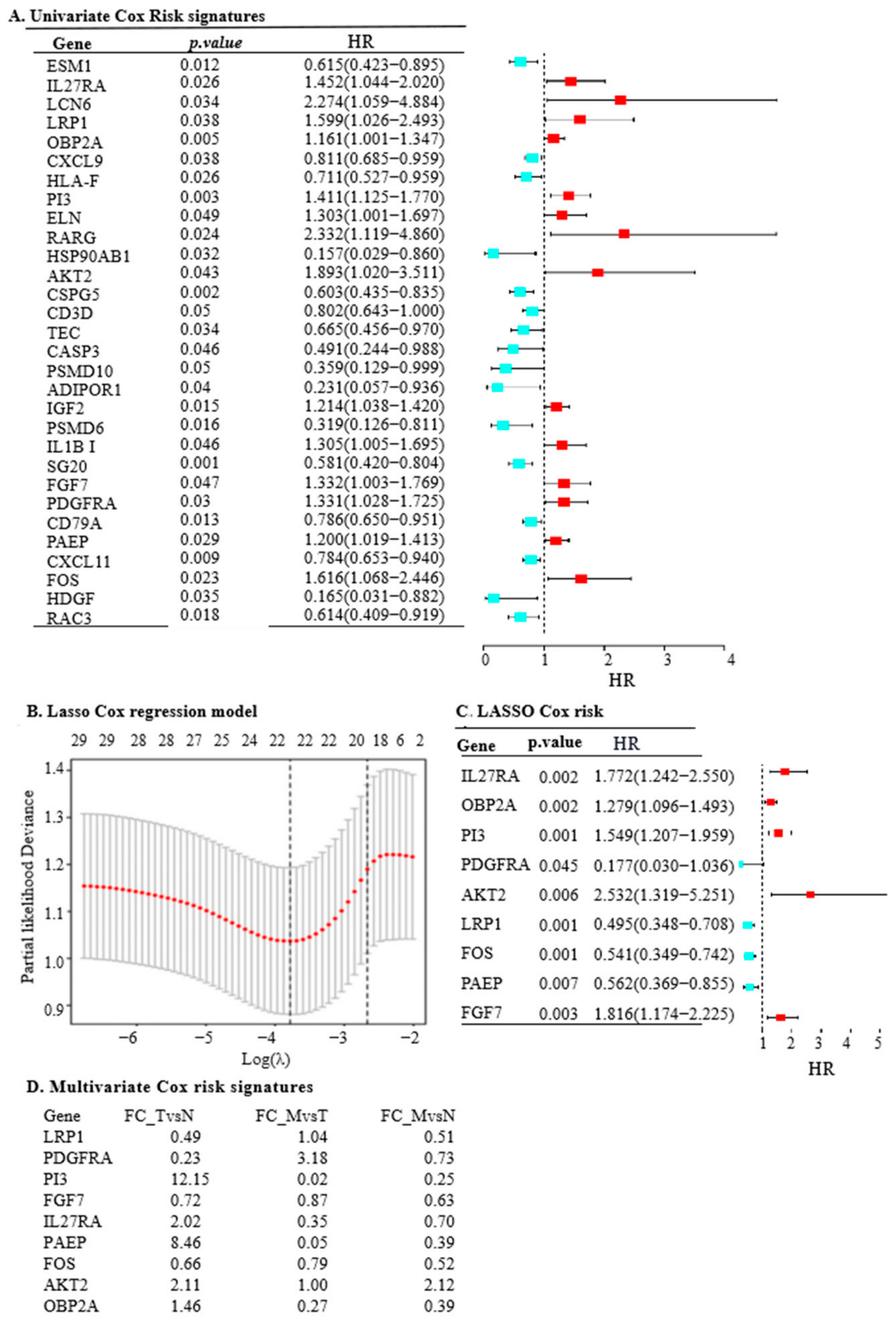
2.1. LRP1, IL27RA, and FGF7 Emerge as Key Cancer-Related IRGs in OC
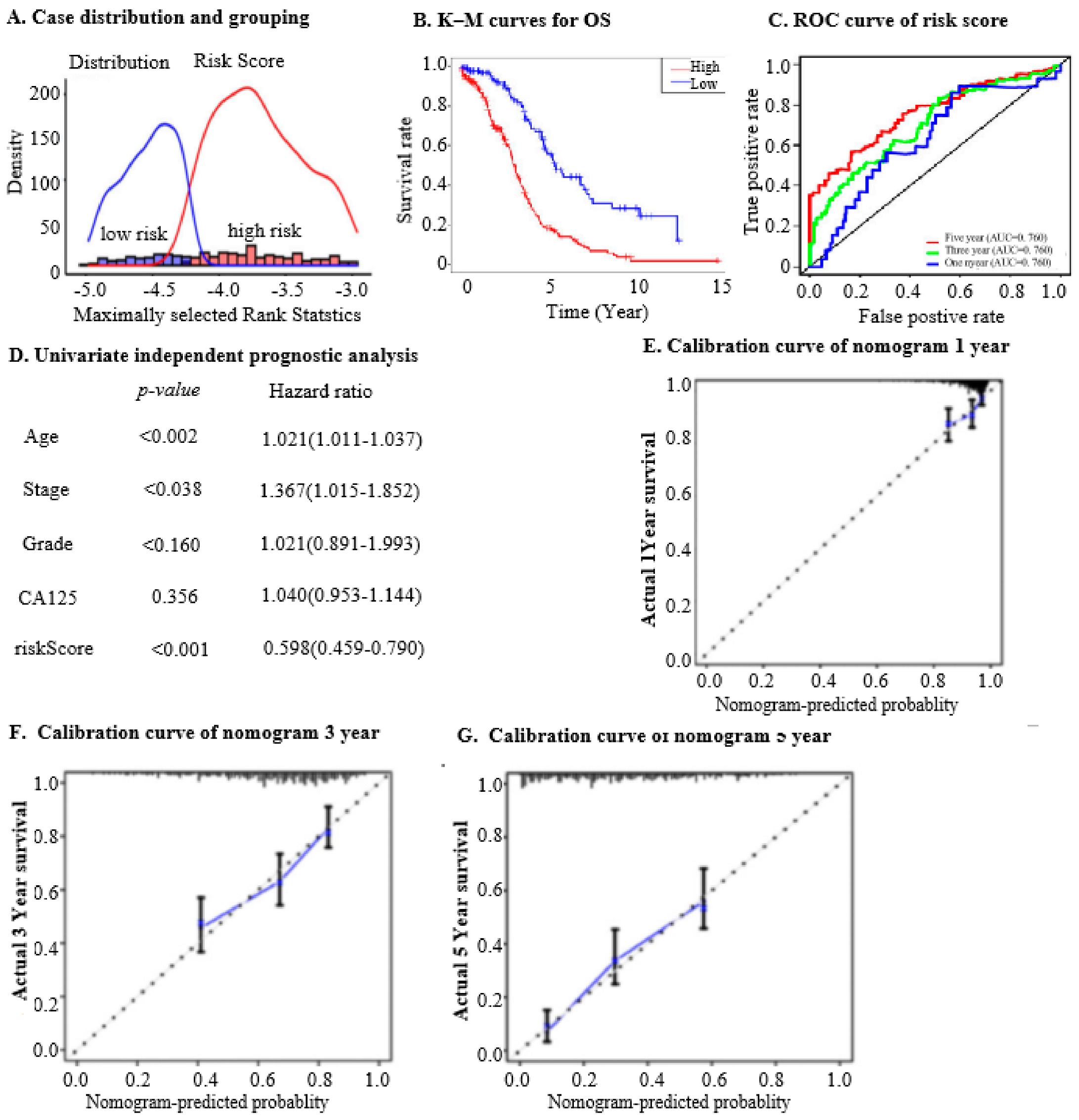
2.2. The Nine IRGs Reflect Poor Survival and Tumour Progression in OC
2.3. The Nine IRGs Identified as a Prognostic Marker in OC
2.4. Prognostic Implications of Nine IRGs in OC
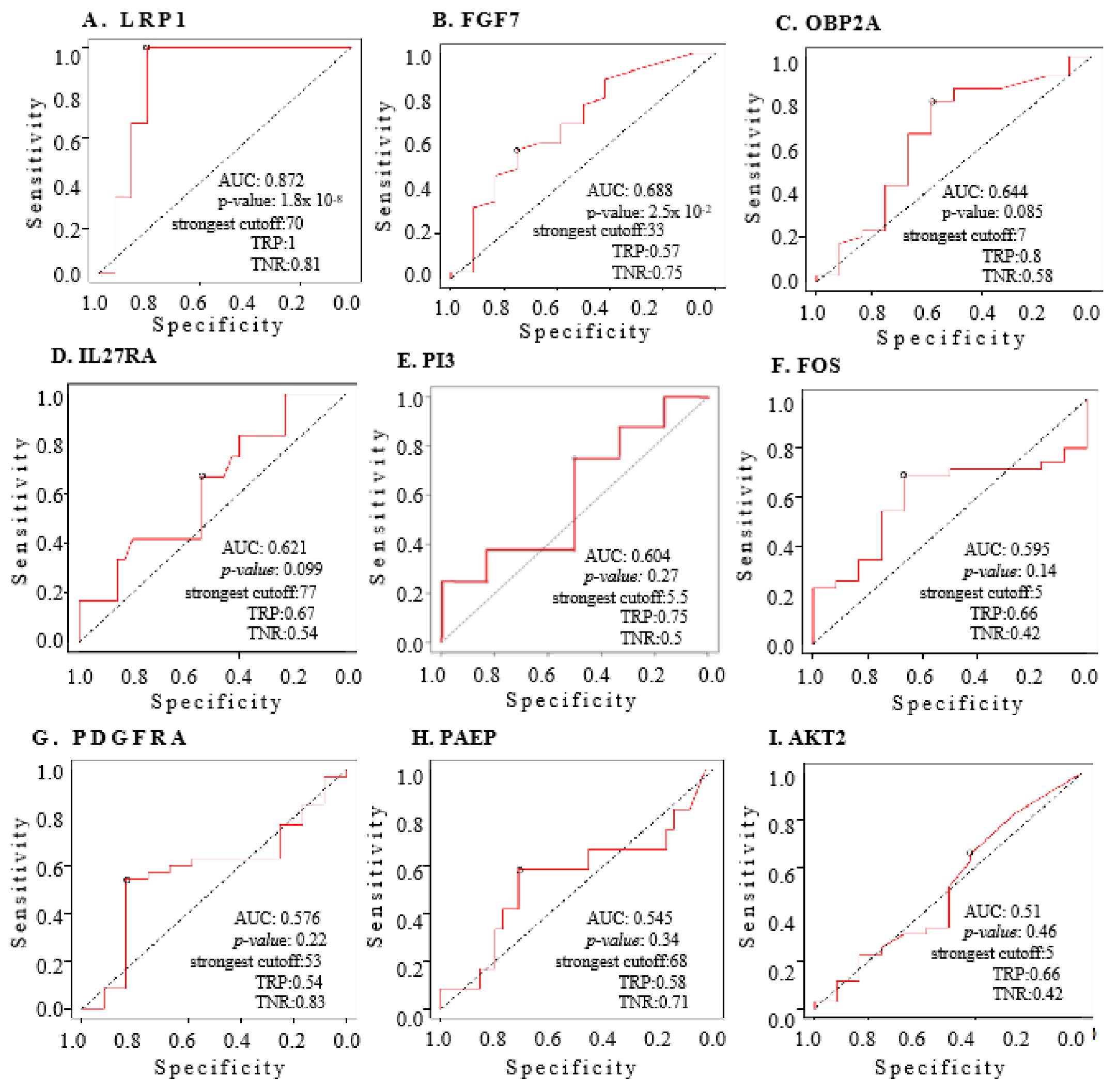
2.5. The Nine IRGs as Indices for Diagnosing OC
2.6. The Nine IRGs Are Enriched to Indicate Enhanced Oncogenic Activities in OC
2.7. The Nine IRGs Reflect Immunosuppression in High-Risk OC Patients
2.8. The Nine IRGs Expression Signatures Predict a Worse Response to Immunotherapy in OC
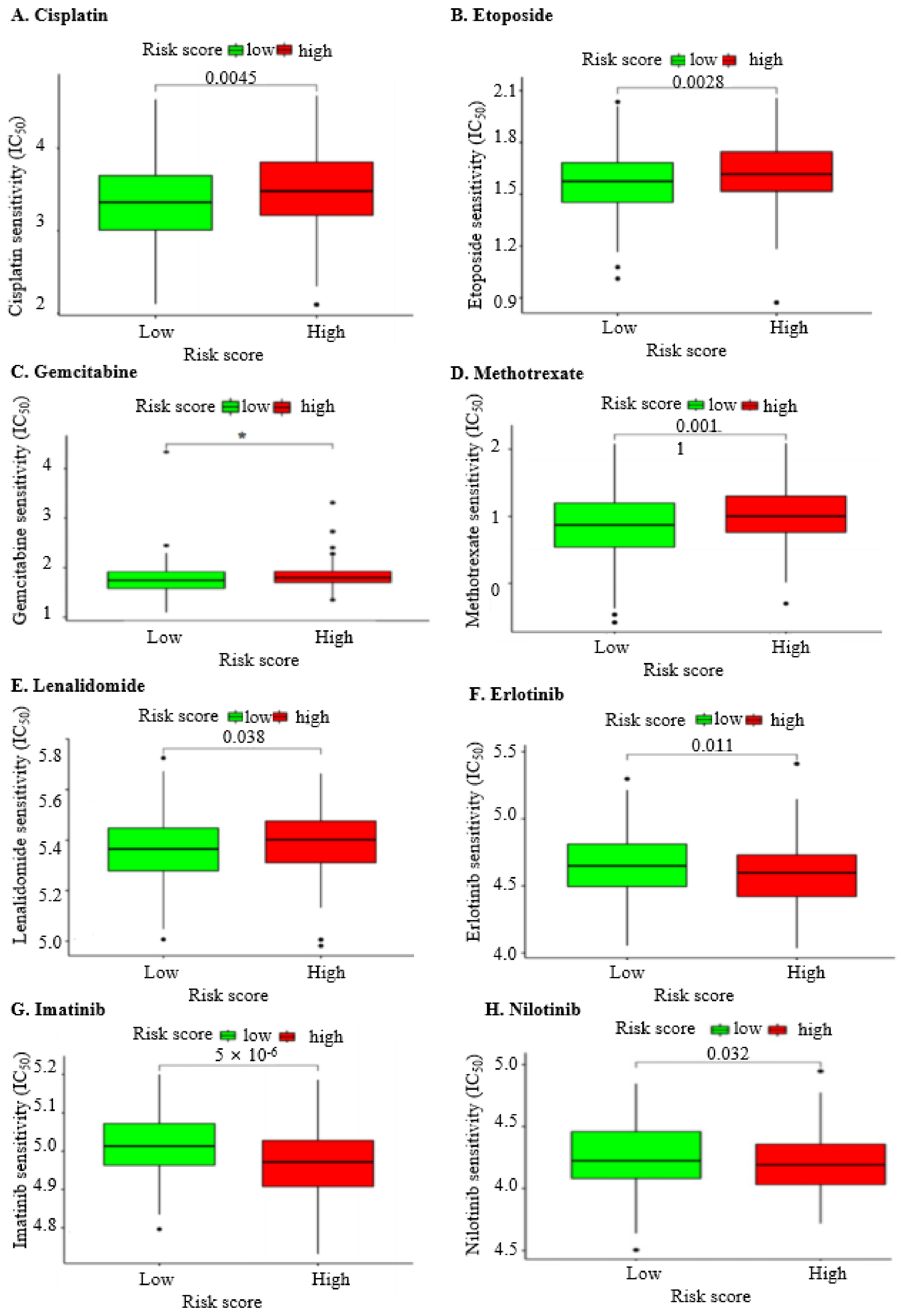
2.9. The Nine IRGs Indicate a Poorer Response to Chemotherapy in OC
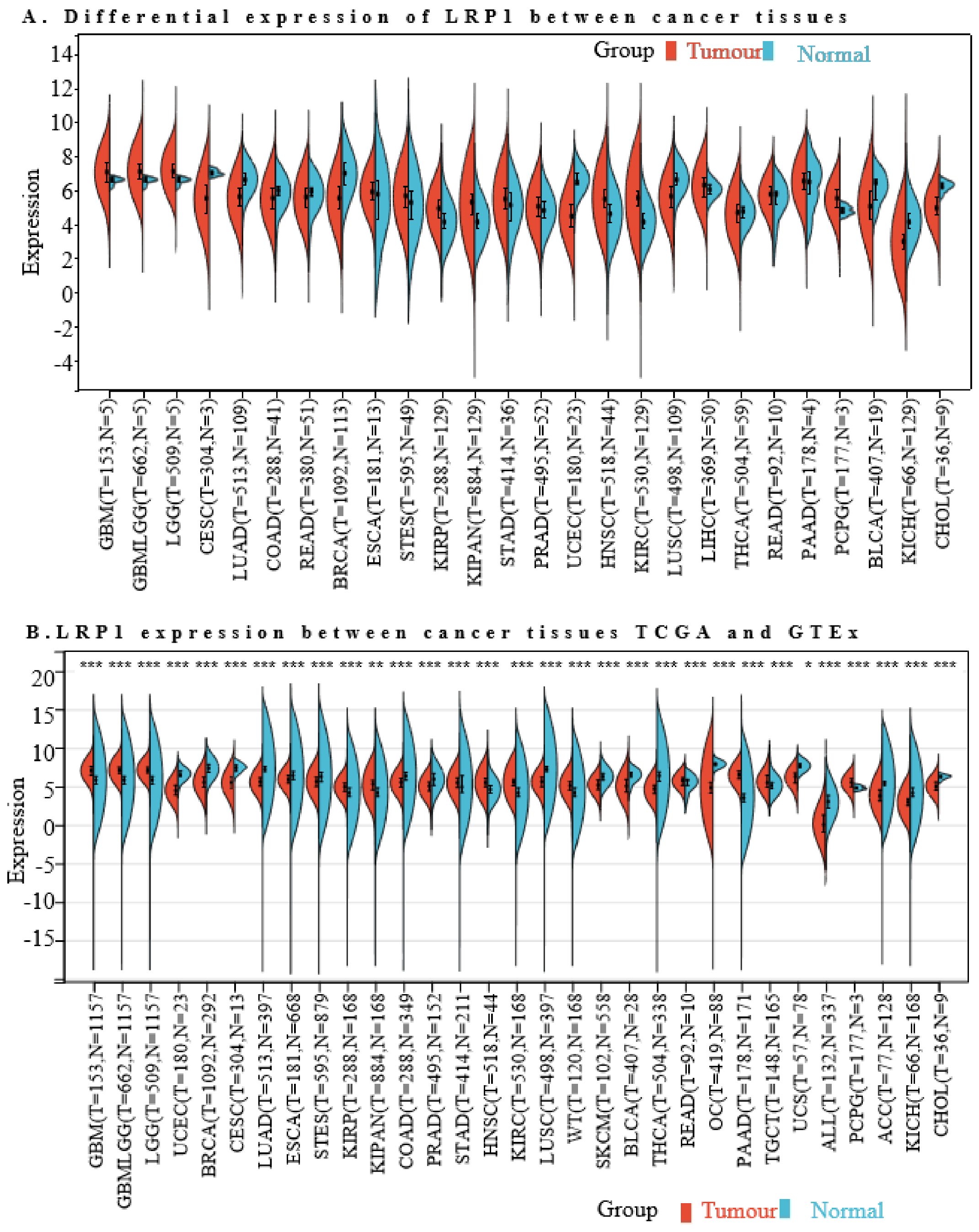
2.10. The LRP1 mRNA Levels in Cancer Types
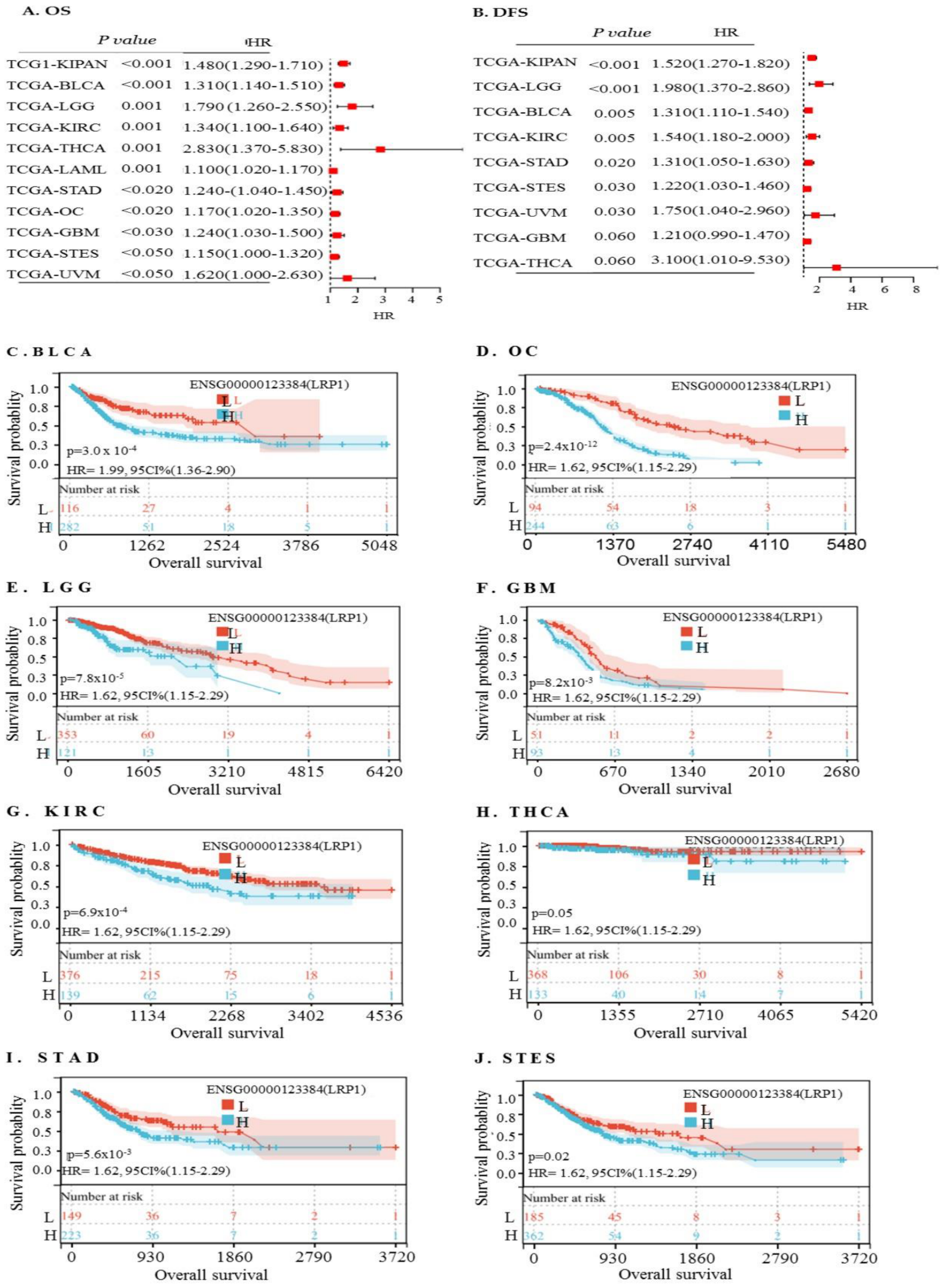
2.11. High LRP1 Expression Predicts Poorer Prognosis in Rare Cancer Types
2.12. The Relationship between LRP1 Expression and Immune Infiltration across Solid Tumours
2.13. Correlation between LRP1 Expression and Tumour Purity in Cancers
2.14. Functional Enrichment Analysis of LRP1 across Cancers
3. Discussion
Shortcomings of the Current Study
4. Materials and Methods
4.1. Dataset Selection
4.2. Differential Gene Analysis and Functional Enrichment of IRGs
4.3. Prognostic Risk Model Construction and Validation
4.4. Prognostic Potential Assessment of Key Genes in OC Patients
4.5. Tumour-Infiltrating Immune Cells Fraction Calculation
4.6. Estimation of the Immunoreactivity
4.7. Detection of the Diagnostic Values of Risk Indicators in OC
4.8. Drug Susceptibility Testing
4.9. Analysis of IRG Expression and Survival Analysis in OC
4.10. Relationship between the Expression of Nine IRGs and Immunity
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, S.; Cheng, C.; Lin, Z.; Xiao, L.; Su, X.; Zheng, L.; Mu, Y.; Liao, M.; Ouyang, R.; Li, W.; et al. The global burden and associated factors of ovarian cancer in 1990–2019: Findings from the Global Burden of Disease Study 2019. BMC Public Health 2022, 22, 1455. [Google Scholar] [CrossRef] [PubMed]
- Webb, P.M.; Jordan, S.J. Global epidemiology of epithelial ovarian cancer. Nat. Rev. Clin. Oncol. 2024, 21, 389–400. [Google Scholar] [CrossRef]
- Chandra, A.; Pius, C.; Nabeel, M.; Nair, M.; Vishwanatha, J.K.; Ahmad, S.; Basha, R. Ovarian cancer: Current status and strategies for improving therapeutic outcomes. Cancer Med. 2019, 8, 7018–7031. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Zhang, X.; Pandey, V.; Garg, M. Neo-vascularization-based therapeutic perspectives in advanced ovarian cancer. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188888. [Google Scholar] [CrossRef]
- Zuckerbrot-Schuldenfrei, M.; Aviel-Ronen, S.; Zilberberg, A.; Efroni, S. Ovarian cancer is detectable from peripheral blood using machine learning over T-cell receptor repertoires. Brief. Bioinform. 2024, 25, bbae075. [Google Scholar] [CrossRef]
- Zou, R.; Jiang, Q.; Luo, X.; Chen, M.; Yuan, L.; Yao, L. Cytoreductive surgery is feasible in patients with limited regional platinum-resistant recurrent ovarian cancer. World J. Surg. Oncol. 2023, 21, 375. [Google Scholar] [CrossRef] [PubMed]
- Millert-Kalińska, S.; Przybylski, M.; Pruski, D.; Stawicka-Niełacna, M.; Mądry, R. Epithelial Ovarian Cancer—Varied Treatment Results. Healthcare 2023, 11, 2043. [Google Scholar] [CrossRef]
- Bachmann, C. New Achievements from Molecular Biology and Treatment Options for Refractory/Relapsed Ovarian Cancer—A Systematic Review. Cancers 2023, 15, 5356. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Tan, Y.Q.; Wu, M.M.; Ma, L.; Zhu, T.; Lobie, P.E.; Pandey, V. Long non-coding RNAs in recurrent ovarian cancer: Theranostic perspectives. Cancer Lett. 2021, 502, 97–107. [Google Scholar] [CrossRef]
- Yu, L.; Ding, Y.; Wan, T.; Deng, T.; Huang, H.; Liu, J. Significance of CD47 and Its Association with Tumor Immune Microenvironment Heterogeneity in Ovarian Cancer. Front. Immunol. 2021, 12, 768115. [Google Scholar] [CrossRef]
- Worzfeld, T.; Von Strandmann, E.P.; Huber, M.; Adhikary, T.; Wagner, U.; Reinartz, S.; Müller, R. The Unique Molecular and Cellular Microenvironment of Ovarian Cancer. Front. Oncol. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shen, M.; Wu, L.; Yang, H.; Yao, Y.; Yang, Q.; Du, J.; Liu, L.; Li, Y.; Bai, Y. Stromal cells in the tumor microenvironment: Accomplices of tumor progression? Cell Death Dis. 2023, 14, 587. [Google Scholar] [CrossRef] [PubMed]
- Meurette, O.; Mehlen, P. Notch Signaling in the Tumor Microenvironment. Cancer Cell 2018, 34, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Sun, H.; Liu, S.; Liang, B.; Bai, H.; Wang, S.; Li, S. Identification of a Prognostic Signature for Ovarian Cancer Based on the Microenvironment Genes. Front. Genet. 2021, 12, 680413. [Google Scholar] [CrossRef] [PubMed]
- Şenbabaoğlu, Y.; Gejman, R.S.; Winer, A.G.; Van Allen, E.M.; de Velasco, G.; Miao, D.; Ostrovnaya, I.; Drill, E.; Luna, A.; Weinhold, N.; et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016, 17, 231. [Google Scholar] [CrossRef] [PubMed]
- Winslow, S.; Lindquist, K.E.; Edsjö, A.; Larsson, C. The expression pattern of matrix-producing tumor stroma is of prognostic importance in breast cancer. BMC Cancer 2016, 16, 841. [Google Scholar] [CrossRef] [PubMed]
- Goode, E.L.; Block, M.S.; Kalli, K.R.; Vierkant, R.A.; Chen, W.; Fogarty, Z.C.; Gentry-Maharaj, A.; Toloczko, A.; Hein, A.; Bouligny, A.L.; et al. Dose-Response Association of CD8+ Tumor-Infiltrating Lymphocytes and Survival Time in High-Grade Serous Ovarian Cancer. JAMA Oncol. 2017, 3, e173290. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, X.; Chen, X.; Shi, X.; Chen, S. Prognostic and predictive value of immune/stromal-related gene biomarkers in renal cell carcinoma. Oncol. Lett. 2020, 20, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Olalekan, S.; Xie, B.; Back, R.; Eckart, H.; Basu, A. Characterizing the tumor microenvironment of metastatic ovarian cancer by single-cell transcriptomics. Cell Rep. 2021, 35, 109165. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Huang, Y.-F.; Chang, T.-H.; Chen, C.-C.; Wu, P.-Y.; Huang, S.-C.; Chou, C.-Y. COL11A1 activates cancer-associated fibroblasts by modulating TGF-β3 through the NF-κB/IGFBP2 axis in ovarian cancer cells. Oncogene 2021, 40, 4503–4519. [Google Scholar] [CrossRef]
- Shen, X.; Gu, X.; Ma, R.; Li, X.; Wang, J. Identification of the Immune Signatures for Ovarian Cancer Based on the Tumor Immune Microenvironment Genes. Front. Cell Dev. Biol. 2022, 10, 772701. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.A.; Orozco, C.; Xiao, J.; Javellana, M.; Lengyel, E. The Effects of Chemotherapeutics on the Ovarian Cancer Microenvironment. Cancers 2021, 13, 3136. [Google Scholar] [CrossRef]
- Salama, Y.; Takahashi, S.; Tsuda, Y.; Okada, Y.; Hattori, K.; Heissig, B. YO2 Induces Melanoma Cell Apoptosis through p53-Mediated LRP1 Downregulation. Cancers 2022, 15, 288. [Google Scholar] [CrossRef]
- Au, D.T.; Strickland, D.K.; Muratoglu, S.C. The LDL Receptor-Related Protein 1: At the Crossroads of Lipoprotein Metabolism and Insulin Signaling. J. Diabetes Res. 2017, 2017, 8356537. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-C.; A Seo, J.; Lee, H.; Uner, A.; Yang, W.-M.; Rodrigues, K.C.d.C.; Kim, H.J.; Li, W.; Campbell, J.N.; Dagon, Y.; et al. LRP1 regulates food intake and energy balance in GABAergic neurons independently of leptin action. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E379–E389. [Google Scholar] [CrossRef] [PubMed]
- Byrne, J.C.; Gabhann, J.N.; Stacey, K.B.; Coffey, B.M.; McCarthy, E.; Thomas, W.; Jefferies, C.A. Bruton’s Tyrosine Kinase Is Required for Apoptotic Cell Uptake via Regulating the Phosphorylation and Localization of Calreticulin. J. Immunol. 2013, 190, 5207–5215. [Google Scholar] [CrossRef]
- He, L.; Guo, Z.; Wang, W.; Tian, S.; Lin, R. FUT2 inhibits the EMT and metastasis of colorectal cancer by increasing LRP1 fucosylation. Cell Commun. Signal. 2023, 21, 63. [Google Scholar] [CrossRef]
- Xing, P.; Liao, Z.; Ren, Z.; Zhao, J.; Song, F.; Wang, G.; Chen, K.; Yang, J. Roles of low-density lipoprotein receptor-related protein 1 in tumors. Chin. J. Cancer 2016, 35, 6. [Google Scholar] [CrossRef]
- Kang, H.S.; Kim, J.; Lee, H.-J.; Kwon, B.-M.; Lee, D.-K.; Hong, S.-H. LRP1-dependent pepsin clearance induced by 2′-hydroxycinnamaldehyde attenuates breast cancer cell invasion. Int. J. Biochem. Cell Biol. 2014, 53, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Sizova, O.; John, L.S.; Ma, Q.; Molldrem, J.J. Multi-faceted role of LRP1 in the immune system. Front. Immunol. 2023, 14, 1166189. [Google Scholar] [CrossRef]
- Chen, K.; Martens, Y.A.; Meneses, A.; Ryu, D.H.; Lu, W.; Raulin, A.C.; Li, F.; Zhao, J.; Chen, Y.; Jin, Y.; et al. LRP1 is a neuronal receptor for α-synuclein uptake and spread. Mol. Neurodegener. 2022, 17, 57. [Google Scholar] [CrossRef] [PubMed]
- Mitteer, D.R.; Greer, B.D. Using GraphPad Prism’s Heat Maps for Efficient, Fine-Grained Analyses of Single-Case Data. Behav. Anal. Pract. 2022, 15, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Song, Z.; Huang, L.; Guo, Z.; Tong, B.; Sun, M.; Zhao, J.; Zhang, H.; Zhang, Z.; Li, G. Tumor purity as a prognosis and immunotherapy relevant feature in cervical cancer. Aging 2021, 13, 24768–24785. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.-F.; Liu, C.-J.; Liu, L.-L.; Zhang, Q.; Guo, A.-Y. Expression profile of immune checkpoint genes and their roles in predicting immunotherapy response. Brief. Bioinform. 2021, 22, bbaa176. [Google Scholar] [CrossRef] [PubMed]
- De Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]
- Caruntu, A.; Moraru, L.; Lupu, M.; Vasilescu, F.; Dumitrescu, M.; Cioplea, M.; Popp, C.; Dragusin, A.; Caruntu, C.; Zurac, S. Prognostic Potential of Tumor-Infiltrating Immune Cells in Resectable Oral Squamous Cell Carcinoma. Cancers 2021, 13, 2268. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Sharma, A.; Parambath, S.V.; Gul, I.; Zhang, X.; Lobie, P.E.; Qin, P.; Pandey, V. Machine Learning for Endometrial Cancer Prediction and Prognostication. Front. Oncol. 2022, 12, 852746. [Google Scholar] [CrossRef] [PubMed]
- Jelovac, D.; Armstrong, D.K. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J. Clin. 2011, 61, 183–203. [Google Scholar] [CrossRef]
- Rampes, S.; Choy, S.-P. Early diagnosis of symptomatic ovarian cancer in primary care in the UK: Opportunities and challenges. Prim. Health Care Res. Dev. 2022, 23, e52. [Google Scholar] [CrossRef]
- Alatise, K.L.; Gardner, S.; Alexander-Bryant, A. Mechanisms of Drug Resistance in Ovarian Cancer and Associated Gene Targets. Cancers 2022, 14, 6246. [Google Scholar] [CrossRef] [PubMed]
- Kandalaft, L.E.; Odunsi, K.; Coukos, G. Immunotherapy in Ovarian Cancer: Are We There Yet? J. Clin. Oncol. 2019, 37, 2460–2471. [Google Scholar] [CrossRef]
- Nallasamy, P.; Nimmakayala, R.K.; Parte, S.; Are, A.C.; Batra, S.K.; Ponnusamy, M.P. Tumor microenvironment enriches the stemness features: The architectural event of therapy resistance and metastasis. Mol. Cancer 2022, 21, 225. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Su, X.; Wang, C.; Xu, M. Integrated analysis of prognostic immune-related genes in the tumor microenvironment of ovarian cancer. Ann. Transl. Med. 2022, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Zhang, P.; Zhao, F.; Zhang, S. A novel immune-related prognostic signature in epithelial ovarian carcinoma. Aging 2021, 13, 10289–10311. [Google Scholar] [CrossRef] [PubMed]
- Quan, Q.; Xiong, X.; Wu, S.; Yu, M. Identification of Immune-Related Key Genes in Ovarian Cancer Based on WGCNA. Front. Genet. 2021, 12, 760225. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ma, J.; Zhao, H.; Wang, Q.; Guo, X.; Chen, L.; Cao, Z.; Xu, J.; Zhang, B.; Zhou, X. Serum Exosomes from Epithelial Ovarian Cancer Patients Contain LRP1, Which Promotes the Migration of Epithelial Ovarian Cancer Cell. Mol. Cell. Proteom. 2023, 22, 100520. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Ding, B.; Dai, Z.; Yin, H.; Ding, Y.; Liu, S.; Zhang, K.; Lin, H.; Xiao, Z.; Shen, Y. Cancer-associated fibroblast-secreted FGF7 as an ovarian cancer progression promoter. J. Transl. Med. 2024, 22, 280. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Zhang, Y.; Qu, K.; Yang, X.; Fu, J.; Chen, W.; Li, X. MicroRNA-29B (mir-29b) regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3. Oncotarget 2015, 6, 40799–40814. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Yang, L.; Xu, T.; Si, L.; Cui, C.; Sheng, X.; Chi, Z.; Mao, L.; Lian, B.; Tang, B.; et al. A Functional Synonymous Variant in PDGFRA Is Associated with Better Survival in Acral Melanoma. J. Cancer 2020, 11, 2945–2956. [Google Scholar] [CrossRef]
- Zou, J.; Li, Y.; Liao, N.; Liu, J.; Zhang, Q.; Luo, M.; Xiao, J.; Chen, Y.; Wang, M.; Chen, K.; et al. Identification of key genes associated with polycystic ovary syndrome (PCOS) and ovarian cancer using an integrated bioinformatics analysis. J. Ovarian Res. 2022, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Ferrer, L.; Rößler, K.; Haustein, V.; Schröder, C.; Wicklein, D.; Maltseva, D.; Khaustova, N.; Samatov, T.; Tonevitsky, A.; Mahner, S.; et al. c-FOS suppresses ovarian cancer progression by changing adhesion. Br. J. Cancer 2014, 110, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, J.-S.; Yu, J.-C.; Cui, Q.-C.; Zhou, W.-X.; Kang, W.-M.; Ma, Z.-Q. Negative Association of c-fos Expression as a Favorable Prognostic Indicator in Gastric Cancer. Arch. Med Res. 2010, 41, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Wichert, B.; Milde-Langosch, K.; Galatenko, V.; Schmalfeldt, B.; Oliveira-Ferrer, L. Prognostic role of the sialyltransferase ST6GAL1 in ovarian cancer. Glycobiology 2018, 28, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Au, D.T.; Ying, Z.; Hernández-Ochoa, E.O.; Fondrie, W.E.; Hampton, B.; Migliorini, M.; Galisteo, R.; Schneider, M.F.; Daugherty, A.; Rateri, D.L.; et al. LRP1 (Low-Density Lipoprotein Receptor-Related Protein 1) Regulates Smooth Muscle Contractility by Modulating Ca2+ Signaling and Expression of Cytoskeleton-Related Proteins. Arter. Thromb. Vasc. Biol. 2018, 38, 2651–2664. [Google Scholar] [CrossRef] [PubMed]
- Nikolakopoulou, A.M.; Wang, Y.; Ma, Q.; Sagare, A.P.; Montagne, A.; Huuskonen, M.T.; Rege, S.V.; Kisler, K.; Dai, Z.; Körbelin, J.; et al. Endothelial LRP1 protects against neurodegeneration by blocking cyclophilin A. J. Exp. Med. 2021, 218, e20202207. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Strickland, D.K. LRP: A multifunctional scavenger and signaling receptor. J. Clin. Investig. 2001, 108, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Boucher, P.; Herz, J. Signaling through LRP1: Protection from atherosclerosis and beyond. Biochem. Pharmacol. 2011, 81, 1–5. [Google Scholar] [CrossRef]
- May, P.; Woldt, E.; Matz, R.L.; Boucher, P. The LDL receptor-related protein (LRP) family: An old family of proteins with new physiological functions. Ann. Med. 2007, 39, 219–228. [Google Scholar] [CrossRef]
- He, Z.; Wang, G.; Wu, J.; Tang, Z.; Luo, M. The molecular mechanism of LRP1 in physiological vascular homeostasis and signal transduction pathways. Biomed. Pharmacother. 2021, 139, 111667. [Google Scholar] [CrossRef]
- Xu, X.; Wang, Y.; Bryce, N.S.; Tang, K.; Meagher, N.S.; Kang, E.Y.; Kelemen, L.E.; Köbel, M.; Ramus, S.J.; Friedlander, M.; et al. Targeting the actin/tropomyosin cytoskeleton in epithelial ovarian cancer reveals multiple mechanisms of synergy with anti-microtubule agents. Br. J. Cancer 2021, 125, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Laumont, C.M.; Nelson, B.H. IgA transcytosis: A new weapon in the immune response to cancer? Cancer Cell 2021, 39, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zheng, Y.; Liu, X.; Ji, R.; Chen, Z.; Guo, Q.; Wu, G.; Wang, Y.; Zhou, Y. Comprehensive analysis of gene regulation network and immune signatures of prognostic biomarker YAP1 in pancreatic cancer. J. Cancer 2020, 11, 6960–6969. [Google Scholar] [CrossRef] [PubMed]
- Lippitz, B.E. Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol. 2013, 14, e218–e228. [Google Scholar] [CrossRef] [PubMed]
- Llano-León, M.; Martínez-Enriquez, L.C.; Rodríguez-Bohórquez, O.M.; Velandia-Vargas, E.A.; Lalinde-Ruíz, N.; Villota-Álava, M.A.; Rodríguez-Rodríguez, I.J.; Montilla-Velásquez, M.d.P.; Parra-López, C.A. Effect of neoadjuvant chemotherapy on tumor immune infiltration in breast cancer patients: Systematic review and meta-analysis. PLoS ONE 2023, 18, e0277714. [Google Scholar] [CrossRef] [PubMed]
- Laganà, A.S.; Sofo, V.; Vitale, S.G.; Triolo, O. Epithelial ovarian cancer inherent resistance: May the pleiotropic interaction between reduced immunosurveillance and drug-resistant cells play a key role? Gynecol. Oncol. Rep. 2016, 18, 57–58. [Google Scholar] [CrossRef] [PubMed]
- Laganà, A.S.; Colonese, F.; Colonese, E.; Sofo, V.; Salmeri, F.M.; Granese, R.; Chiofalo, B.; Ciancimino, L.; Triolo, O. Cytogenetic analysis of epithelial ovarian cancer’s stem cells: An overview on new diagnostic and therapeutic perspectives. Eur. J. Gynaecol. Oncol. 2015, 36, 495–505. [Google Scholar] [PubMed]
- Barra, F.; Laganà, A.S.; Ghezzi, F.; Casarin, J.; Ferrero, S. Nintedanib for Advanced Epithelial Ovarian Cancer: A Change of Perspective? Summary of Evidence from a Systematic Review. Gynecol. Obstet. Investig. 2018, 84, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.L.; Cummings, M.; Thangavelu, A.; Theophilou, G.; de Jong, D.; Orsi, N.M. Barriers to Immunotherapy in Ovarian Cancer: Metabolic, Genomic, and Immune Perturbations in the Tumour Microenvironment. Cancers 2021, 13, 6231. [Google Scholar] [CrossRef]
- Lo, C.S.; Sanii, S.; Kroeger, D.R.; Milne, K.; Talhouk, A.; Chiu, D.S.; Rahimi, K.; Shaw, P.A.; Clarke, B.A.; Nelson, B.H. Neoadjuvant Chemotherapy of Ovarian Cancer Results in Three Patterns of Tumor-Infiltrating Lymphocyte Response with Distinct Implications for Immunotherapy. Clin. Cancer Res. 2017, 23, 925–934. [Google Scholar] [CrossRef]
- Le Saux, O.; Ray-Coquard, I.; Labidi-Galy, S.I. Challenges for immunotherapy for the treatment of platinum resistant ovarian cancer. Semin. Cancer Biol. 2021, 77, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhang, J.; Zeng, D.; Sun, H.; Rong, X.; Shi, M.; Bin, J.; Liao, Y.; Liao, W. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I–III colon cancer. Cancer Immunol. Immunother. 2019, 68, 433–442. [Google Scholar] [CrossRef]
- Burugu, S.; Asleh-Aburaya, K.; Nielsen, T.O. Immune infiltrates in the breast cancer microenvironment: Detection, characterization and clinical implication. Breast Cancer 2017, 24, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Kostova, I.; Mandal, R.; Becker, S.; Strebhardt, K. The role of caspase-8 in the tumor microenvironment of ovarian cancer. Cancer Metastasis Rev. 2021, 40, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Garcia, A.; Lynn, R.C.; Poussin, M.; Eiva, M.A.; Shaw, L.C.; O’connor, R.S.; Minutolo, N.G.; Casado-Medrano, V.; Lopez, G.; Matsuyama, T.; et al. CAR-T cell-mediated depletion of immunosuppressive tumor-associated macrophages promotes endogenous antitumor immunity and augments adoptive immunotherapy. Nat. Commun. 2021, 12, 877. [Google Scholar] [CrossRef] [PubMed]
- Brunell, A.E.; Lahesmaa, R.; Autio, A.; Thotakura, A.K. Exhausted T cells hijacking the cancer-immunity cycle: Assets and liabilities. Front. Immunol. 2023, 14, 1151632. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.P.; Alizadeh, A.A.; Tang, C.; Myklebust, J.H.; Varghese, B.; Gill, S.; Jan, M.; Cha, A.C.; Chan, C.K.; Tan, B.T.; et al. Anti-CD47 Antibody Synergizes with Rituximab to Promote Phagocytosis and Eradicate Non-Hodgkin Lymphoma. Cell 2010, 142, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Konishi, I. Molecular Histopathology for Establishing Diagnostic Method and Clinical Therapy for Ovarian Carcinoma. J. Clin. Med. Res. 2023, 15, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Bougen, N.; Amiry, N.; Yuan, Y.; Kong, X.; Pandey, V.; Vidal, L.; Perry, J.; Zhu, T.; Lobie, P. Trefoil factor 1 suppression of E-CADHERIN enhances prostate carcinoma cell invasiveness and metastasis. Cancer Lett. 2013, 332, 19–29. [Google Scholar] [CrossRef]
- Wang, X.-N.; Wang, S.-J.; Pandey, V.; Chen, P.; Li, Q.; Wu, Z.-S.; Wu, Q.; Lobie, P.E. Trefoil Factor 3 as a Novel Biomarker to Distinguish between Adenocarcinoma and Squamous Cell Carcinoma. Medicine 2015, 94, e860. [Google Scholar] [CrossRef]
- Vidal, L.J.-P.; Perry, J.K.; Vouyovitch, C.M.; Pandey, V.; Brunet-Dunand, S.E.; Mertani, H.C.; Liu, D.-X.; Lobie, P.E. PAX5α Enhances the Epithelial Behavior of Human Mammary Carcinoma Cells. Mol. Cancer Res. 2010, 8, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Yuan, Y.; Chong, Q.-Y.; Yang, Y.; Li, R.; Li, X.; Kong, X.; Qian, P.; Xiong, Z.; Pandey, V.; et al. Autocrine Prolactin Stimulates Endometrial Carcinoma Growth and Metastasis and Reduces Sensitivity to Chemotherapy. Endocrinology 2017, 158, 1595–1611. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; You, M.-L.; Chong, Q.-Y.; Pandey, V.; Zhuang, Q.-S.; Liu, D.-X.; Ma, L.; Zhu, T.; Lobie, P.E. Autocrine Human Growth Hormone Promotes Invasive and Cancer Stem Cell-Like Behavior of Hepatocellular Carcinoma Cells by STAT3 Dependent Inhibition of CLAUDIN-1 Expression. Int. J. Mol. Sci. 2017, 18, 1274. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hao, J.; Tan, Y.Q.; Zhu, T.; Pandey, V.; Lobie, P.E. CXC Chemokine Signaling in Progression of Epithelial Ovarian Cancer: Theranostic Perspectives. Int. J. Mol. Sci. 2022, 23, 2642. [Google Scholar] [CrossRef] [PubMed]
- Aldaqal, S.M.; Bakhsh, T.M.; Almarhabi, Y.A.; Fakiha, M.G. Thoracic actinomycosis presented with tracheoesophageal fistula and fatal pul-monary infection. Saudi Med. J. 2004, 25, 1471–1473. [Google Scholar] [PubMed]
- Tung, C.S.; Mok, S.C.; Tsang, Y.T.M.; Zu, Z.; Song, H.; Liu, J.; Deavers, M.T.; Malpica, A.; Wolf, J.K.; Lu, K.H.; et al. PAX2 expression in low malignant potential ovarian tumors and low-grade ovarian serous carcinomas. Mod. Pathol. 2009, 22, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Vathipadiekal, V.; Wang, X.V.; Wei, W.; Waldron, L.; Drapkin, R.; Gillette, M.; Skates, S.J.; Birrer, M.J. Creation of a Human Secretome: A Novel Composite Library of Human Secreted Proteins: Validation Using Ovarian Cancer Gene Expression Data and a Virtual Secretome Array. Clin. Cancer Res. 2015, 21, 4960–4969. [Google Scholar] [CrossRef] [PubMed]
- Stany, M.P.; Vathipadiekal, V.; Ozbun, L.; Stone, R.L.; Mok, S.C.; Xue, H.; Kagami, T.; Wang, Y.; McAlpine, J.N.; Bowtell, D.; et al. Identification of Novel Therapeutic Targets in Microdissected Clear Cell Ovarian Cancers. PLoS ONE 2011, 6, e21121. [Google Scholar] [CrossRef]
- Harris, A.L. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br. J. Cancer 2005, 93, 385–386. [Google Scholar] [CrossRef]
- Xue, J.-M.; Liu, Y.; Wan, L.-H.; Zhu, Y.-X. Comprehensive Analysis of Differential Gene Expression to Identify Common Gene Signatures in Multiple Cancers. Med Sci. Monit. 2020, 26, e919953. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Huang, J.; Wang, G.; Liao, K.; Xie, N.; Deng, K. UCP1 modulates immune infiltration level and survival outcome in ovarian cancer patients. J. Ovarian Res. 2022, 15, 16. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. The logrank test. BMJ 2004, 328, 1073. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]



| Gene | p-Value | HR (95% CI) | Figure |
|---|---|---|---|
| PAEP | 0.0036 | 0.80 (0.69–0.93) | Figure S3A |
| IL27RA | 0.033 | 0.86 (0.76–0.99) | Figure S3B |
| LRP1 | 0.00031 | 1.28 (1.12–1.47) | Figure S3C |
| FGF7 | 0.016 | 1.18 (1.03–1.36) | Figure S3D |
| PDGFRA | 0.00023 | 1.32 (1.14–1.53) | Figure S3E |
| PI3 | 7.2 × 10−5 | 1.34 (1.16–1.55) | Figure S3F |
| Gene | Correlation Coefficient | Correlation Coefficient (r) | p-Value | Figure |
|---|---|---|---|---|
| LRP1 | Positive | 0.34 | 8.9 × 10−13 | Figure S13A |
| PI3 | Positive | 0.19 | 8.7 × 10−5 | Figure S13B |
| PAEP | Positive | 0.2 | 4.0 × 10−5 | Figure S13C |
| FOS | Positive | 0.21 | 1.1 × 10−5 | Figure S13D |
| FGF7 | Positive | 0.66 | 7.4 × 10−54 | Figure S13E |
| PDGFRA | Positive | 0.46 | 9.9 × 10−24 | Figure S13F |
| Gene | Correlation Coefficient (r) | p-Value | Figure |
|---|---|---|---|
| LRP1 | 0.49 | 1.2 × 10−26 | Figure S14A |
| FGF7 | 0.81 | 7.4 × 10−97 | Figure S14B |
| FOS | 0.28 | 6.5 × 10−9 | Figure S14C |
| PI3 | 0.12 | 0.02 | Figure S14D |
| PAEP | 0.19 | 9.9 × 10−5 | Figure S14E |
| PDGFRA | 0.64 | 6.1 × 10−49 | Figure S14F |
| Drug | Risk Group | p-Value | |
|---|---|---|---|
| Low | High | ||
| Cisplatin | 3.3 | 3.52 | 0.0045 |
| Etoposide | 1.56 | 1.65 | 0.0028 |
| Gemcitabine | 1.78 | 1.83 | 0.0011 |
| Methotrexate | 0.9 | 1.04 | 0.0011 |
| Lenalidomide | 5.37 | 5.41 | 0.038 |
| Erlotinib | 4.6 | 4.56 | 0.011 |
| Imatinib | 5.04 | 4.97 | 5 × 10−6 |
| Nilotinib | 4.2 | 4.12 | 0.032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolde, T.; Bhardwaj, V.; Reyad-ul-Ferdous, M.; Qin, P.; Pandey, V. The Integrated Bioinformatic Approach Reveals the Prognostic Significance of LRP1 Expression in Ovarian Cancer. Int. J. Mol. Sci. 2024, 25, 7996. https://doi.org/10.3390/ijms25147996
Wolde T, Bhardwaj V, Reyad-ul-Ferdous M, Qin P, Pandey V. The Integrated Bioinformatic Approach Reveals the Prognostic Significance of LRP1 Expression in Ovarian Cancer. International Journal of Molecular Sciences. 2024; 25(14):7996. https://doi.org/10.3390/ijms25147996
Chicago/Turabian StyleWolde, Tesfaye, Vipul Bhardwaj, Md. Reyad-ul-Ferdous, Peiwu Qin, and Vijay Pandey. 2024. "The Integrated Bioinformatic Approach Reveals the Prognostic Significance of LRP1 Expression in Ovarian Cancer" International Journal of Molecular Sciences 25, no. 14: 7996. https://doi.org/10.3390/ijms25147996





