Enhancement of Dendrobine Production by CRISPR/Act3.0-Mediated Transcriptional Activation of Multiple Endogenous Genes in Dendrobium Plants
Abstract
1. Introduction
2. Results
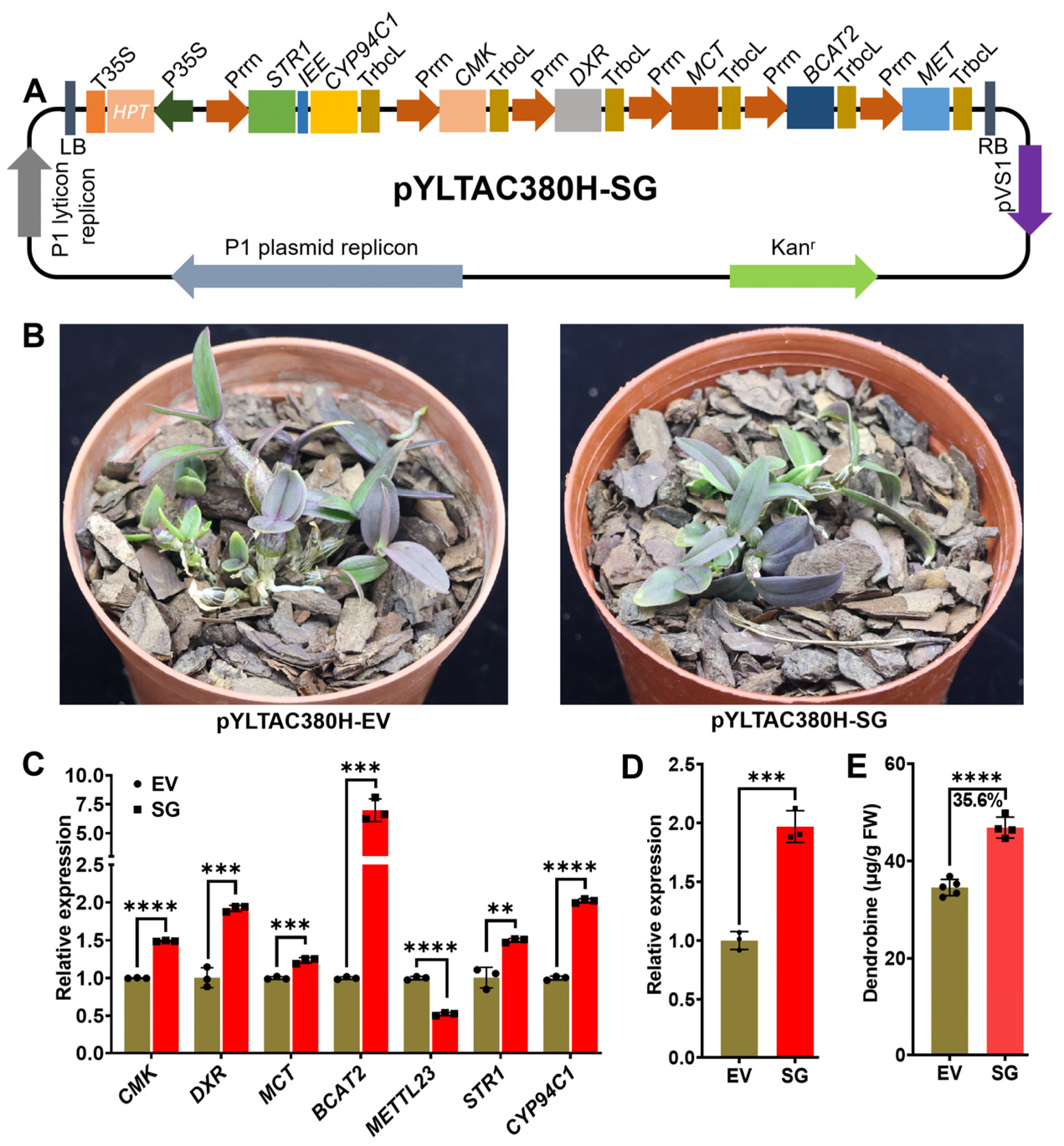
2.1. Overexpression of Multiple Stacked Genes Enhanced Dendrobine Production in Transgenic D. catenatum
2.2. Transcriptional Activation of MCT (2-C-Methyl-d-Erythritol 4-Phosphate Cytidylyltransferase) by CRISPR/Act3.0 Enhanced Dendrobine Production in D. catenatum Leaves
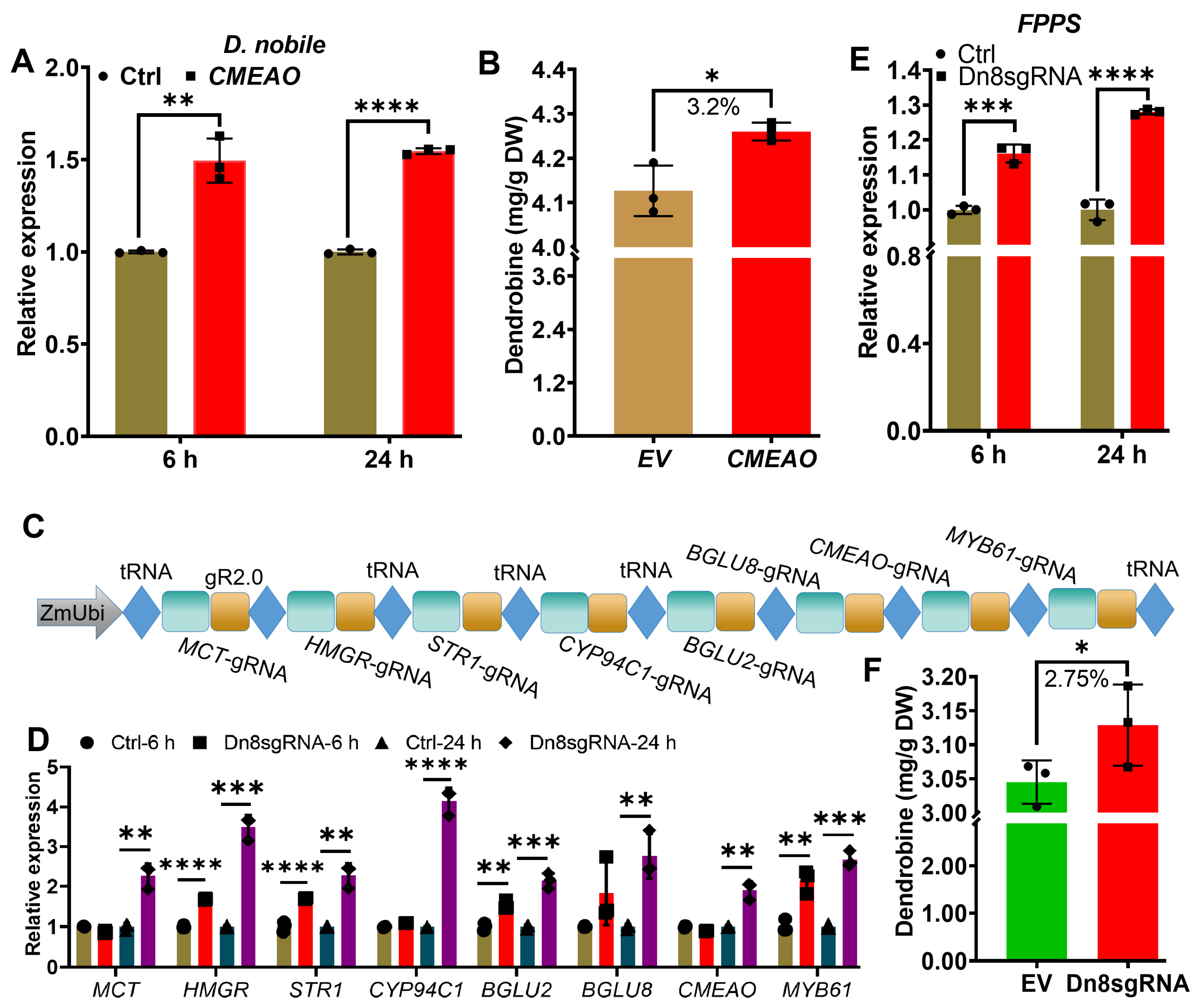
2.3. Transcriptional Activation of Multiple Endogenous Genes by CRISPR/Act3.0 Enhanced Dendrobine Production in D. nobile Leaves
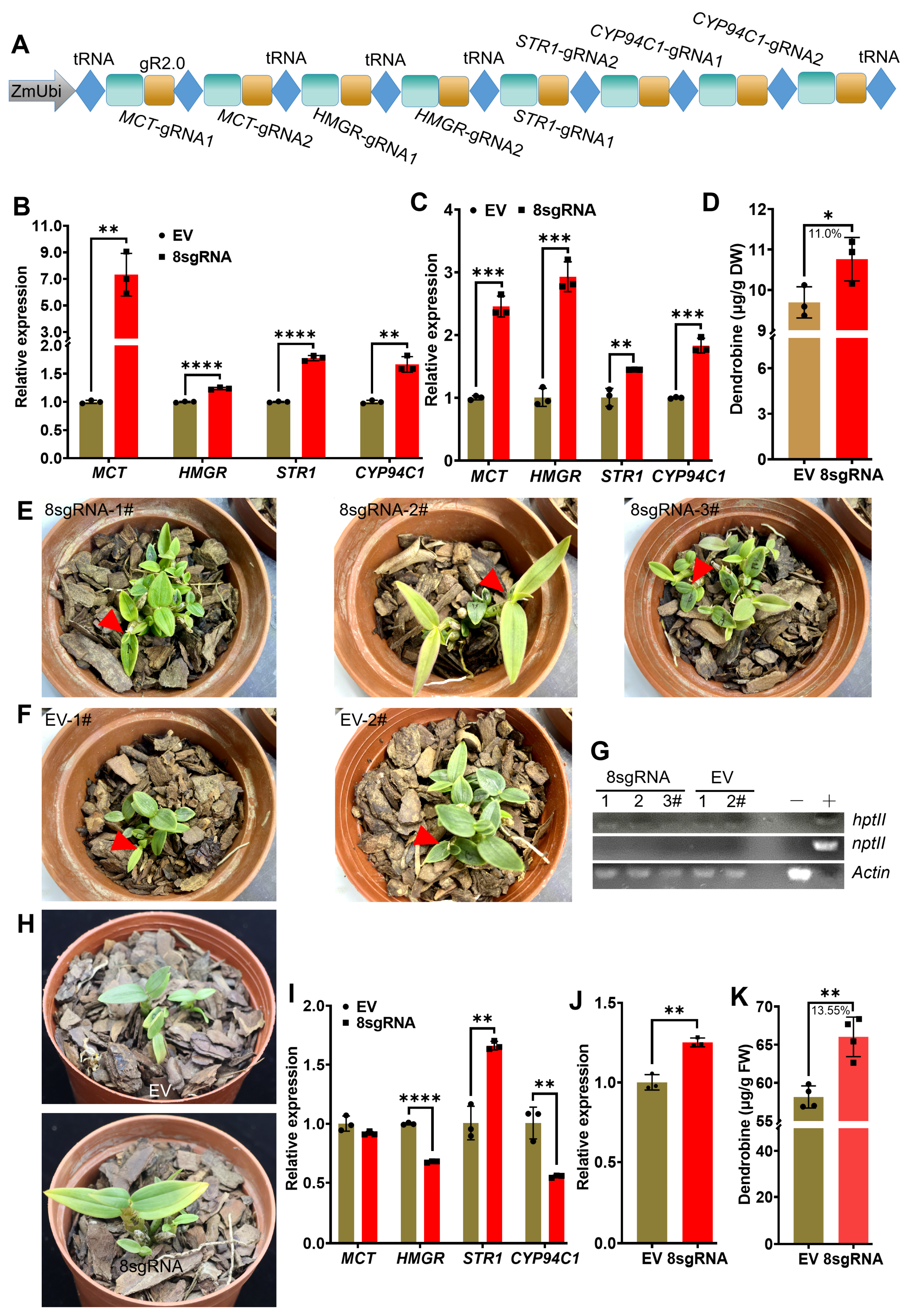
2.4. Transcriptional Activation of Multiple Endogenous Genes by CRISPR/Act3.0 Improved Dendrobine Production in Transgenic D. catenatum Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Genomic PCR
4.3. LC/MS
4.4. Dual-Luciferase Assay
4.5. Vector Construction
4.6. Genetic Transformation
4.7. Quantitative Real-Time PCR (qRT-PCR) Validation of Gene Expression
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tikendra, L.; Potshangbam, A.; Amom, T.; Dey, A.; Nongdam, P. Understanding the genetic diversity and population structure of Dendrobium chrysotoxum Lindl.-An endangered medicinal orchid and implication for its conservation. S. Afr. J. Bot. 2021, 138, 364–376. [Google Scholar] [CrossRef]
- Yan, S.; Zhao, T.; Li, Y.; Hu, Y.; Chun, Z. Agronomic and quality traits of different Dendrobium nobile in Hejiang. Chin. J. Exp. Tradit. Med. Formulae 2018, 24, 73–77. [Google Scholar]
- Liu, Y.; Zhang, J.; Zhan, R.; Chen, Y. Isopentenylated bibenzyls and phenolic compounds from Dendrobium chrysotoxum Lindl. Chem. Biodivers. 2022, 19, e202200259. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, M.; Cui, H.; Li, J.; Wang, M. Transcriptomic landscape of medicinal Dendrobium reveals genes associated with the biosynthesis of bioactive components. Front. Plant Sci. 2020, 11, 391. [Google Scholar] [CrossRef]
- Zhang, G.; Bi, Z.; Wang, Z.; Xu, L.; Xu, G. Progress on chemical composition of Dendrobium Sw. Chin. Tradit. Herb. Drugs 2003, 6, 5–8. [Google Scholar]
- Adejobi, O.; Guan, J.; Yang, L.; Hu, J.-M.; Yu, A.; Muraguri, S.; Liu, A. Transcriptomic analyses shed light on critical genes associated with bibenzyl biosynthesis in Dendrobium officinale. Plants 2021, 10, 633. [Google Scholar] [CrossRef]
- Morita, H.; Fujiwara, M.; Yoshida, N.; Kobayashi, J. New picrotoxinin-type and dendrobine-type sesquiterpenoids from Dendrobium snowflake ’Red Star’. Tetrahedron 2000, 56, 5801–5805. [Google Scholar] [CrossRef]
- Zhang, C.; Kong, Y.; Zhang, M.; Wu, Q.; Shi, J. Two new alkaloids from Dendrobium nobile Lindl. exhibited neuroprotective activity, and dendrobine alleviated Aβ1-42-induced apoptosis by inhibiting CDK5 activation in PC12 cells. Drug Dev. Res. 2023, 84, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Zhao, Y.; Ye, F.; Shi, Y.; Kennelly, E.J.; Chen, S.; Zhao, D. Identification, biological activities and biosynthetic pathway of Dendrobium alkaloids. Front. Pharmacol. 2021, 12, 605994. [Google Scholar] [CrossRef]
- Gong, D.; Chen, X.; Guo, S.; Wang, B.; Li, B. Recent advances and new insights in biosynthesis of dendrobine and sesquiterpenes. Appl. Microbiol. Biotechnol. 2021, 105, 6597–6606. [Google Scholar] [CrossRef] [PubMed]
- Selma, S.; Ceulemans, E.; Goossens, A.; Lacchini, E. Clustered regularly interspaced short palindromic repeats tools for plant metabolic engineering: Achievements and perspectives. Curr. Opin. Biotechnol. 2023, 79, 102856. [Google Scholar] [CrossRef]
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 2021, 7, 942–953. [Google Scholar] [CrossRef]
- Yao, T.; Yuan, G.; Lu, H.; Liu, Y.; Zhang, J.; Tuskan, G.A.; Muchero, W.; Chen, J.-G.; Yang, X. CRISPR/Cas9-based gene activation and base editing in Populus. Hortic. Res. 2023, 10, uhad085. [Google Scholar] [CrossRef]
- Pan, C.; Li, G.; Malzahn, A.; Cheng, Y.; Leyson, B.; Sretenovic, S.; Gurel, F.; Coleman, G.D.; Qi, Y. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat. Plants 2022, 8, 513–525. [Google Scholar] [CrossRef]
- Xiong, X.; Liang, J.; Li, Z.; Gong, B.; Li, J. Multiplex and optimization of dCas9-TV-mediated gene activation in plants. J. Integr. Plant Biol. 2021, 63, 634–645. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, T.; Xing, J.; Wang, R.; Zhao, Y. Positional effects on efficiency of CRISPR/Cas9-based transcriptional activation in rice plants. Abiotech 2020, 1, 1–5. [Google Scholar] [CrossRef]
- Zhao, M.; Zhao, Y.; Yang, Z.; Ming, F.; Li, J.; Kong, D.; Wang, Y.; Chen, P.; Wang, M.; Wang, Z. Metabolic pathway engineering improves dendrobine production in Dendrobium catenatum. Int. J. Mol. Sci. 2023, 25, 397. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, M.; Zhang, X.; Gu, L.; Li, J.; Ming, F.; Wang, M.; Wang, Z. MIR396-GRF/GIF enhances in planta shoot regeneration of Dendrobium catenatum. BMC Genom. 2024, 25, 543. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Zhang, J.; Zhu, G.F.; Liang, C.L.; Ye, Q.S. Comparison of polysaccharide and alkaloid contents in Dendrobium. Chin. Agric Sci. Bull. 2015, 31, 242–246. [Google Scholar]
- Jin, R.; Sun, J.; Zhang, Y. A determination of total alkaloids in eleven species of Dendrobium. J. China Pharm. Univ. 1981, 1, 9–13. [Google Scholar]
- Wang, Z.; Zhao, M.; Zhang, X.; Deng, X.; Li, J.; Wang, M. Genome-wide identification and characterization of active ingredients related β-glucosidases in Dendrobium catenatum. BMC Genom. 2022, 23, 612. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, J.; Tan, J.; Yang, Y.; Li, S.; Gou, Y.; Luo, Y.; Li, T.; Xiao, W.; Xue, Y.; et al. Efficient assembly of long DNA fragments and multiple genes with improved nickase-based cloning and Cre/loxP recombination. Plant Biotechnol. J. 2022, 20, 1983–1995. [Google Scholar] [CrossRef]
- Wang, D.; Qu, Z.; Adelson, D.; Zhu, J.; Timmis, J. Transcription of nuclear organellar DNA in a model plant system. Genome Biol. Evol. 2014, 6, 1327–1334. [Google Scholar] [CrossRef]
- Liu, N.; Sun, Z.; Liao, X.; Yuan, M.; Li, X.; Zhang, H. Mass fraction variation of alkaloids and polysaccharides in Dendrobium nobile Lindl with different harvest time. J. Jilin Univ. 2010, 48, 511–515. [Google Scholar]
- Lu, A.; Jiang, Y.; Wu, J.; Tan, D.; Qin, L.; Lu, Y.; Qian, Y.; Bai, C.; Yang, J.; Ling, H. Opposite trends of glycosides and alkaloids in Dendrobium nobile of different age based on UPLC-Q/TOF-MS combined with multivariate statistical analyses. Phytochem. Anal. 2022, 33, 619–634. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Han, B.; Yang, Y.; Wang, Z.; Sun, Z. Research progress on constituents of alkaloids in plants from Dendrobium Sw. Chin. Tradit. Herb. Drugs 2019, 50, 3246–3254. [Google Scholar]
- Concordet, J.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Pan, C.; Qi, Y. CRISPR-Combo-mediated orthogonal genome editing and transcriptional activation for plant breeding. Nat. Protoc. 2023, 18, 1760–1794. [Google Scholar] [CrossRef]
- Li, Q.; Ding, G.; Li, B.; Guo, S. Transcriptome analysis of genes involved in dendrobine biosynthesis in Dendrobium nobile Lindl. infected with mycorrhizal fungus MF23 (Mycena sp.). Sci. Rep. 2017, 7, 316. [Google Scholar] [CrossRef]
- Gong, D.; Wu, B.; Qin, H.; Fu, D.; Guo, S.; Wang, B.; Li, B. Functional characterization of a farnesyl diphosphate synthase from Dendrobium nobile Lindl. ABM Express 2022, 12, 129. [Google Scholar] [CrossRef]
- Demirer, G.; Zhang, H.; Matos, J.; Goh, N.S.; Cunningham, F.J.; Sung, Y.; Chang, R.; Aditham, A.J.; Chio, L.; Cho, M.-J.; et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019, 14, 456–464. [Google Scholar] [CrossRef]
- Su, W.; Xu, M.; Radani, Y.; Yang, L. Technological development and application of plant genetic transformation. Int. J. Mol. Sci. 2023, 24, 10646. [Google Scholar] [CrossRef]
- Bhowmik, S.; Cheng, A.; Long, H.; Tan, G.Z.H.; Hoang, T.M.L.; Karbaschi, M.R.; Williams, B.; Higgins, T.J.V.; Mundree, S.G. Robust Genetic Transformation System to Obtain Non-chimeric Transgenic Chickpea. Front. Plant Sci. 2019, 10, 524. [Google Scholar] [CrossRef]
- Xu, D.; Chu, S.; Xiao, S.; Zhu, F.; Qian, G.; Li, L. Comparison of the content of polysaccharides, dendrobine and amino acids in Dendrobium ironum and Dendrobium chrysogenum of different origins. Lishizhen Med. Mater. Med. Res. 2016, 27, 2738–2740. [Google Scholar]
- Zhang, T.; Lian, H.; Zhou, C.; Xu, L.; Jiao, Y.; Wang, J. A two-step model for de novo activation of Wuchel during plant shoot regeneration. Plant Cell 2017, 29, 1073–1087. [Google Scholar] [CrossRef]
- Yang, F.; Lu, C.; Wei, Y.; Wu, J.; Ren, R.; Gao, J.; Ahmad, S.; Jin, J.; Xv, Y.; Liang, G.; et al. Organ-specific gene expression reveals the role of the cymbidium ensifolium-miR396/Growth-Regulating Factors module in flower development of the orchid plant Cymbidium ensifolium. Front. Plant Sci. 2022, 12, 799778. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Yang, Z.; Li, J.; Ming, F.; Kong, D.; Xu, H.; Wang, Y.; Chen, P.; Duan, X.; Wang, M.; et al. Enhancement of Dendrobine Production by CRISPR/Act3.0-Mediated Transcriptional Activation of Multiple Endogenous Genes in Dendrobium Plants. Int. J. Mol. Sci. 2025, 26, 1487. https://doi.org/10.3390/ijms26041487
Zhao M, Yang Z, Li J, Ming F, Kong D, Xu H, Wang Y, Chen P, Duan X, Wang M, et al. Enhancement of Dendrobine Production by CRISPR/Act3.0-Mediated Transcriptional Activation of Multiple Endogenous Genes in Dendrobium Plants. International Journal of Molecular Sciences. 2025; 26(4):1487. https://doi.org/10.3390/ijms26041487
Chicago/Turabian StyleZhao, Meili, Zhenyu Yang, Jian Li, Feng Ming, Demin Kong, Haifeng Xu, Yu Wang, Peng Chen, Xiaojuan Duan, Meina Wang, and et al. 2025. "Enhancement of Dendrobine Production by CRISPR/Act3.0-Mediated Transcriptional Activation of Multiple Endogenous Genes in Dendrobium Plants" International Journal of Molecular Sciences 26, no. 4: 1487. https://doi.org/10.3390/ijms26041487
APA StyleZhao, M., Yang, Z., Li, J., Ming, F., Kong, D., Xu, H., Wang, Y., Chen, P., Duan, X., Wang, M., & Wang, Z. (2025). Enhancement of Dendrobine Production by CRISPR/Act3.0-Mediated Transcriptional Activation of Multiple Endogenous Genes in Dendrobium Plants. International Journal of Molecular Sciences, 26(4), 1487. https://doi.org/10.3390/ijms26041487





