AdNAC20 Regulates Lignin and Coumarin Biosynthesis in the Roots of Angelica dahurica var. formosana
Abstract
1. Introduction
2. Results
2.1. Physiological Changes during Early Bolting of ADF
2.2. Cloning, Sequence Analysis and Expression Profiles of AdNAC20 in ADF
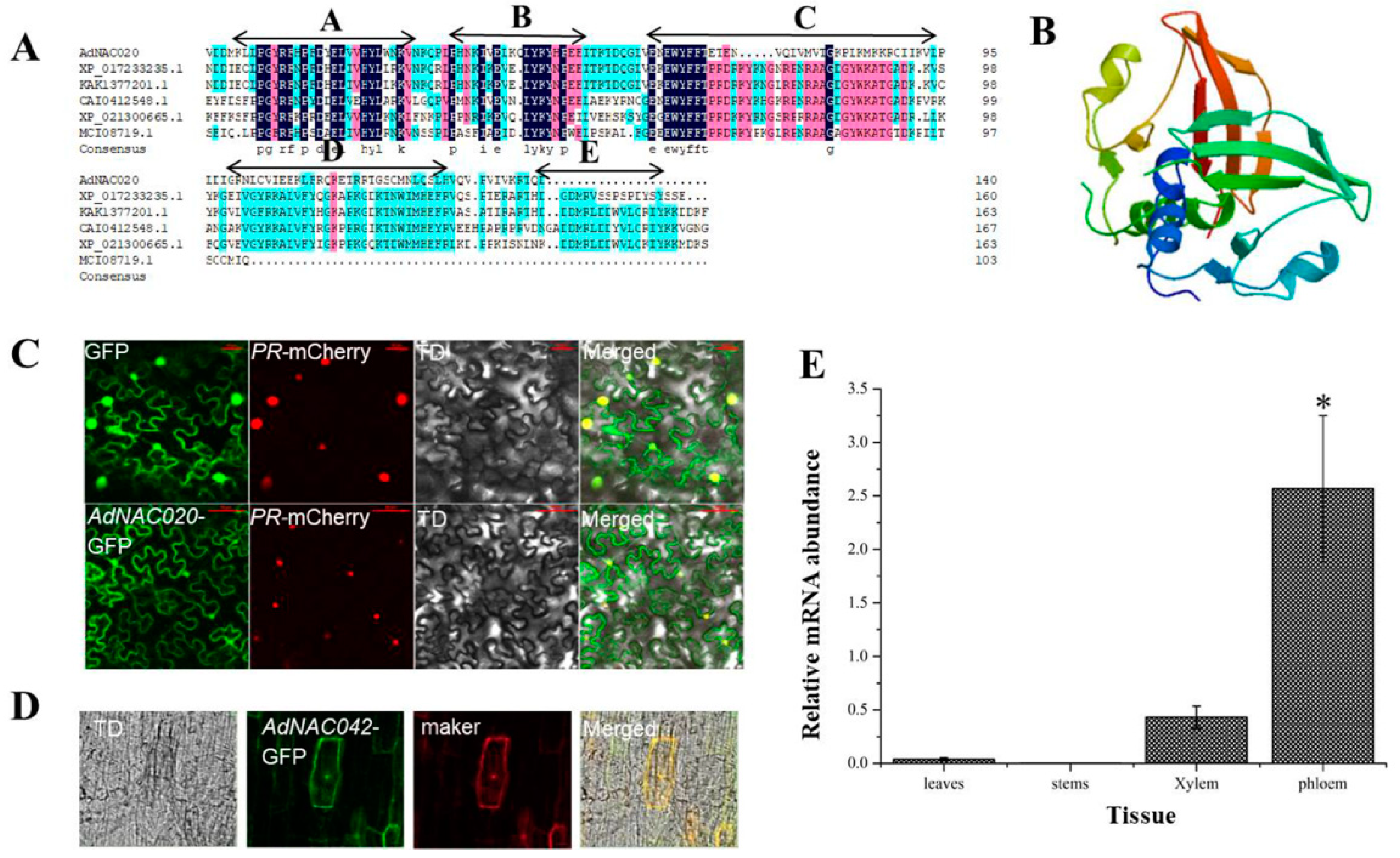
2.2.1. Cloning and Sequence Analysis of AdNAC20
2.2.2. Transient AdNAC20 Expression in ADF Roots
2.3. Heterogeneous Expression of AdNAC20 in A. thaliana
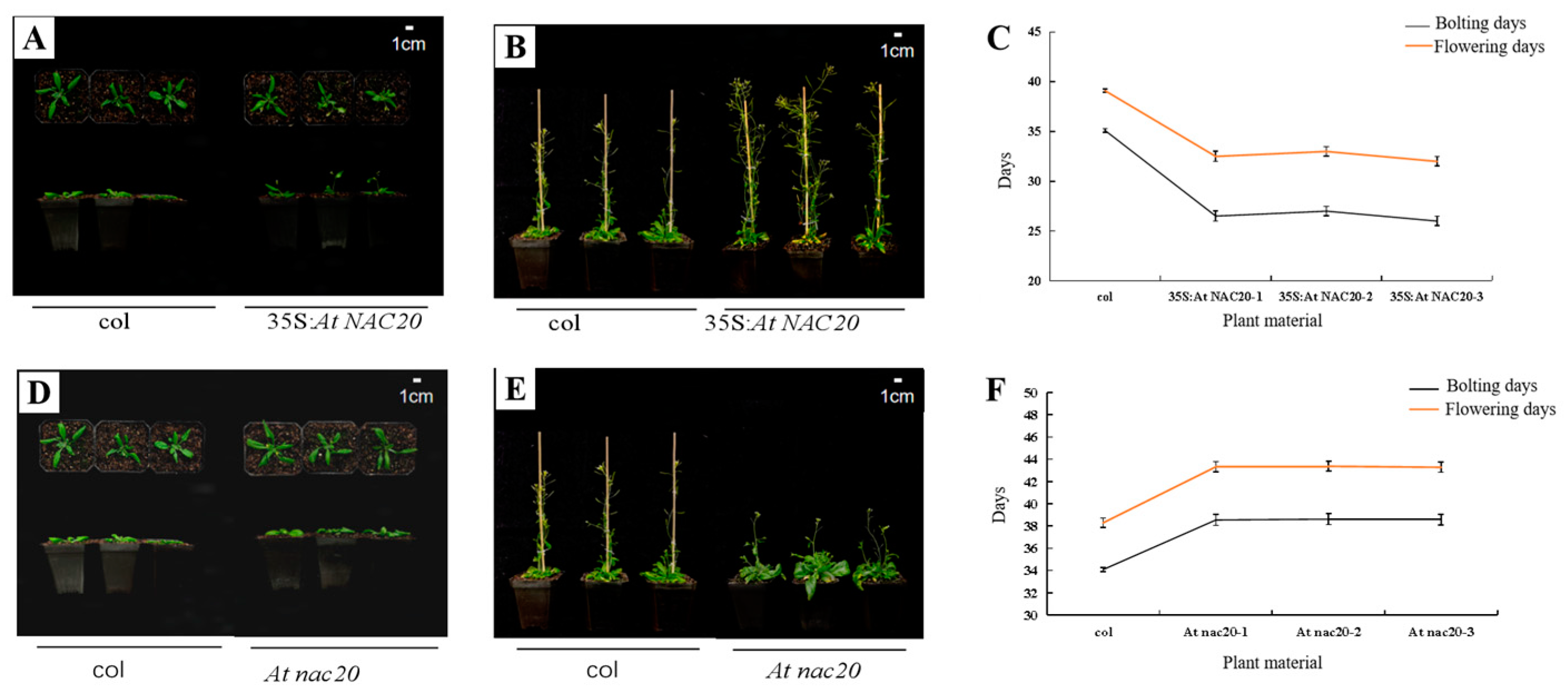
2.3.1. AdNAC20 Regulates Plant Height and Other Phenotypes in the Aerial Parts of A. thaliana
2.3.2. AdNAC20 Regulates Root Phenotype and Lignin Content in A. thaliana
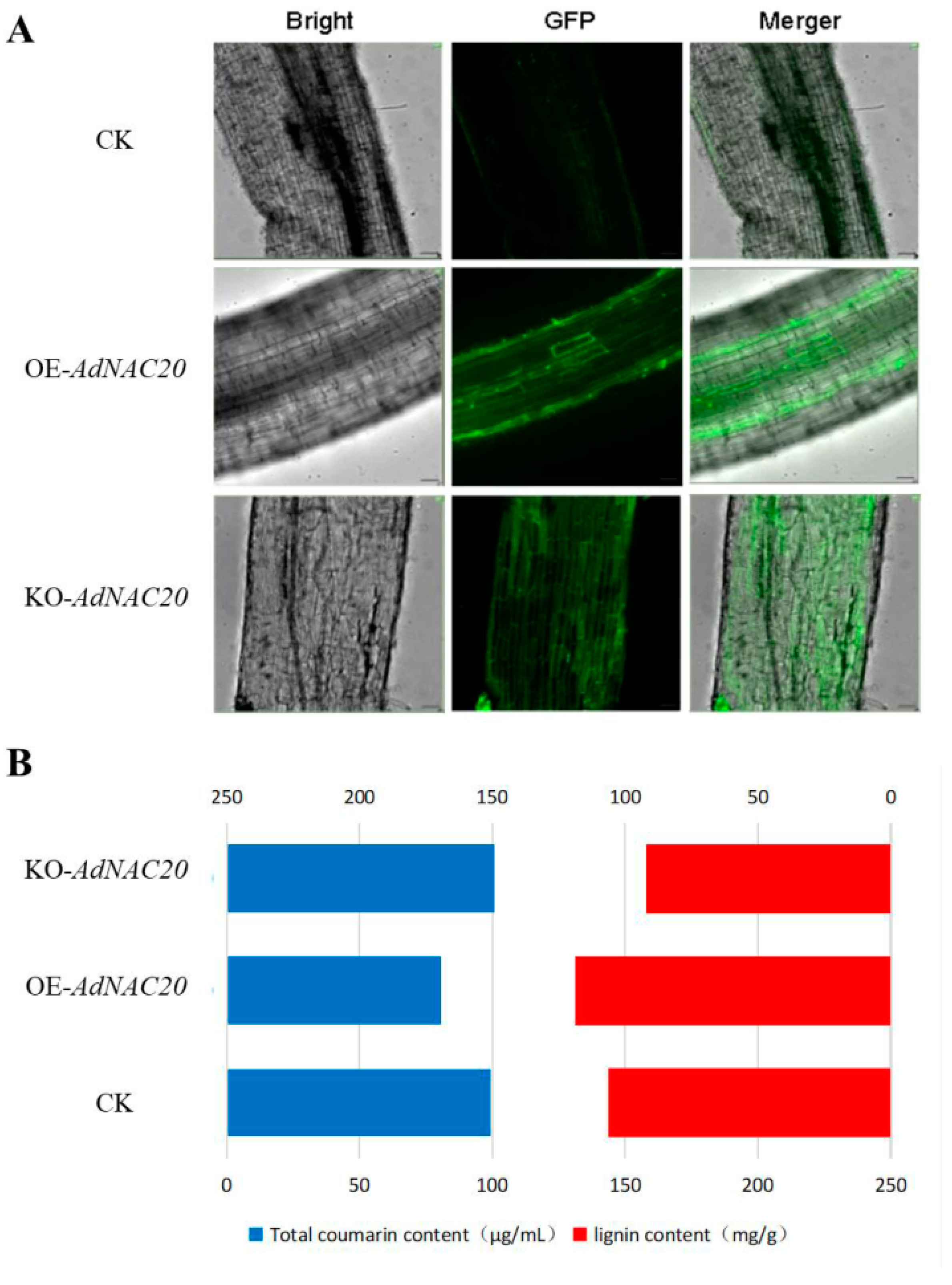
2.4. AdNAC20 Increases Lignin and Reduces Coumarin in Transgenic ADF
2.4.1. Generation of AdNAC20-Overexpressing Plants and Mutant Plants
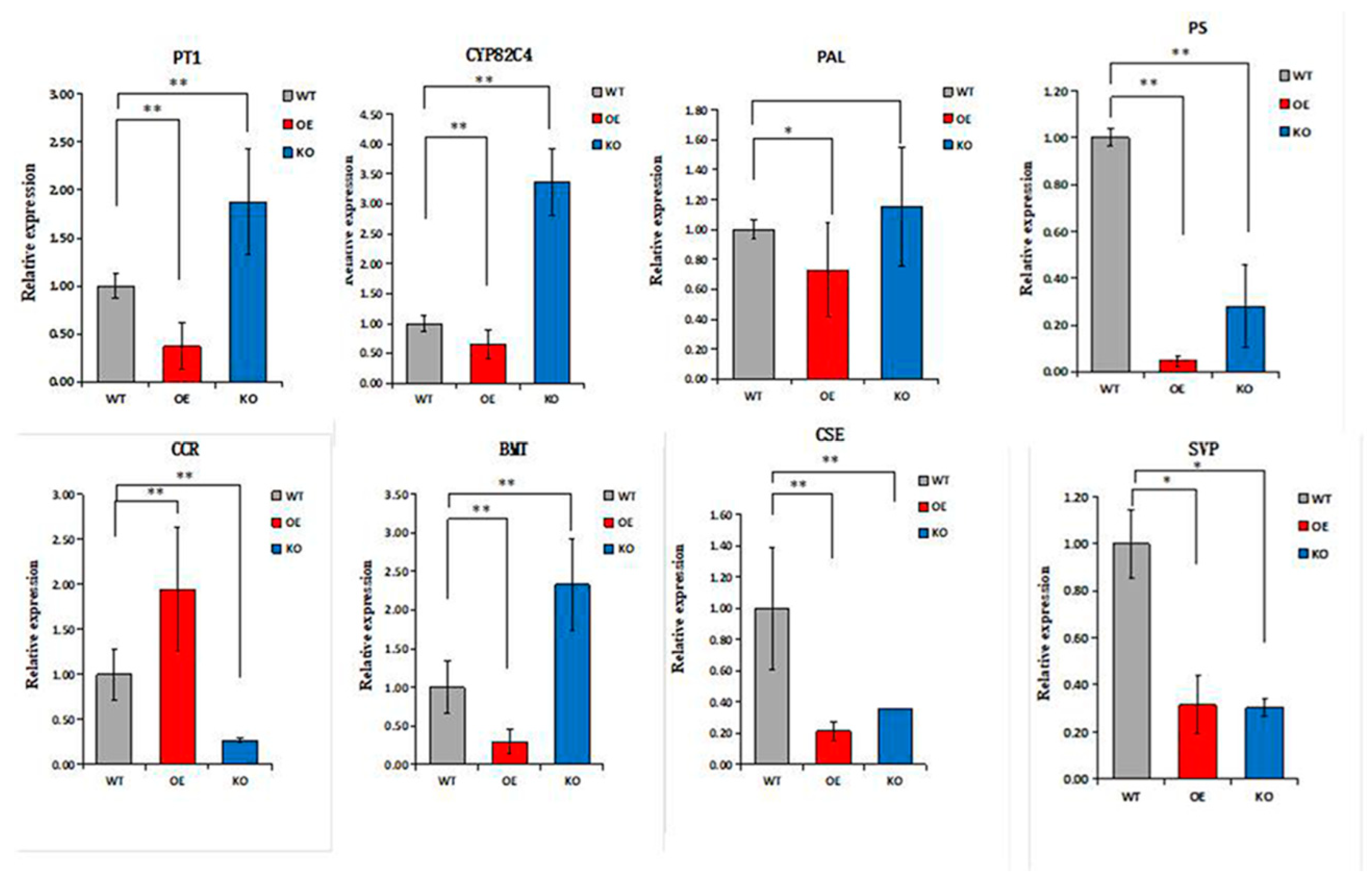
2.4.2. Comparative Phenotypes, Coumarin and Lignin Determination of Transgenic ADF
2.5. Transcriptome Data Analysis of Transgenic ADF Plants
2.5.1. Functional Annotation and Classification of ADF Unigenes
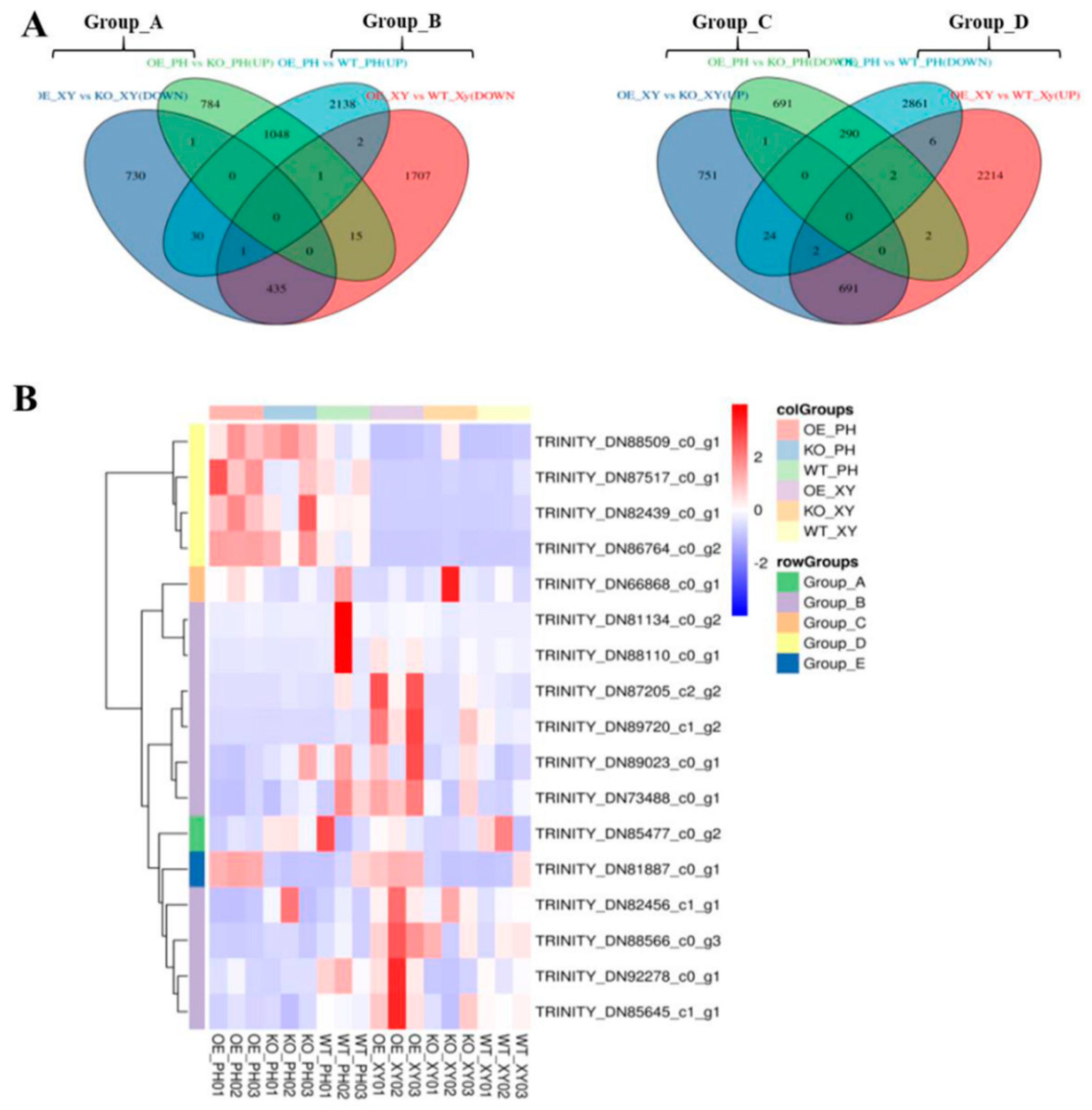
2.5.2. DEG Analysis
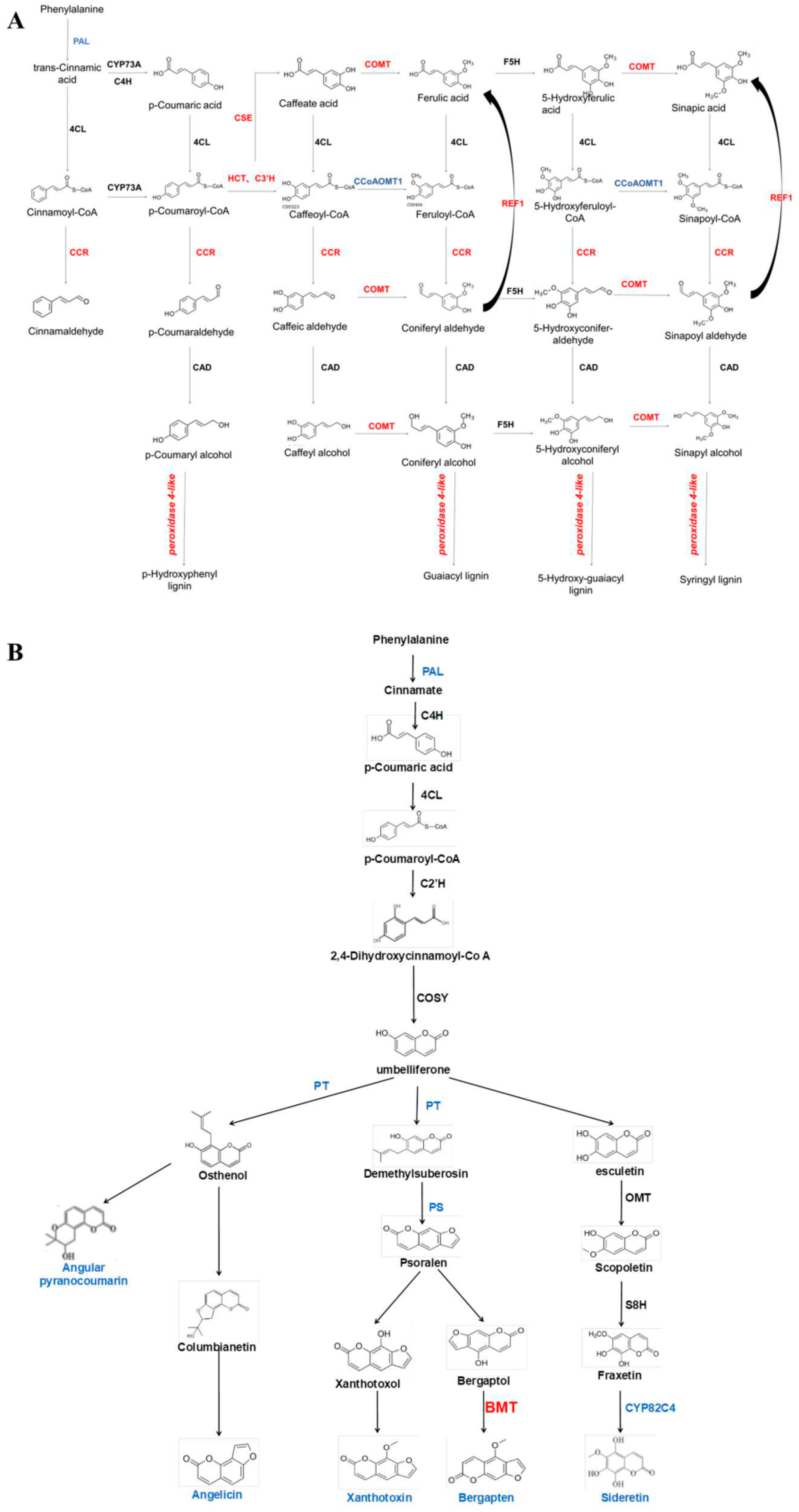
Analysis of DEGs Associated with the Biosynthesis of Coumarin and Lignin Metabolism in ADF Phloem
Analysis of DEGs with Opposite Expression Patterns in the Xylem and Phloem of ADF
Protein-Protein Interaction Analysis of AdNAC20
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Histological Analysis
4.3. Coumarin and Lignin Determination
4.4. RNA Extraction, Gene Cloning and qRT-PCR
4.5. Plasmid Construction
4.6. Subcellular Localization Analysis
4.7. Transient Expression of ADF Roots
4.8. Plant Transformation
4.9. Illumina Sequencing and Differentially Expressed Gene (DEG) Analysis
4.10. HPLC Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; Chemical Medical Science Press: Beijing, China, 2020; Volume 1, pp. 105–106. [Google Scholar]
- Kang, J.; Zhou, L.; Sun, J.; Han, J.; Guo, D.-A. Chromatographic fingerprint analysis and characterization of furocoumarins in the roots of Angelica dahurica by HPLC/DAD/ESI-MSn technique. J. Pharm. Biomed. Anal. 2008, 47, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Zhong, S.H.; Jia, M.R.; Xiong, Y.; Jiang, G.H.; Tang, S.W. Comparasion of macroscopic and microscopic characteris-tics of Chuan Baizhi and Gong Baizhi. Lishizhen Med. Mater. Med. Res. 2005, 9, 833–834. [Google Scholar]
- Wu, P.; Wang, X.; Guo, J.; Zhang, S.; Li, Q.; Zhang, M.; Fang, Q.M.; Luo, B.; Wang, H.S.; He, W. Analysis of the difference be-tween early-bolting and non-bolting roots of Angelica dahurica based on transcriptome sequencing. Sci. Rep. 2023, 13, 7847. [Google Scholar] [CrossRef]
- Chen, C.; Huang, W.; Hou, K.; Wu, W. Bolting, an Important Process in Plant Development, Two Types in Plants. J. Plant Biol. 2019, 62, 161–169. [Google Scholar] [CrossRef]
- Song, C.; Li, X.; Jia, B.; Liu, L.; Wei, P.; Manzoor, M.A.; Wang, F.; Li, B.Y.; Wang, G.; Chen, C.; et al. Comparative Transcriptomics Unveil the Crucial Genes Involved in Coumarin Biosynthesis in Peucedanum praeruptorum Dunn. Front. Plant Sci. 2022, 13, 899819. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, W.; Yang, X.-W. Simultaneous Quantification of Nine New Furanocoumarins in Angelicae Dahuricae Radix Using Ultra-Fast Liquid Chromatography with Tandem Mass Spectrometry. Molecules 2017, 22, 322. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Rahim, A.; Zhang, X.; Busatto, N. Editorial: Phenylpropanoid biosynthesis in plants. Front. Plant Sci. 2023, 14, 1230664. [Google Scholar] [CrossRef]
- Do, C.-T.; Pollet, B.; Thévenin, J.; Sibout, R.; Denoue, D.; Barrière, Y.; Lapierre, C.; Jouanin, L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 2007, 226, 1117–1129. [Google Scholar] [CrossRef]
- Kai, K.; Mizutani, M.; Shimizu, B. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 2008, 55, 989–999. [Google Scholar] [CrossRef]
- Song, C.; Li, X.; Jia, B.; Liu, L.; Ou, J.; Han, B. De novo Transcriptome Sequencing Coupled With Co-expression Analysis Reveal the Transcriptional Regulation of Key Genes Involved in the Formation of Active Ingredients in Peucedanum praeruptorum Dunn Under Bolting Period. Front. Genet. 2021, 12, 683037. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Xu, C.; Alejos-Gonzalez, F.; Wang, H.; Yang, J.; Judd, R.; Xie, D.-Y. Overexpression of Artemisia annua Cinnamyl Alcohol Dehydrogenase Increases Lignin and Coumarin and Reduces Artemisinin and Other Sesquiterpenes. Front. Plant Sci. 2018, 9, 828. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Yang, S.H.; Park, A.H.; Lerouxel, O.; Han, K. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 2007, 50, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhong, R.; Ye, Z.H. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of sec-ondary wall biosynthesis in vessels. PLoS ONE 2014, 9, e105726. [Google Scholar]
- Souer, E.; Houwelingen, A.V.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia is required for pattern for-mation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Yamaguchi, M.; Tamura, T.; Nakano, Y.; Nishikubo, N.; Yoneda, A.; Kato, K.; Kubo, M.; Kajita, S.; Katayama, Y.; et al. Multiple Classes of Transcription Factors Regulate the Expression of VASCULAR-RELATED NAC-DOMAIN7, a Master Switch of Xylem Vessel Differentiation. Plant Cell Physiol. 2014, 56, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC Do-main Transcription Factor, Negatively Regulates Xylem Vessel Formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Avci, U.; Grant, E.H.; Haigler, C.H.; Beers, E.P. XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J. 2007, 53, 425–436. [Google Scholar] [CrossRef]
- Huang, D.; Wang, S.; Zhang, B.; Shang-Guan, K.; Shi, Y.; Zhang, D.; Liu, X.; Wu, K.; Xu, Z.; Fu, X.; et al. A gibberellin-mediated DELLA-NAC signaling cas-cade regulates cellulose synthesis in Rice. Plant Cell 2015, 27, 1681–1696. [Google Scholar] [CrossRef]
- Ning, Y.Q.; Ma, Z.Y.; Huang, H.W.; Mo, H.X.; Zhao, T.T.; Li, L.; Cai, T.; Chen, S.; Ma, L.; He, X.-J. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 2015, 43, 1469–1484. [Google Scholar] [CrossRef] [PubMed]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUN-NEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2017, 16, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-J.; Xu, X.; Chen, J.-S.; Jiang, Y.-J.; Yao, F.; Jiang, M.-Y.; Zhang, H.-H.; Wu, W. Bioinformatics analysis and expression pattern of NAC transcription factor family of Angelica dahurica var. formosana from Sichuan province. China J. Chin. Mater. Medica 2021, 46, 1769–1782. [Google Scholar]
- Nakamura, M.; Claes, A.R.; Grebe, T.; Hermkes, R.; Viotti, C.; Ikeda, Y.; Grebe, M. Auxin and ROP GTPase signaling of polar nu-clear migration in root epidermal hair cells. Plant Physiol. 2017, 176, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Cho, H.-T. PINOID Positively Regulates Auxin Efflux in Arabidopsis Root Hair Cells and Tobacco Cells. Plant Cell 2006, 18, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, R.; Zhao, S.; Lu, C.; Zhu, Z.; Li, H. Transporter NRT1.5/NPF7.3 suppresses primary root growth under low K+ stress by regulating the degradation of PIN-FORMED2. BMC Plant Biol. 2022, 22, 330. [Google Scholar] [CrossRef] [PubMed]
- Grones, P.; Abas, M.; Hajný, J.; Jones, A.; Waidmann, S.; Kleine-Vehn, J.; Friml, J. PID/WAG-mediated phosphorylation of the Arabidopsis PIN3 auxin transporter mediates polarity switches during gravitropism. Sci. Rep. 2018, 8, 10279. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Runciman, B.; Tasker-Brown, W.; Østergaard, L. Two Auxin Response Elements Fine-Tune PINOID Expression During Gynoecium Development in Arabidopsis thaliana. Biomolecules 2019, 9, 526. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Lee, J.; Moosa, M.M.; Nian, Y.; Hu, L.; Xu, Z.; McCoy, J.G.; Ferreon, A.C.M.; Im, W.; Zhou, M. Structure of an EIIC sugar transporter trapped in an inward-facing conformation. Proc. Natl. Acad. Sci. USA 2018, 115, 5962–5967. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yao, G.; Li, L.; Li, T.; Zhao, Y.; Hu, K. E3 ligase BRG3 persulfidation delays tomato ripening by reducing ubiqui-tination of the repressor WRKY71. Plant Physiol. 2023, 192, 616–632. [Google Scholar] [CrossRef]
- Park, J.; Nguyen, K.T.; Park, E.; Jeon, J.S.; Choi, G. DELLA proteins and their interacting RING Finger proteins repress gibberel-lin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 2013, 25, 927–943. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Qiu, C.; Qian, W.; Wang, Y.; Sun, L.; Li, Y.; Ding, Z. Ammonium triggered the response mechanism of lysine crotonylome in tea plants. BMC Genom. 2019, 20, 340. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.B.; Bastress, K.L.; Ruegger, M.O.; Denault, J.W.; Chapple, C. The Arabidopsis thaliana REDUCED EPIDERMAL FLU-ORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 2004, 16, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, N.; Zeng, Z.; Xu, S.; Huang, C.; Wang, W.; Liu, T.; Luo, J.; Kong, L. Cloning, functional characterization, and catalytic mecha-nism of a Bergaptol O-Methyltransferase from Peucedanum praeruptorum Dunn. Front. Plant Sci. 2016, 7, 722. [Google Scholar] [CrossRef]
- Hehmann, M.; Lukačin, R.; Ekiert, H.; Matern, U. Furanocoumarin biosynthesis in Ammi majus L. Cloning of bergaptol O-methyltransferase. Eur. J. Biochem. 2004, 271, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Song, J.N.; Hou, F.J.; Wang, Q.; Huang, K.; Sun, J.H.; Zheng, Y.G. Analysis of accumulation of coumarins in Angelica dahurica by histochemical technique. Guangzhou Chem. Ind. 2017, 45, 132–134. [Google Scholar]
- Zhao, D.Y.; Hao, Q.X.; Kang, L.P.; Zhang, Y.; Chen, M.L.; Wang, T.L.; Guo, L.P. Advance in studying early bolting of Umbelliferae medicinal plant. China J. Chin. Mater. Med. 2015, 41, 20–23. [Google Scholar]
- Xue, C.; Yao, J.-L.; Xue, Y.-S.; Su, G.-Q.; Wang, L.; Lin, L.-K.; Allan, A.C.; Zhang, S.-L.; Wu, J. PbrMYB169 positively regulates lignification of stone cells in pear fruit. J. Exp. Bot. 2019, 70, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, N.; Sui, Z.; Huang, C.; Zeng, Z.; Kong, L. The Molecular and Structural Basis of O-methylation Reaction in Coumarin Biosynthesis in Peucedanum praeruptorum Dunn. Int. J. Mol. Sci. 2019, 20, 1533. [Google Scholar] [CrossRef]
- Tamura, T.; Endo, H.; Suzuki, A.; Sato, Y.; Kato, K.; Ohtani, M.; Yamaguchi, M.; Demura, T. Affinity-based high-resolution analysis of DNA binding by VASCULAR-RELATED NAC-DOMAIN7 via fluorescence correlation spectroscopy. Plant J. 2019, 100, 298–313. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. MYB46 and MYB83 Bind to the SMRE Sites and Directly Activate a Suite of Transcription Factors and Secondary Wall Biosynthetic Genes. Plant Cell Physiol. 2011, 53, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Demura, T. The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 2018, 56, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.; Tak, H.; Ganapathi, T.R. Xylem specific activation of 5′upstream regulatory region of two NAC transcription factors (MusaVND6 and MusaVND7) in banana is regulated by SNBE-like sites. PLoS ONE 2018, 13, e0192852. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. DNA-binding specificity and molecular functions of NAC transcrip-tion factors. Plant Sci. 2005, 169, 785–797. [Google Scholar] [CrossRef]
- Yang, L.; Wang, H.; Liu, J.; Li, L.; Fan, Y.; Song, Y.; Sun, S.; Wang, L.; Zhu, X.; Wang, X.; et al. A simple and effective system for foreign gene expression in plants via root absorption of agrobacterial suspension. J. Biotechnol. 2008, 134, 320–324. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Li, X.; Wang, N.; She, W.; Guo, Z.; Pan, H.; Yu, Y.; Ye, J.; Pan, D.; Pan, T. Identification and functional analysis of the CgNAC043 gene involved in lignin synthesis from Citrus grandis “San Hong”. Plants 2021, 11, 403. [Google Scholar] [CrossRef]





| Tissue | OE vs. WT | KO vs. WT | OE vs. KO | |||
|---|---|---|---|---|---|---|
| UP | DOWN | UP | DOWN | UP | DOWN | |
| Leaf | 1664 | 1420 | 1613 | 944 | 913 | 1002 |
| Root phloem | 3220 | 3185 | 1377 | 1644 | 1849 | 966 |
| Root xylem | 2917 | 2161 | 998 | 1132 | 1469 | 1197 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, W.; Huang, W.; Chen, C.; Chen, J.; Zhao, L.; Jiang, Y.; Du, X.; Liu, R.; Chen, Y.; Hou, K.; et al. AdNAC20 Regulates Lignin and Coumarin Biosynthesis in the Roots of Angelica dahurica var. formosana. Int. J. Mol. Sci. 2024, 25, 7998. https://doi.org/10.3390/ijms25147998
Qu W, Huang W, Chen C, Chen J, Zhao L, Jiang Y, Du X, Liu R, Chen Y, Hou K, et al. AdNAC20 Regulates Lignin and Coumarin Biosynthesis in the Roots of Angelica dahurica var. formosana. International Journal of Molecular Sciences. 2024; 25(14):7998. https://doi.org/10.3390/ijms25147998
Chicago/Turabian StyleQu, Wenjie, Wenjuan Huang, Chen Chen, Jinsong Chen, Lin Zhao, Yijie Jiang, Xuan Du, Renlang Liu, Yinyin Chen, Kai Hou, and et al. 2024. "AdNAC20 Regulates Lignin and Coumarin Biosynthesis in the Roots of Angelica dahurica var. formosana" International Journal of Molecular Sciences 25, no. 14: 7998. https://doi.org/10.3390/ijms25147998
APA StyleQu, W., Huang, W., Chen, C., Chen, J., Zhao, L., Jiang, Y., Du, X., Liu, R., Chen, Y., Hou, K., Xu, D., & Wu, W. (2024). AdNAC20 Regulates Lignin and Coumarin Biosynthesis in the Roots of Angelica dahurica var. formosana. International Journal of Molecular Sciences, 25(14), 7998. https://doi.org/10.3390/ijms25147998







