The Establishment of Complement System Is from Gene Duplication and Domain Shuffling
Abstract
:1. Introduction
2. Results
2.1. The Distribution, Structural Domain, and Phylogenetic Tree Analysis of C1qs, Ficolins, and MBLs
2.2. The Distribution, Structural Domain, and Phylogenetic Tree Analysis of C1rs, C1ss, and MASPs
2.3. The Distribution, Structural Domain, and Phylogenetic Tree Analysis of C2s, BFs, and DFs
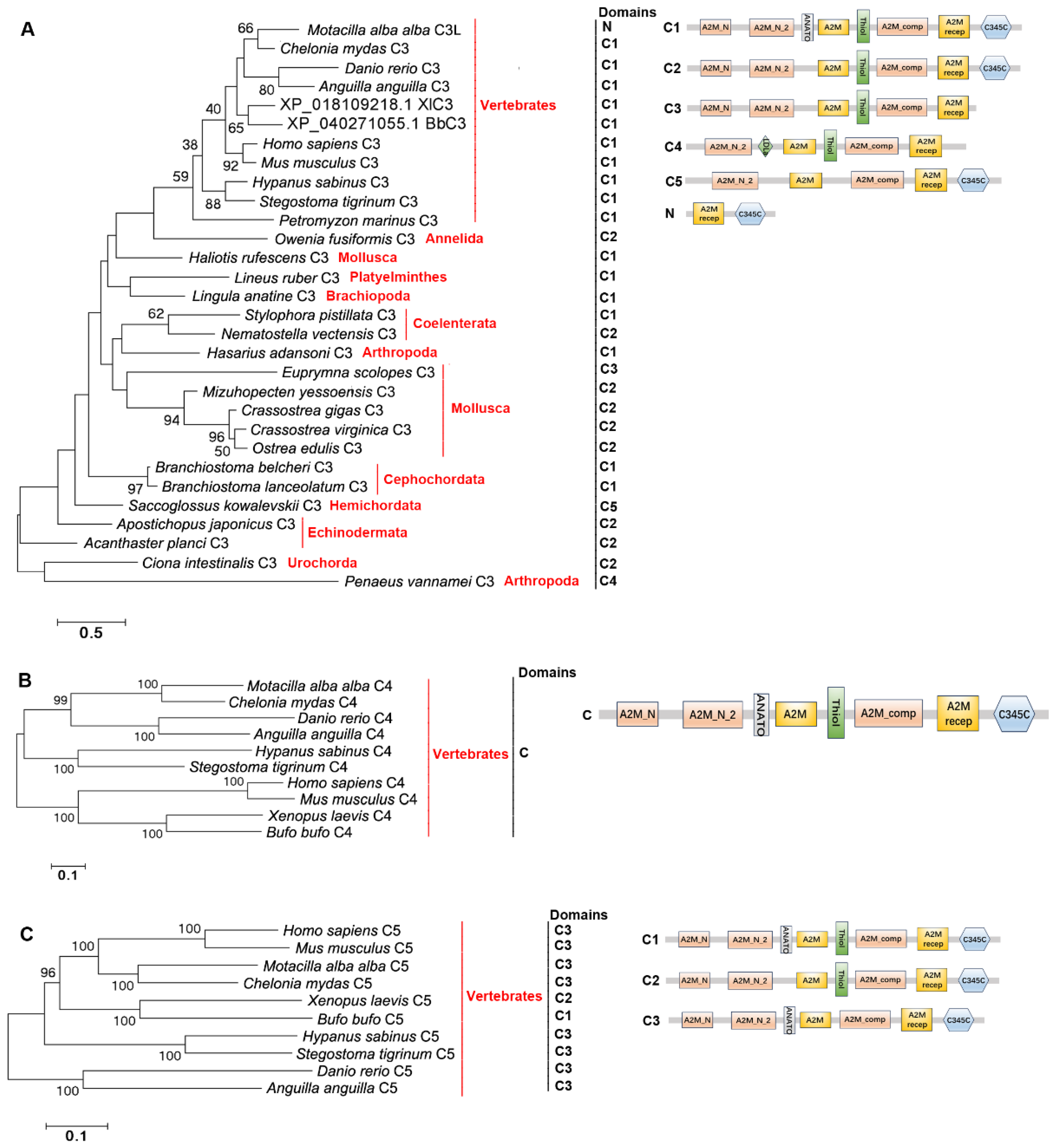
2.4. The Distribution, Structural Domain, and Phylogenetic Tree Analysis of C3s, C4s, and C5s
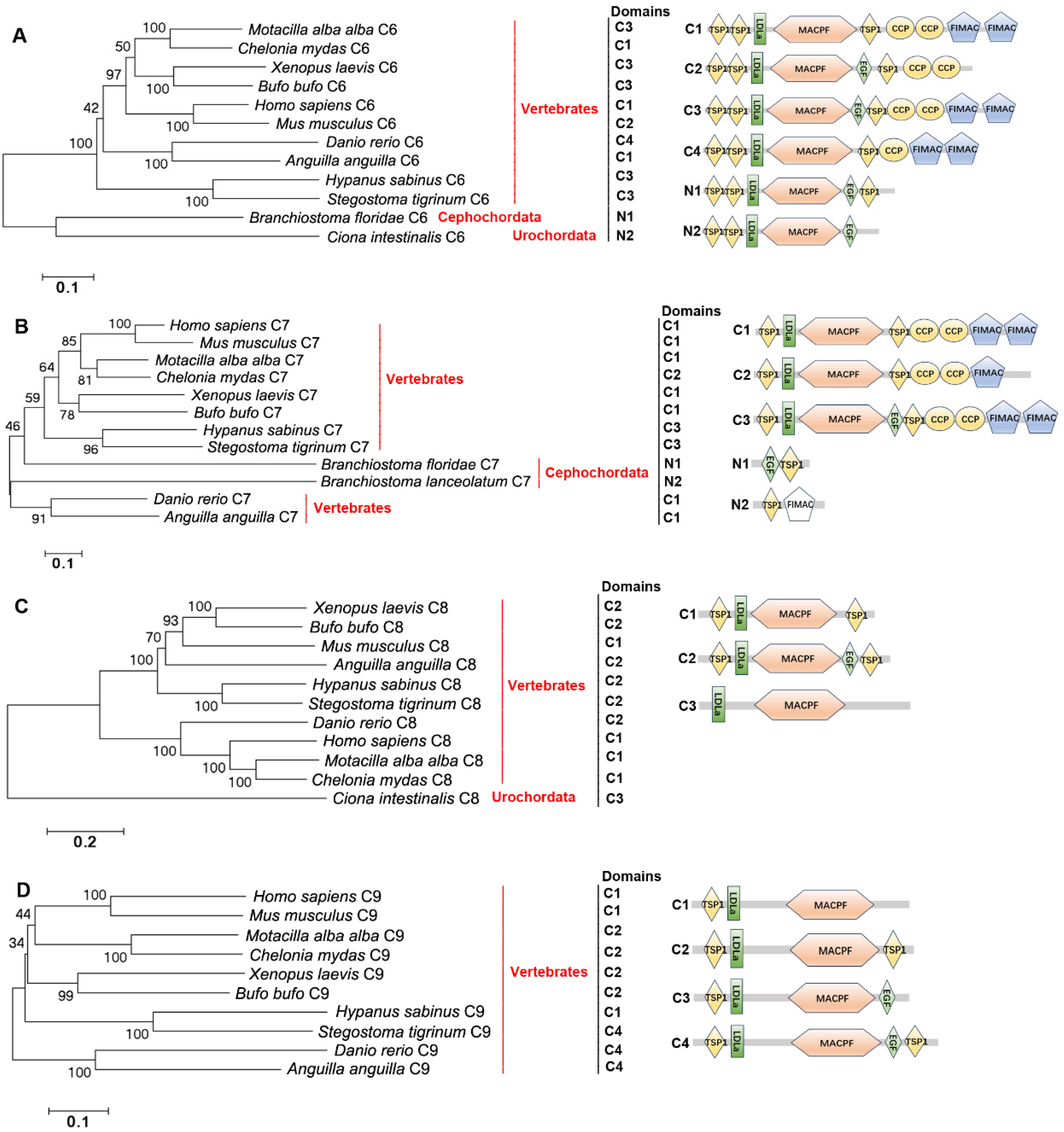
2.5. The Distribution, Structural Domain, and Phylogenetic Tree Analysis of C6s, C7s, C8s, and C9s
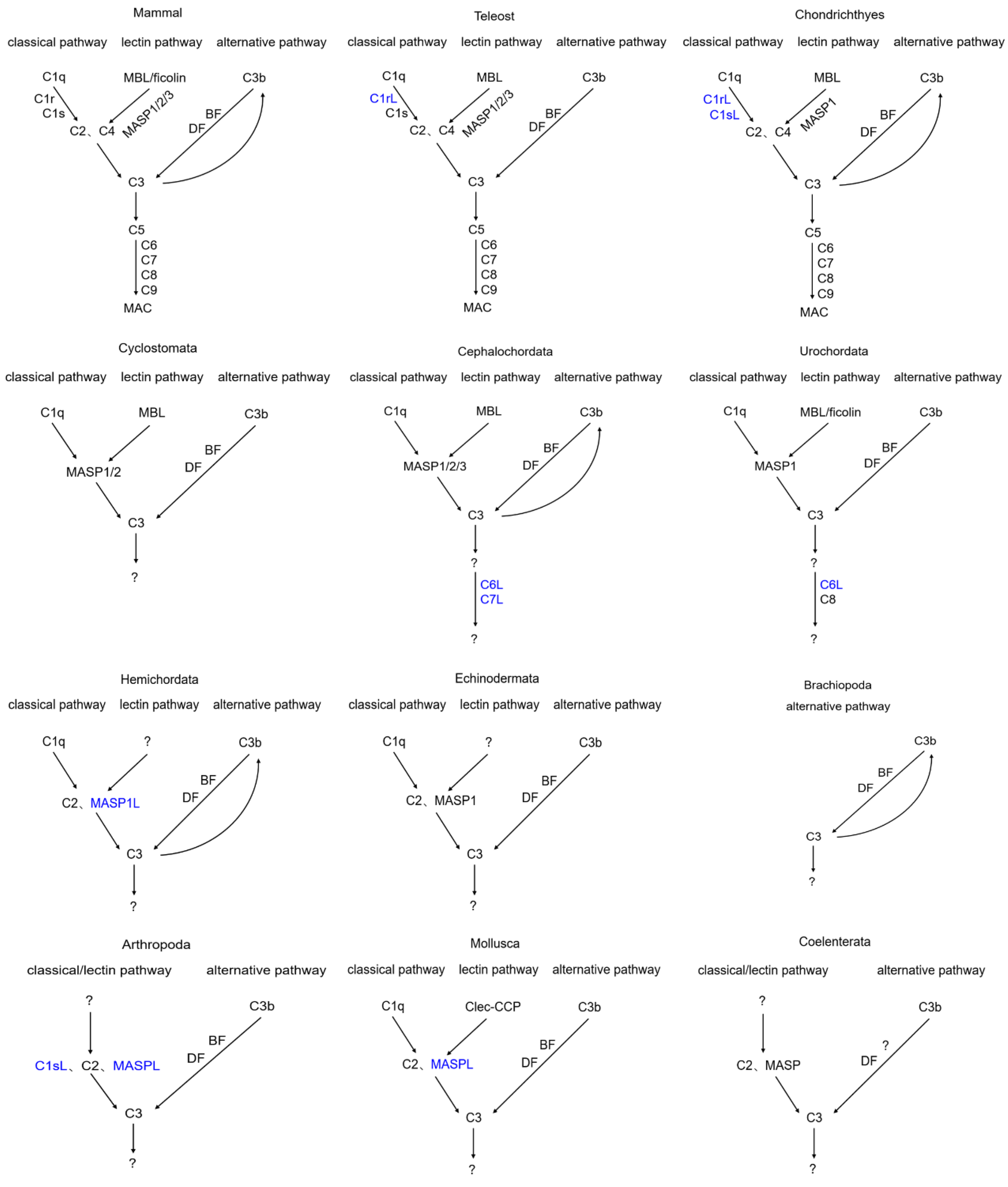
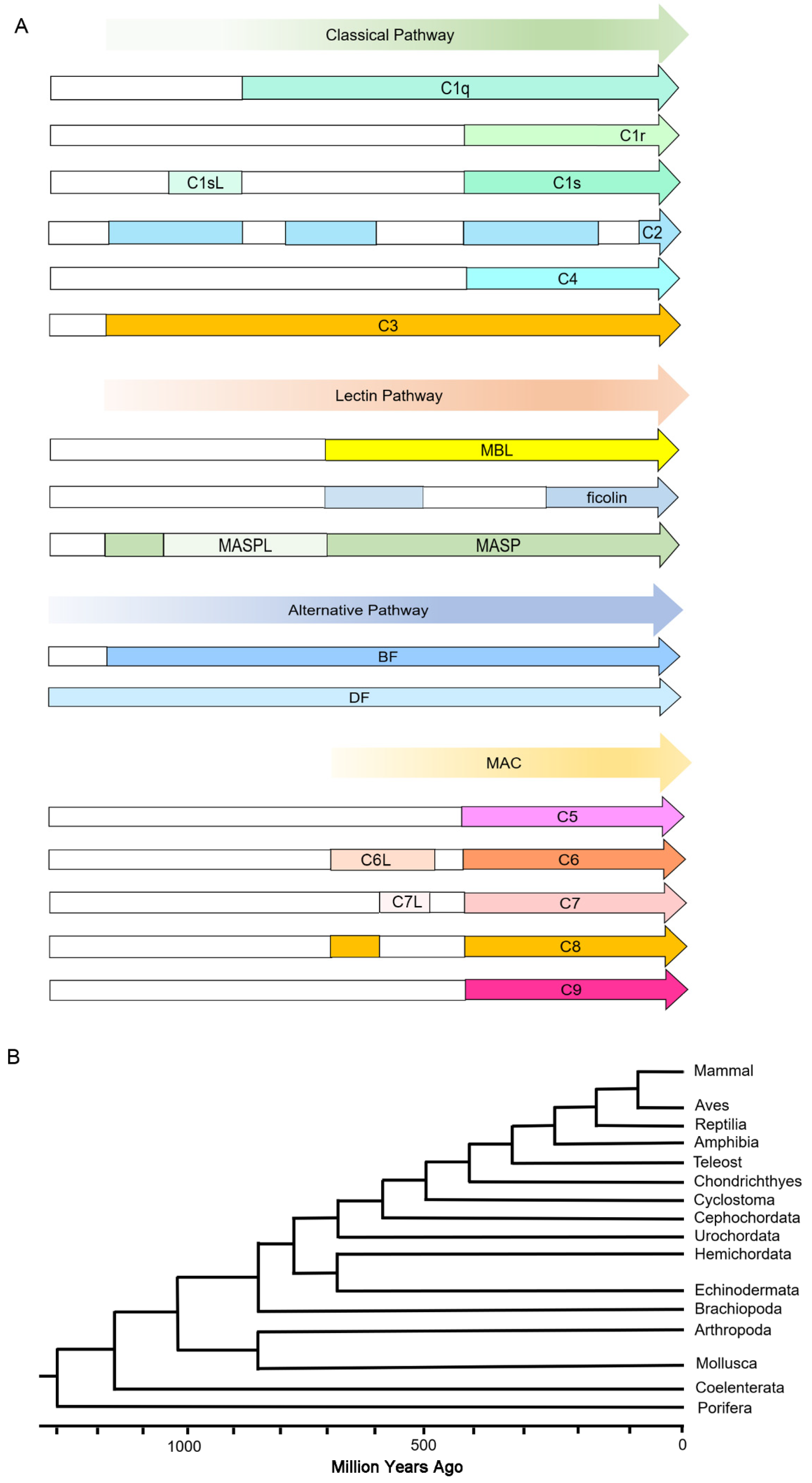
3. Discussion
4. Materials and Methods
4.1. The Amino Acid Sequences of Complement Components
4.2. BLASTp Analysis of Complement Components
4.3. The Evolutionary Analysis of Complement Components
4.4. The Structural Domain Analysis of Complement Components
4.5. The Evolutionary Tree of Metazoan
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nonaka, M. Evolution of the complement system. Sub-cell. Biochem. 2014, 80, 31–43. [Google Scholar]
- Nonaka, M.; Kimura, A. Genomic view of the evolution of the complement system. Immunogenetics 2006, 58, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004, 5, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shen, Y.; Xu, X.; Yang, W.; Li, J. Tracing and exploring the evolutionary origin and systematic function of fish complement C9. Mol. Genet. Genom. (MGG) 2021, 296, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Najafpour, B.; Cardoso, J.C.R.; Canario, A.V.M.; Power, D.M. Specific Evolution and Gene Family Expansion of Complement 3 and Regulatory Factor H in Fish. Front. Immunol. 2020, 11, 568631. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shen, Y.; Xu, X.; Yang, W.; Li, J. Fish complement C4 gene evolution and gene/protein regulatory network analyses and simulated stereo conformation of C4-MASP-2 protein complex. Fish Shellfish Immunol. 2020, 107 Pt A, 54–63. [Google Scholar] [CrossRef]
- Zhu, Y.; Thangamani, S.; Ho, B.; Ding, J.L. The ancient origin of the complement system. EMBO J. 2005, 24, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Sunyer, J.O.; Boshra, H.; Lorenzo, G.; Parra, D.; Freedman, B.; Bosch, N. Evolution of complement as an effector system in innate and adaptive immunity. Immunol. Res. 2003, 27, 549–564. [Google Scholar] [CrossRef]
- Noris, M.; Remuzzi, G. Overview of complement activation and regulation. Semin. Nephrol. 2013, 33, 479–492. [Google Scholar] [CrossRef]
- Dunkelberger, J.R.; Song, W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef]
- Pinto, M.R.; Melillo, D.; Giacomelli, S.; Sfyroera, G.; Lambris, J.D. Ancient origin of the complement system: Emerging invertebrate models. Adv. Exp. Med. Biol. 2007, 598, 372–388. [Google Scholar] [PubMed]
- Li, H.; Kong, N.; Sun, J.; Wang, W.; Li, M.; Gong, C.; Dong, M.; Wang, M.; Wang, L.; Song, L. A C1qDC (CgC1qDC-6) with a collagen-like domain mediates hemocyte phagocytosis and migration in oysters. Dev. Comp. Immunol. 2019, 98, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Nilojan, J.; Bathige, S.; Kugapreethan, R.; Yang, H.; Kim, M.J.; Nam, B.H.; Lee, J. Molecular features and the transcriptional and functional delineation of complement system activators C1r and C1s from Sebastes schlegelii. Dev. Comp. Immunol. 2018, 81, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chen, M.; Ding, M.; Zhong, M.; Li, B.; Wang, Y.; Fu, S.; Yin, X.; Guo, Z.; Ye, J. C1r and C1s from Nile tilapia (Oreochromis niloticus): Molecular characterization, transcriptional profiling upon bacterial and IFN-gamma inductions and potential role in response to bacterial infection. Fish Shellfish Immunol. 2017, 70, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, R.; Xu, T. Genomic characterization and expression pattern of Bf/C2 and C4 in miiuy croaker and molecular evolution analysis on mammals and fishes. Fish Shellfish Immunol. 2014, 39, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Ourth, D.D.; Rose, W.M.; Siefkes, M.J. Isolation of mannose-binding C-type lectin from sea lamprey (Petromyzon marinus) plasma and binding to Aeromonas salmonicida. Vet. Immunol. Immunopathol. 2008, 126, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Skjoedt, M.O.; Palarasah, Y.; Rasmussen, K.; Vitved, L.; Salomonsen, J.; Kliem, A.; Hansen, S.; Koch, C.; Skjodt, K. Two mannose-binding lectin homologues and an MBL-associated serine protease are expressed in the gut epithelia of the urochordate species Ciona intestinalis. Dev. Comp. Immunol. 2010, 34, 59–68. [Google Scholar] [CrossRef]
- Sekine, H.; Kenjo, A.; Azumi, K.; Ohi, G.; Takahashi, M.; Kasukawa, R.; Ichikawa, N.; Nakata, M.; Mizuochi, T.; Matsushita, M.; et al. An ancient lectin-dependent complement system in an ascidian: Novel lectin isolated from the plasma of the solitary ascidian, Halocynthia roretzi. J. Immunol. 2001, 167, 4504–4510. [Google Scholar] [CrossRef]
- Huang, H.; Huang, S.; Yu, Y.; Yuan, S.; Li, R.; Wang, X.; Zhao, H.; Yu, Y.; Li, J.; Yang, M.; et al. Functional characterization of a ficolin-mediated complement pathway in amphioxus. J. Biol. Chem. 2011, 286, 36739–36748. [Google Scholar] [CrossRef]
- Kenjo, A.; Takahashi, M.; Matsushita, M.; Endo, Y.; Nakata, M.; Mizuochi, T.; Fujita, T. Cloning and characterization of novel ficolins from the solitary ascidian, Halocynthia roretzi. J. Biol. Chem. 2001, 276, 19959–19965. [Google Scholar] [CrossRef]
- Miller, D.J.; Hemmrich, G.; Ball, E.E.; Hayward, D.C.; Khalturin, K.; Funayama, N.; Agata, K.; Bosch, T.C. The innate immune repertoire in cnidaria-ancestral complexity and stochastic gene loss. Genome Biol. 2007, 8, R59. [Google Scholar] [CrossRef] [PubMed]
- Dehal, P.; Satou, Y.; Campbell, R.K.; Chapman, J.; Degnan, B.; De Tomaso, A.; Davidson, B.; Di Gregorio, A.; Gelpke, M.; Goodstein, D.M.; et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 2002, 298, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 1998, 282, 2012–2018. [CrossRef] [PubMed]
- Nakao, M.; Uemura, T.; Yano, T. Terminal components of carp complement constituting a membrane attack complex. Mol. Immunol. 1996, 33, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Nonaka, M. Molecular cloning of the terminal complement components C6 and C8β of cartilaginous fish. Fish Shellfish Immunol. 2009, 27, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Rus, H.; Cudrici, C.; Niculescu, F. The role of the complement system in innate immunity. Immunol. Res. 2005, 33, 103–112. [Google Scholar] [CrossRef]
- Kulkarni, D.H.; Starick, M.; Alburquerque, R.A.; Kulkarni, H.S. Local complement activation and modulation in mucosal immunity. Mucosal Immunol. 2024, 4, S1933-0219(24)00047-3. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.S.; Wijeyewickrema, L.C.; Hor, L.; Tan, S.; Lameignere, E.; Conway, E.M.; Blom, A.M.; Mohlin, F.C.; Liu, X.; Payne, R.J.; et al. The Structural Basis for Complement Inhibition by Gigastasin, a Protease Inhibitor from the Giant Amazon Leech. J. Immunol. 2017, 199, 3883–3891. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Endo, Y.; Nonaka, M. Primitive complement system—Recognition and activation. Mol. Immunol. 2004, 41, 103–111. [Google Scholar] [CrossRef]
- Freeley, S.; Kemper, C.; Le Friec, G. The “ins and outs” of complement-driven immune responses. Immunol. Rev. 2016, 274, 16–32. [Google Scholar] [CrossRef]
- Giang, J.; Seelen, M.A.J.; van Doorn, M.B.A.; Rissmann, R.; Prens, E.P.; Damman, J. Complement Activation in Inflammatory Skin Diseases. Front. Immunol. 2018, 9, 639. [Google Scholar] [CrossRef] [PubMed]
- Walport, M.J. Complement. First of two parts. N. Engl. J. Med. 2001, 344, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Gorbushin, A.M. Derivatives of the lectin complement pathway in Lophotrochozoa. Dev. Comp. Immunol. 2019, 94, 35–58. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.R.; Wang, X.X.; Liu, P.P.; Wang, X.W. Complement-related proteins in crustacean immunity. Dev. Comp. Immunol. 2023, 139, 104577. [Google Scholar] [CrossRef]
- Sun, J.; Wang, L.; Song, L. The primitive complement system in molluscs. Dev. Comp. Immunol. 2023, 139, 104565. [Google Scholar] [CrossRef]
- Xiao, K.; Zhang, S.; Li, C. The complement system and complement-like factors in sea cucumber. Dev. Comp. Immunol. 2022, 136, 104511. [Google Scholar] [CrossRef]
- Sun, J.; Wang, L.; Yang, W.; Li, Y.; Jin, Y.; Wang, L.; Song, L. A novel C-type lectin activates the complement cascade in the primitive oyster Crassostrea gigas. J. Biol. Chem. 2021, 297, 101352. [Google Scholar] [CrossRef]
- Nicola, F.; Loriano, B. Morula cells as key hemocytes of the lectin pathway of complement activation in the colonial tunicate Botryllus schlosseri. Fish Shellfish Immunol. 2017, 63, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Almitairi, J.O.M.; Girija, U.V.; Furze, C.M.; Simpson-Gray, X.; Badakshi, F.; Marshall, J.E.; Schwaeble, W.J.; Mitchell, D.A.; Moody, P.C.E.; Wallis, R. Structure of the C1r-C1s interaction of the C1 complex of complement activation. Proc. Natl. Acad. Sci. USA 2018, 115, 768–773. [Google Scholar] [CrossRef]
- Hobart, M.J. Evolution of the terminal complement genes: Ancient and modern. Exp. Clin. Immunogenet. 1998, 15, 235–243. [Google Scholar] [CrossRef]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). BioEssays News Rev. Mol. Cell. Dev. Biol. 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Dehal, P.; Boore, J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M. The Complement System of Agnathans. Front. Immunol. 2018, 9, 1405. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Schartl, M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999, 11, 699–704. [Google Scholar] [CrossRef]
- Matsushita, M.; Matsushita, A.; Endo, Y.; Nakata, M.; Kojima, N.; Mizuochi, T.; Fujita, T. Origin of the classical complement pathway: Lamprey orthologue of mammalian C1q acts as a lectin. Proc. Natl. Acad. Sci. USA 2004, 101, 10127–10131. [Google Scholar] [CrossRef]








| Domain Abbreviation | Complete Name | Domain Abbreviation | Complete Name |
|---|---|---|---|
| Collagen | Collagen domain | CRD | Carbohydrate-recognition domain |
| C1q | C1q domain | CUB | C1r/C1s-Uegf-BMP domain |
| FREP | Fibrinogen-related domain | EGF | Epidermal growth factor |
| CCP | Complement control protein | Tryp_SPc | Trypsin-like serine protease |
| IG | Immunoglobulin | LDLa | Low density lipoprotein A |
| VWA | Von willebrand factor | A2M | Alpha-2-macroglobulin region |
| ANATO | Anaphylatoxin homologous domain | Thiol | Alpha-macro-globulin thiol-ester bond-forming region |
| A2M recep | Receptor domain region of the alpha-2-macroglobulin family | C345C | Netrin C-terminal Domain |
| TSP1 | Thrombospondin type 1 | MACPF | Membrane attack complex/perforin |
| FIMAC | Factor I membrane attack complex |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Liu, C.; Wang, L.; Song, L. The Establishment of Complement System Is from Gene Duplication and Domain Shuffling. Int. J. Mol. Sci. 2024, 25, 8119. https://doi.org/10.3390/ijms25158119
Sun J, Liu C, Wang L, Song L. The Establishment of Complement System Is from Gene Duplication and Domain Shuffling. International Journal of Molecular Sciences. 2024; 25(15):8119. https://doi.org/10.3390/ijms25158119
Chicago/Turabian StyleSun, Jiejie, Chang Liu, Lingling Wang, and Linsheng Song. 2024. "The Establishment of Complement System Is from Gene Duplication and Domain Shuffling" International Journal of Molecular Sciences 25, no. 15: 8119. https://doi.org/10.3390/ijms25158119





