The Expression Profiles of lncRNAs Are Associated with Neoadjuvant Chemotherapy Resistance in Locally Advanced, Luminal B-Type Breast Cancer
Abstract
:1. Introduction
2. Results
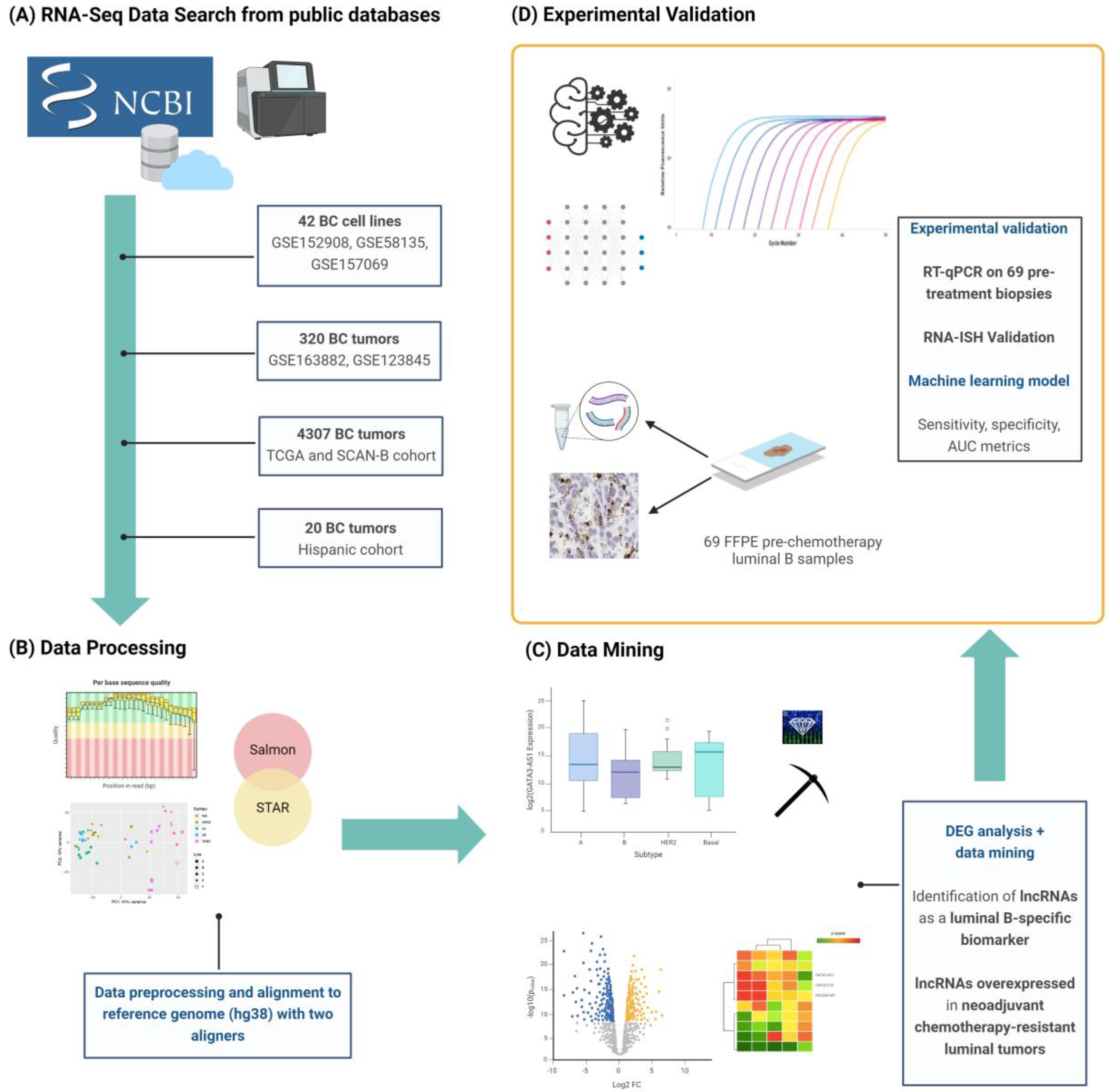
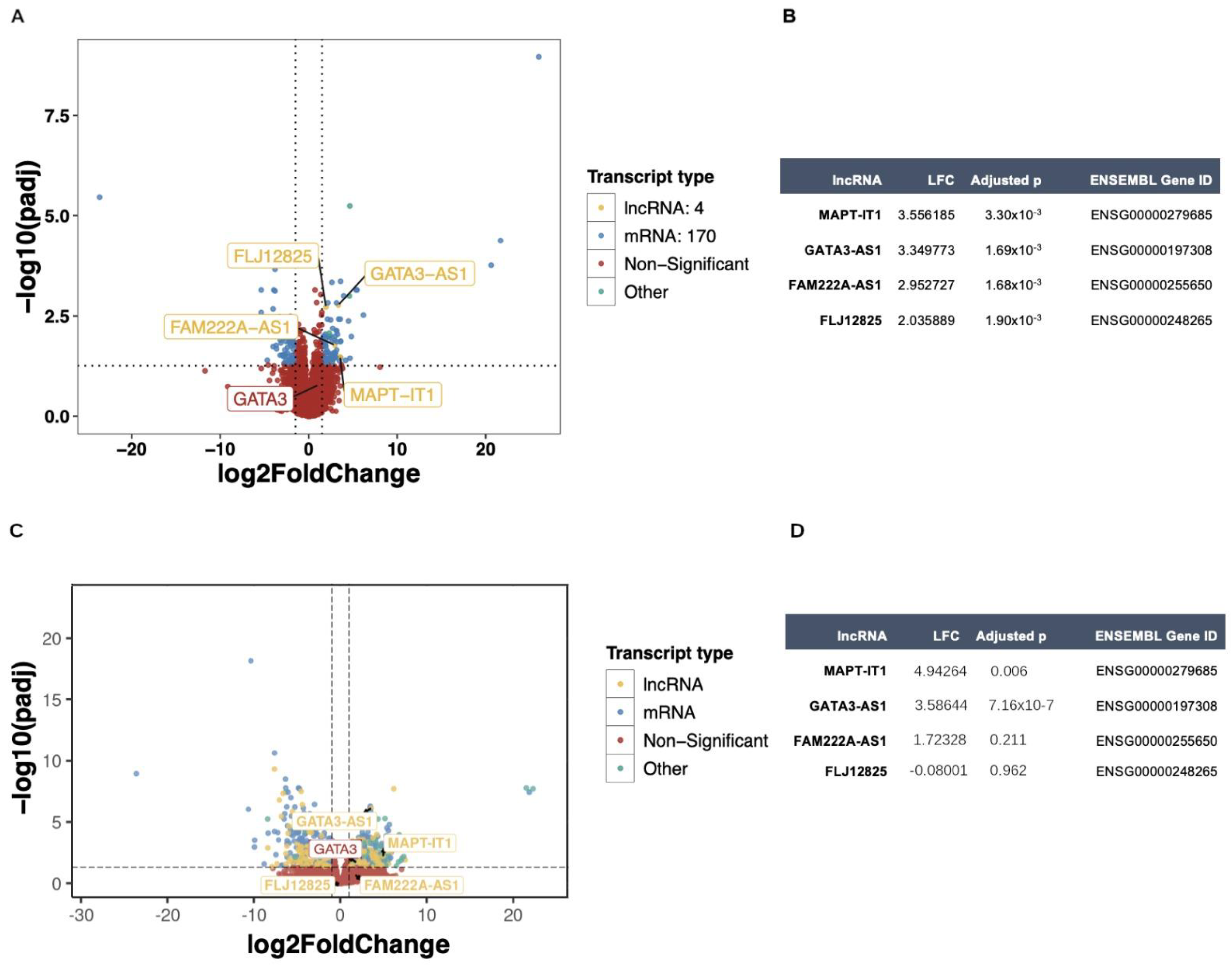
2.1. Transcriptomic Profiling of lncRNAs in Breast Cancer Cell Lines
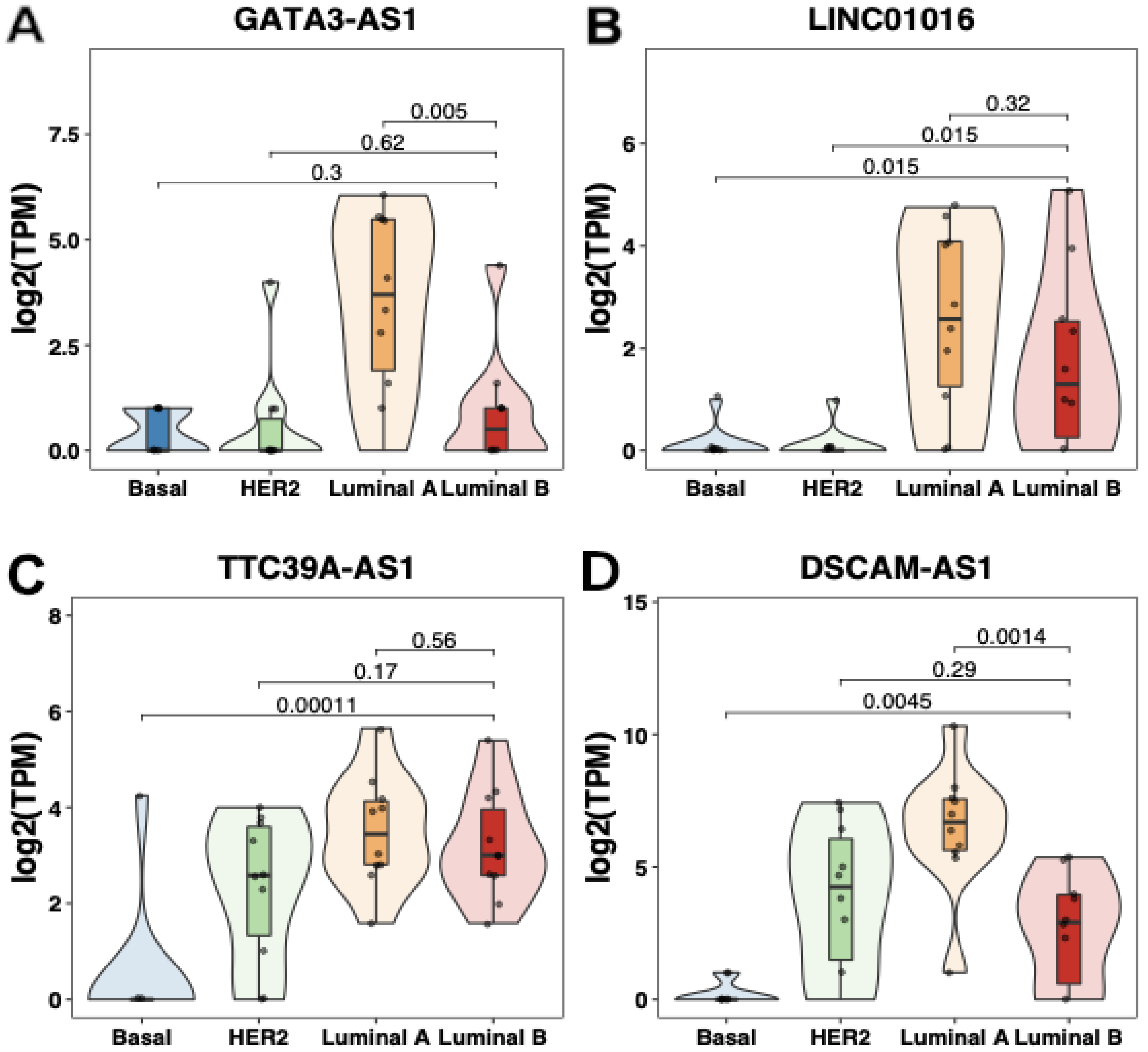
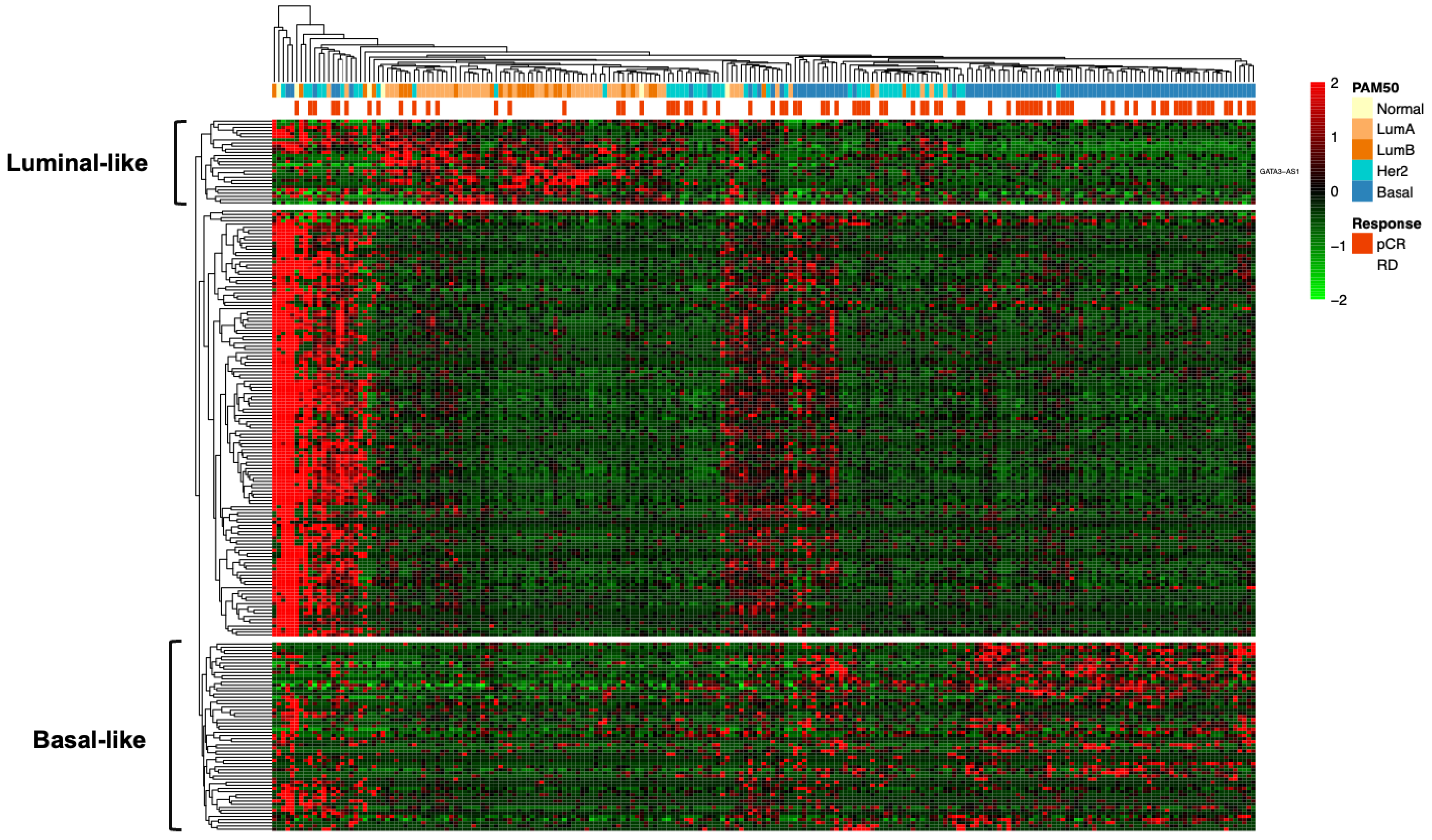
2.2. Transcriptomic Profiling of lncRNAs in Patients with Locally Advanced Breast Cancer
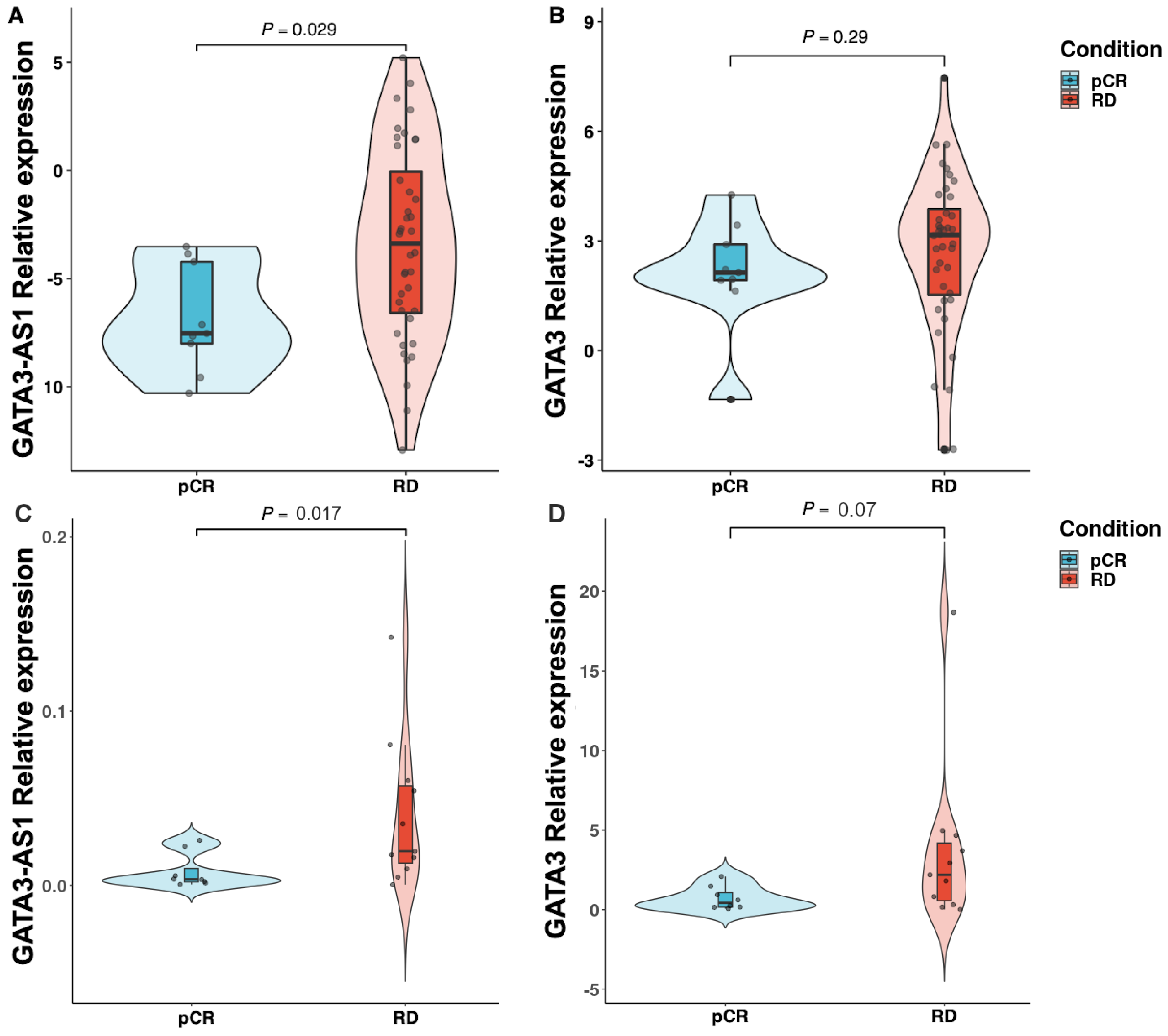
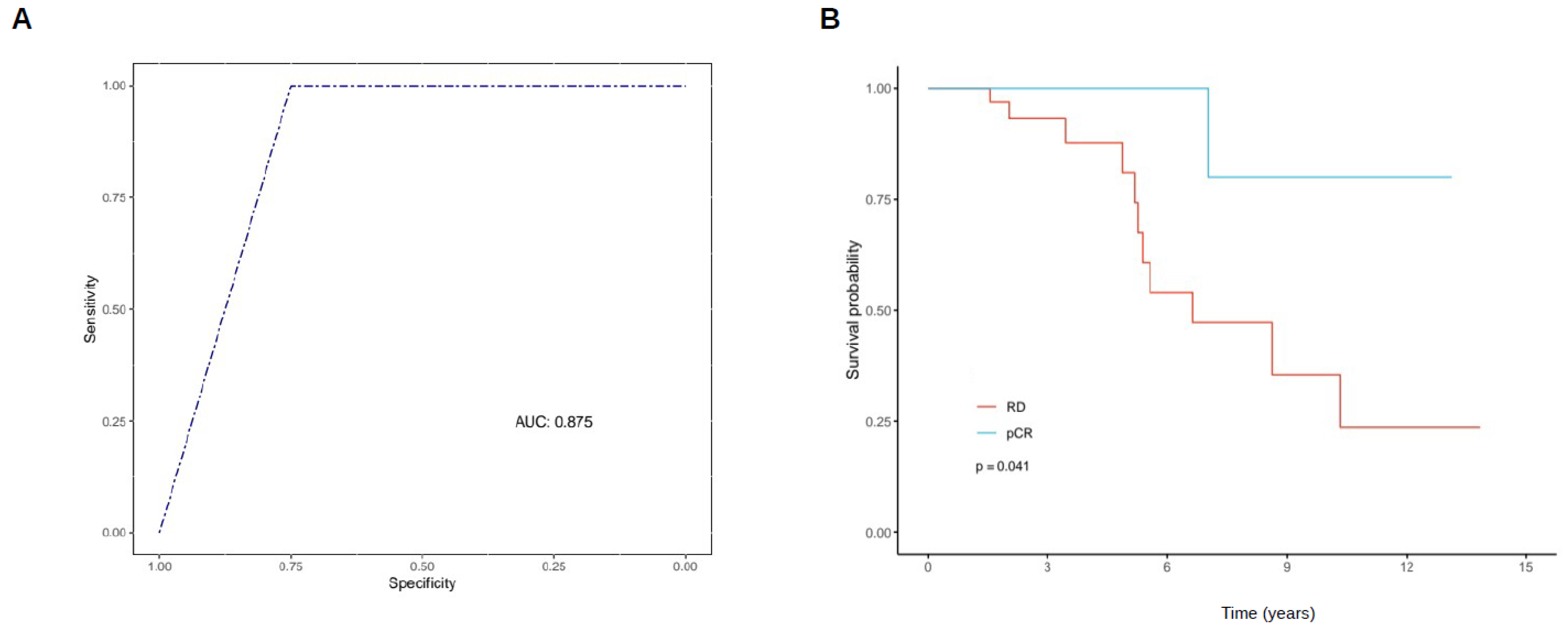
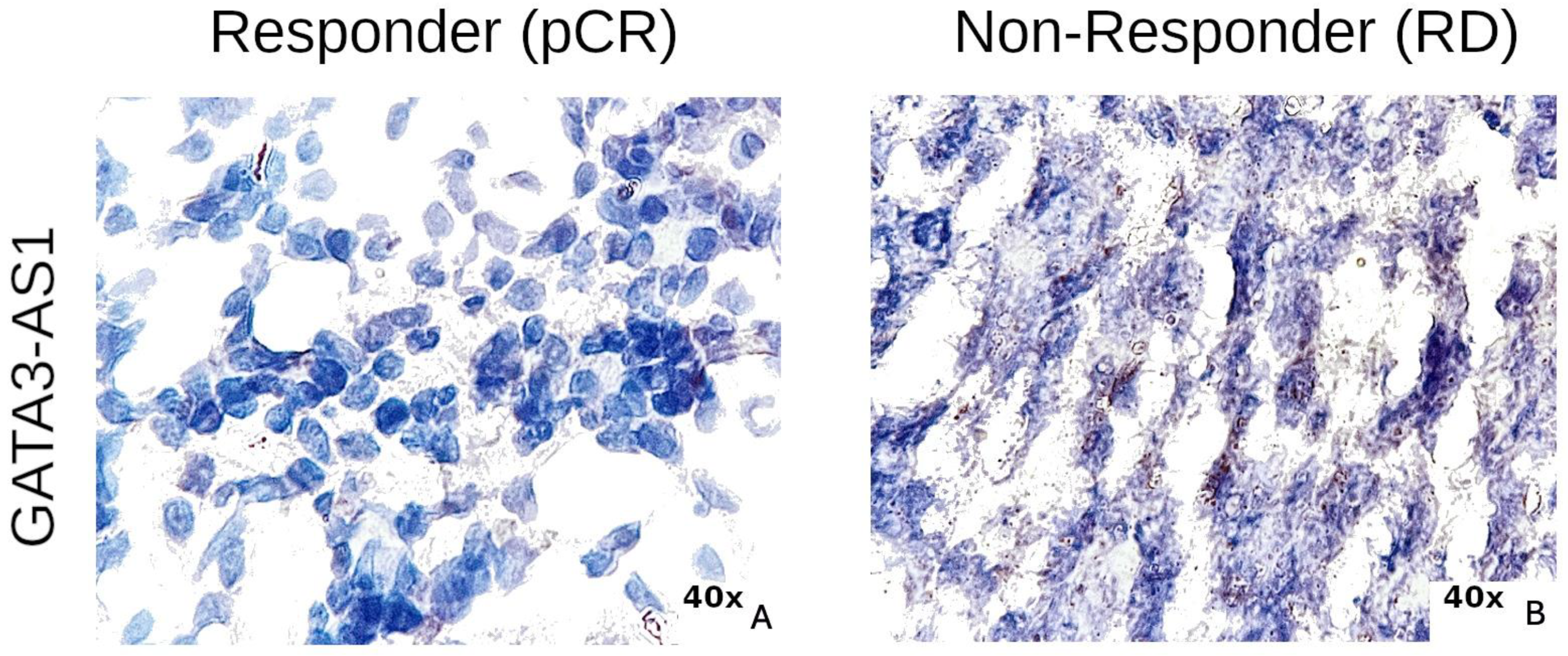
2.3. Experimental Validation Analysis of GATA3-AS1 in Patients with Locally Advanced Breast Cancer
3. Discussion
4. Materials and Methods
4.1. Total RNA Extraction
4.2. Real-Time PCR (RT–qPCR) Validation
4.3. RNA ISH
4.4. RNA Sequencing for Independent Validation Cohort
4.5. Bioinformatic Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Unger-Saldaña, K.; Bandala-Jacques, A.; Huerta-Gutierrez, R.; Zamora-Muñoz, S.; Hernández-Ávila, J.E.; Cabrera-Galeana, P.; Mohar, A.; Lajous, M. Breast Cancer Survival in Mexico between 2007 and 2016 in Women without Social Security: A Retrospective Cohort Study. Lancet Reg. Health Am. 2023, 23, 100541. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; García-Sáenz, J.A.; Fuentes-Ferrer, M.; López Garcia-Asenjo, J.A.; Furió, V.; Román, J.M.; Moreno, A.; de la Hoya, M.; Díaz-Rubio, E.; Martín, M.; et al. Correlation between Response to Neoadjuvant Chemotherapy and Survival in Locally Advanced Breast Cancer Patients. Ann. Oncol. 2013, 24, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Rodenhuis, S.; Mandjes, I.a.M.; Wesseling, J.; van deVijver, M.J.; Peeters, M.-J.T.D.F.V.; Sonke, G.S.; Linn, S.C. A Simple System for Grading the Response of Breast Cancer to Neoadjuvant Chemotherapy. Ann. Oncol. 2010, 21, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef]
- Wallden, B.; Storhoff, J.; Nielsen, T.; Dowidar, N.; Schaper, C.; Ferree, S.; Liu, S.; Leung, S.; Geiss, G.; Snider, J.; et al. Development and Verification of the PAM50-Based Prosigna Breast Cancer Gene Signature Assay. BMC Med. Genom. 2015, 8, 54. [Google Scholar] [CrossRef]
- Cheang, M.C.U.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 Index, HER2 Status, and Prognosis of Patients with Luminal B Breast Cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene Expression Patterns of Breast Carcinomas Distinguish Tumor Subclasses with Clinical Implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Millikan, R.C.; Newman, B.; Tse, C.-K.; Moorman, P.G.; Conway, K.; Dressler, L.G.; Smith, L.V.; Labbok, M.H.; Geradts, J.; Bensen, J.T.; et al. Epidemiology of Basal-like Breast Cancer. Breast Cancer Res. Treat. 2008, 109, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Tse, C.K.; Edmiston, S.; et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, B.L.; Barnard-Tidy, J.; Eeden, R.I.V.; Smit, T.; Nayler, S.; Benn, C. Pathological Complete Response in Early Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy: Focus on Ki-67 and Molecular Subtypes. Ann. Oncol. 2019, 30, iii37. [Google Scholar] [CrossRef]
- Haque, W.; Verma, V.; Hatch, S.; Suzanne Klimberg, V.; Brian Butler, E.; Teh, B.S. Response Rates and Pathologic Complete Response by Breast Cancer Molecular Subtype Following Neoadjuvant Chemotherapy. Breast Cancer Res. Treat. 2018, 170, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Walens, A.; Lin, J.; Damrauer, J.S.; McKinney, B.; Lupo, R.; Newcomb, R.; Fox, D.B.; Mabe, N.W.; Gresham, J.; Sheng, Z.; et al. Adaptation and Selection Shape Clonal Evolution of Tumors during Residual Disease and Recurrence. Nat. Commun. 2020, 11, 5017. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, S.; Xin, Q.; Zhang, Y.; Wang, K.; Li, M. Recent Progress of CDK4/6 Inhibitors’ Current Practice in Breast Cancer. Cancer Gene Ther. 2024. [Google Scholar] [CrossRef]
- Piccart, M.; van ’t Veer, L.J.; Poncet, C.; Lopes Cardozo, J.M.N.; Delaloge, S.; Pierga, J.-Y.; Vuylsteke, P.; Brain, E.; Vrijaldenhoven, S.; Neijenhuis, P.A.; et al. 70-Gene Signature as an Aid for Treatment Decisions in Early Breast Cancer: Updated Results of the Phase 3 Randomised MINDACT Trial with an Exploratory Analysis by Age. Lancet Oncol. 2021, 22, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Derouane, F.; van Marcke, C.; Berlière, M.; Gerday, A.; Fellah, L.; Leconte, I.; Van Bockstal, M.R.; Galant, C.; Corbet, C.; Duhoux, F.P. Predictive Biomarkers of Response to Neoadjuvant Chemotherapy in Breast Cancer: Current and Future Perspectives for Precision Medicine. Cancers 2022, 14, 3876. [Google Scholar] [CrossRef]
- Beitsch, P.D.; Pellicane, J.V.; Pusztai, L.; Baron, P.; Cobain, E.F.; Murray, M.K.; Ashikari, A.; Kelemen, P.R.; Mislowsky, A.M.; Barone, J.; et al. MammaPrint Index as a Predictive Biomarker for Neoadjuvant Chemotherapy Response and Outcome in Patients with HR+HER2- Breast Cancer in NBRST. J. Clin. Oncol. 2023, 41, 521. [Google Scholar] [CrossRef]
- McVeigh, T.P.; Kerin, M.J. Clinical Use of the Oncotype DX Genomic Test to Guide Treatment Decisions for Patients with Invasive Breast Cancer. Breast Cancer 2017, 9, 393–400. [Google Scholar] [CrossRef] [PubMed]
- American Association for Cancer Research. MammaPrint Reduces Breast Cancer Overtreatment. Cancer Discov. 2016, 6, OF4. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Liu, Y.-H.; Martin, T.A.; Jiang, W.G. The Era of Multigene Panels Comes? The Clinical Utility of Oncotype DX and MammaPrint. World J. Oncol. 2017, 8, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Lee, K.H.; Kim, T.R.; Chun, Y.S.; Lee, T.H.; Park, H.K. Ki-67 as a Predictor of Response to Neoadjuvant Chemotherapy in Breast Cancer Patients. J. Breast Cancer 2014, 17, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Liefaard, M.C.; van der Voort, A.; van Seijen, M.; Thijssen, B.; Sanders, J.; Vonk, S.; Mittempergher, L.; Bhaskaran, R.; de Munck, L.; van Leeuwen-Stok, A.E.; et al. Tumor-Infiltrating Lymphocytes in HER2-Positive Breast Cancer Treated with Neoadjuvant Chemotherapy and Dual HER2-Blockade. npj Breast Cancer 2024, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Espinosa, L.; Alcaraz, N.; De La Rosa-Velázquez, I.A.; Díaz-Chávez, J.; Cabrera-Galeana, P.; Vega, R.R.; Reynoso-Noveron, N.; Maldonado-Martínez, H.A.; González-Barrios, R.; Montiel-Manríquez, R.; et al. Transcriptome Analysis Identifies GATA3-AS1 as a Long Noncoding RNA Associated with Resistance to Neoadjuvant Chemotherapy in Locally Advanced Breast Cancer Patients. J. Mol. Diagn. 2021, 23, 1306–1323. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, L.; Cirmena, G.; Garuti, A.; Scabini, S.; Grillo, F.; Mastracci, L.; Isnaldi, E.; Marrone, C.; Gonella, R.; Murialdo, R.; et al. Development of a Long Non-Coding RNA Signature for Prediction of Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Adenocarcinoma. PLoS ONE 2020, 15, e0226595. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Niknafs, Y.S.; Han, S.; Ma, T.; Speers, C.; Zhang, C.; Wilder-Romans, K.; Iyer, M.K.; Pitchiaya, S.; Malik, R.; Hosono, Y.; et al. The lncRNA Landscape of Breast Cancer Reveals a Role for DSCAM-AS1 in Breast Cancer Progression. Nat. Commun. 2016, 7, 12791. [Google Scholar] [CrossRef]
- Moradi, M.-T.; Fallahi, H.; Rahimi, Z. The Clinical Significance of Circulating DSCAM-AS1 in Patients with ER-Positive Breast Cancer and Construction of Its Competitive Endogenous RNA Network. Mol. Biol. Rep. 2020, 47, 7685–7697. [Google Scholar] [CrossRef]
- Huang, N.; Chi, Y.; Xue, J.; Liu, M.; Huang, S.; Mo, M.; Zhou, S.; Wu, J. Long Non-Coding RNA Metastasis Associated in Lung Adenocarcinoma Transcript 1 (MALAT1) Interacts with Estrogen Receptor and Predicted Poor Survival in Breast Cancer. Oncotarget 2016, 7, 37957–37965. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Li, C.; Xing, Z.; Hu, Q.; Liang, K.; Han, L.; Wang, C.; Hawke, D.H.; Wang, S.; Zhang, Y.; et al. The LINK-A lncRNA Activates Normoxic HIF1α Signalling in Triple-Negative Breast Cancer. Nat. Cell Biol. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Huang, J.; Zhou, N.; Watabe, K.; Lu, Z.; Wu, F.; Xu, M.; Mo, Y.-Y. Long Non-Coding RNA UCA1 Promotes Breast Tumor Growth by Suppression of P27 (Kip1). Cell Death Dis. 2014, 5, e1008. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR Induces Epithelial-to-Mesenchymal Transition and Contributes to Breast Cancer Tumorigenesis and Metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The Long Noncoding RNA H19 Promotes Tamoxifen Resistance in Breast Cancer via Autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef]
- Zhou, W.; Pang, Y.; Yao, Y.; Qiao, H. Development of a Ten-lncRNA Signature Prognostic Model for Breast Cancer Survival: A Study with the TCGA Database. Anal. Cell. Pathol. 2020, 2020, e6827057. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Bui, L.T.; Nguyen, C.V.; Do, K.H.; Tran, G.L.; Do, T.A. Neoadjuvant Doxorubicin-Paclitaxel Combined Chemotherapy in Patients with Inoperable Stage III Breast Cancer: A Retrospective Cohort Study with 10 Years of Follow-Up in Vietnam. Oncol. Ther. 2023, 11, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, H.; Ma, R.; Guo, X.; Zhang, G.; Liu, S.; Zhu, W.; Liu, H.; Gao, P. ETS-1-Activated LINC01016 over-Expression Promotes Tumor Progression via Suppression of RFFL-Mediated DHX9 Ubiquitination Degradation in Breast Cancers. Cell Death Dis. 2023, 14, 507. [Google Scholar] [CrossRef] [PubMed]
- Macari, A.; Soberanis-Pina, P.; Varela-Santoyo, E.; Valle-Sanchez, M.A.; Leal-Hidalgo, J.L.; Torres-Guillen, V.M.; Motola-Kuba, D.; Ruiz-Morales, J.M.; Dorantes-Heredia, R. Prevalence and Molecular Profile of Breast Carcinoma Using Immunohistochemistry Markers in Mexican Women. World J. Oncol. 2021, 12, 119–123. [Google Scholar] [CrossRef]
- Arriaga-Canon, C.; Contreras-Espinosa, L.; Aguilar-Villanueva, S.; Bargalló-Rocha, E.; García-Gordillo, J.A.; Cabrera-Galeana, P.; Castro-Hernández, C.; Jiménez-Trejo, F.; Herrera, L.A. The Clinical Utility of lncRNAs and Their Application as Molecular Biomarkers in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 7426. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.H.; Lal, S.; Lee, J.E.; Choi, Y.-L.; Wen, J.; Ram, S.; Ding, Y.; Lee, S.-H.; Powell, E.; Lee, S.K.; et al. Chemotherapy Induces Dynamic Immune Responses in Breast Cancers That Impact Treatment Outcome. Nat. Commun. 2020, 11, 6175. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hao, L.; Qian, X.; Lin, L.; Pan, Y.; Han, X. Machine Learning Models Based on Immunological Genes to Predict the Response to Neoadjuvant Therapy in Breast Cancer Patients. Front. Immunol. 2022, 13, 948601. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Saal, L.H.; Vallon-Christersson, J.; Häkkinen, J.; Hegardt, C.; Grabau, D.; Winter, C.; Brueffer, C.; Tang, M.-H.E.; Reuterswärd, C.; Schulz, R.; et al. The Sweden Cancerome Analysis Network—Breast (SCAN-B) Initiative: A Large-Scale Multicenter Infrastructure towards Implementation of Breast Cancer Genomic Analyses in the Clinical Routine. Genome Med. 2015, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, H.R.; Spurlock, C.F.; Aune, T.M. A Long-Noncoding RNA, Gata3-AS1, Is a Positive Transcriptional Regulator of Transcription Factor Gata3 in TH2 Cells. J. Immunol. 2017, 198, 150.11. [Google Scholar] [CrossRef]
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F.I.; Aune, T.M. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front. Immunol. 2018, 9, 2512. [Google Scholar] [CrossRef] [PubMed]
- Takaku, M.; Grimm, S.A.; Roberts, J.D.; Chrysovergis, K.; Bennett, B.D.; Myers, P.; Perera, L.; Tucker, C.J.; Perou, C.M.; Wade, P.A. GATA3 Zinc Finger 2 Mutations Reprogram the Breast Cancer Transcriptional Network. Nat. Commun. 2018, 9, 1059. [Google Scholar] [CrossRef]
- Yu, X.; Guo, S.; Song, W.; Xiang, T.; Yang, C.; Tao, K.; Zhou, L.; Cao, Y.; Liu, S. Estrogen Receptor α (ERα) Status Evaluation Using RNAscope in Situ Hybridization: A Reliable and Complementary Method for IHC in Breast Cancer Tissues. Hum. Pathol. 2017, 61, 121–129. [Google Scholar] [CrossRef]
- Brown, J.R.; DiGiovanna, M.P.; Killelea, B.; Lannin, D.R.; Rimm, D.L. Quantitative Assessment Ki-67 Score for Prediction of Response to Neoadjuvant Chemotherapy in Breast Cancer. Lab. Investig. 2014, 94, 98–106. [Google Scholar] [CrossRef]
- Seo, J.; Koh, J.; Lee, D.-W.; Kim, J.; Ryu, H.S.; Lee, K.-H.; Kim, T.-Y.; Im, S.-A. HER2 Amplification Level by in Situ Hybridization Predicts Survival Outcome in Advanced HER2-Positive Breast Cancer Treated with Pertuzumab, Trastuzumab, and Docetaxel Regardless of HER2 IHC Results. Breast Cancer Res. 2023, 25, 154. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Liu, C.; Hu, C.; Yuan, J.; Liao, W.; Xiao, Z. Prognostic Factors for Luminal B-like Breast Cancer. Curr. Med. Sci. 2019, 39, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Guarneri, V.; Tosi, A.; Bisagni, G.; Musolino, A.; Spazzapan, S.; Moretti, G.; Vernaci, G.M.; Griguolo, G.; Giarratano, T.; et al. Neoadjuvant Chemotherapy and Immunotherapy in Luminal B-like Breast Cancer: Results of the Phase II GIADA Trial. Clin. Cancer Res. 2022, 28, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Villarreal-Garza, C.; Mohar, A.; Bargallo-Rocha, J.E.; Lasa-Gonsebatt, F.; Reynoso-Noverón, N.; Matus-Santos, J.; Cabrera, P.; Arce-Salinas, C.; Lara-Medina, F.; Alvarado-Miranda, A.; et al. Molecular Subtypes and Prognosis in Young Mexican Women With Breast Cancer. Clin. Breast Cancer 2017, 17, e95–e102. [Google Scholar] [CrossRef] [PubMed]
- Paramita, S.; Raharjo, E.N.; Niasari, M.; Azizah, F.; Hanifah, N.A. Luminal B Is the Most Common Intrinsic Molecular Subtypes of Invasive Ductal Breast Carcinoma Patients in East Kalimantan, Indonesia. Asian Pac. J. Cancer Prev. 2019, 20, 2247–2252. [Google Scholar] [CrossRef]
- Xia, Q.; Yan, Q.; Wang, Z.; Huang, Q.; Zheng, X.; Shen, J.; Du, L.; Li, H.; Duan, S. Disulfidptosis-Associated lncRNAs Predict Breast Cancer Subtypes. Sci. Rep. 2023, 13, 16268. [Google Scholar] [CrossRef]
- Voith von Voithenberg, L.; Fomitcheva Khartchenko, A.; Huber, D.; Schraml, P.; Kaigala, G.V. Spatially Multiplexed RNA in Situ Hybridization to Reveal Tumor Heterogeneity. Nucleic Acids Res. 2020, 48, e17. [Google Scholar] [CrossRef]
- Hirko, K.A.; Rocque, G.; Reasor, E.; Taye, A.; Daly, A.; Cutress, R.I.; Copson, E.R.; Lee, D.-W.; Lee, K.-H.; Im, S.-A.; et al. The Impact of Race and Ethnicity in Breast Cancer—Disparities and Implications for Precision Oncology. BMC Med. 2022, 20, 72. [Google Scholar] [CrossRef]
- Zhao, F.; Miyashita, M.; Hattori, M.; Yoshimatsu, T.; Howard, F.; Kaneva, K.; Jones, R.; Bell, J.S.K.; Fleming, G.F.; Jaskowiak, N.; et al. Racial Disparities in Pathological Complete Response Among Patients Receiving Neoadjuvant Chemotherapy for Early-Stage Breast Cancer. JAMA Netw. Open 2023, 6, e233329. [Google Scholar] [CrossRef]
- Nakshatri, H.; Kumar, B.; Burney, H.N.; Cox, M.L.; Jacobsen, M.; Sandusky, G.E.; D’Souza-Schorey, C.; Storniolo, A.M.V. Genetic Ancestry–Dependent Differences in Breast Cancer–Induced Field Defects in the Tumor-Adjacent Normal Breast. Clin. Cancer Res. 2019, 25, 2848–2859. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Sahoo, S.; Brien, R.; Jung, S.; Humphries, B.; Lee, W.; Cheng, Y.-H.; Zhang, Z.; Luker, K.E.; Wicha, M.S.; et al. Single-Cell RNA-Sequencing of Migratory Breast Cancer Cells: Discovering Genes Associated with Cancer Metastasis. Analyst 2019, 144, 7296–7309. [Google Scholar] [CrossRef] [PubMed]
- Bartoschek, M.; Oskolkov, N.; Bocci, M.; Lövrot, J.; Larsson, C.; Sommarin, M.; Madsen, C.D.; Lindgren, D.; Pekar, G.; Karlsson, G.; et al. Spatially and Functionally Distinct Subclasses of Breast Cancer-Associated Fibroblasts Revealed by Single Cell RNA Sequencing. Nat. Commun. 2018, 9, 5150. [Google Scholar] [CrossRef] [PubMed]
- McKellar, D.W.; Mantri, M.; Hinchman, M.M.; Parker, J.S.L.; Sethupathy, P.; Cosgrove, B.D.; De Vlaminck, I. Spatial Mapping of the Total Transcriptome by in Situ Polyadenylation. Nat. Biotechnol. 2023, 41, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/Help/1%20Introduction/1.1%20What%20is%20FastQC.html (accessed on 17 July 2024).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Pan, B.; Kusko, R.; Xiao, W.; Zheng, Y.; Liu, Z.; Xiao, C.; Sakkiah, S.; Guo, W.; Gong, P.; Zhang, C.; et al. Similarities and Differences between Variants Called with Human Reference Genome HG19 or HG38. BMC Bioinform. 2019, 20, 101. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon: Fast and Bias-Aware Quantification of Transcript Expression Using Dual-Phase Inference. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Kronthaler, F.; Zöllner, S. R and RStudio. In Data Analysis with RStudio: An Easygoing Introduction; Kronthaler, F., Zöllner, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–12. ISBN 978-3-662-62518-7. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast Gene Set Enrichment Analysis. bioRxiv 2021, 060012. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, T.-Y.; Anderton, L.C.; Cao, Q.; Yang, R. LncGSEA: A Versatile Tool to Infer lncRNA Associated Pathways from Large-Scale Cancer Transcriptome Sequencing Data. BMC Genom. 2021, 22, 574. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Aria, M.; Cuccurullo, C.; Gnasso, A. A Comparison among Interpretative Proposals for Random Forests. Mach. Learn. Appl. 2021, 6, 100094. [Google Scholar] [CrossRef]



| Clinical Characteristic | Resistant Disease (RD) | Pathological Complete Response (pCR) | p Value |
|---|---|---|---|
| Mean age, years (SD) | 47 (9.9) | 44 (10.3) | 0.38 † |
| Menopause, n (%) | 0.86 ƒ | ||
| Premenopause | 24 (60%) | 4 (44.4%) | |
| Postmenopause | 16 (40%) | 5 (55.6%) | |
| Histology, n (%) | 0.52 ƒ | ||
| Infiltrating ductal carcinoma | 25 (64.1%) | 6 (75%) | |
| Infiltrating canalicular carcinoma | 9 (23.1%) | 1 (12.5%) | |
| Infiltrating lobular carcinoma | 5 (12.8%) | 1 (12.5%) | |
| T, n (%) | 0.52 ƒ | ||
| 1 | 1 (2.5%) | 1 (11.1%) | |
| 2 | 15 (37.5%) | 4 (44.4%) | |
| 3 | 15 (37.5%) | 3 (33.3%) | |
| 4 | 9 (22.5%) | 1 (11.1%) | |
| N, n (%) | |||
| 0 | 0 | 2 (22.2%) | 0.05 ƒ |
| 1 | 18 (45%) | 2 (22.2%) | |
| 2 | 12 (30%) | 3 (33.3%) | |
| 3 | 10 (25%) | 2 (22.2%) | |
| Clinical stage, n (%) | 0.47 ƒ | ||
| IIB | 8 (20%) | 4 (44.4%) | |
| IIIA | 16 (40%) | 3 (33.3%) | |
| IIIB | 5 (12.5%) | 0 | |
| IIIC | 11 (27.5%) | 2 (22.2%) | |
| Ki67 expression %, median (range) | 30 (20–40) | 30 (20–40) | 0.73 § |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Woge, M.; Contreras-Espinosa, L.; García-Gordillo, J.A.; Aguilar-Villanueva, S.; Bargallo-Rocha, E.; Cabrera-Galeana, P.; Vasquez-Mata, T.; Cervantes-López, X.; Vargas-Lías, D.S.; Montiel-Manríquez, R.; et al. The Expression Profiles of lncRNAs Are Associated with Neoadjuvant Chemotherapy Resistance in Locally Advanced, Luminal B-Type Breast Cancer. Int. J. Mol. Sci. 2024, 25, 8077. https://doi.org/10.3390/ijms25158077
González-Woge M, Contreras-Espinosa L, García-Gordillo JA, Aguilar-Villanueva S, Bargallo-Rocha E, Cabrera-Galeana P, Vasquez-Mata T, Cervantes-López X, Vargas-Lías DS, Montiel-Manríquez R, et al. The Expression Profiles of lncRNAs Are Associated with Neoadjuvant Chemotherapy Resistance in Locally Advanced, Luminal B-Type Breast Cancer. International Journal of Molecular Sciences. 2024; 25(15):8077. https://doi.org/10.3390/ijms25158077
Chicago/Turabian StyleGonzález-Woge, Miguel, Laura Contreras-Espinosa, José Antonio García-Gordillo, Sergio Aguilar-Villanueva, Enrique Bargallo-Rocha, Paula Cabrera-Galeana, Tania Vasquez-Mata, Ximena Cervantes-López, Diana Sofía Vargas-Lías, Rogelio Montiel-Manríquez, and et al. 2024. "The Expression Profiles of lncRNAs Are Associated with Neoadjuvant Chemotherapy Resistance in Locally Advanced, Luminal B-Type Breast Cancer" International Journal of Molecular Sciences 25, no. 15: 8077. https://doi.org/10.3390/ijms25158077






