Discordant ALK Status in Non-Small Cell Lung Carcinoma: A Detailed Reevaluation Comparing IHC, FISH, and NGS Analyses
Abstract
1. Introduction
2. Results
2.1. Concordance and Discordance between FISH and IHC
2.2. Demographic Distribution
2.3. Histopathology
2.4. Comparison of ALK Copy Number Gains in ALK Concordant and Discordant Samples
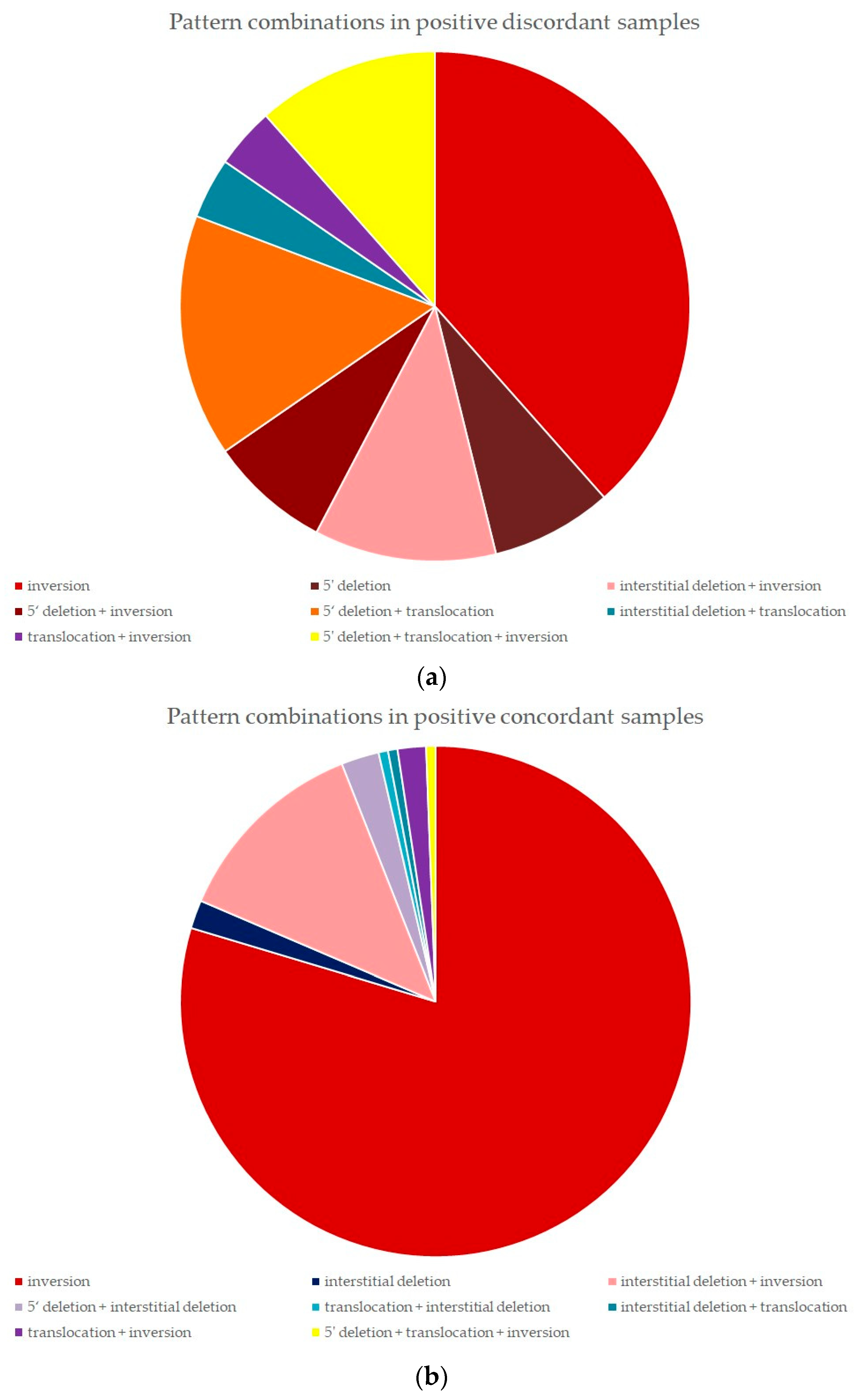
2.5. FISH Pattern Distribution in Positive Discordant and Positive Concordant Samples
2.5.1. Positive Discordant Samples
2.5.2. Positive Concordant Samples
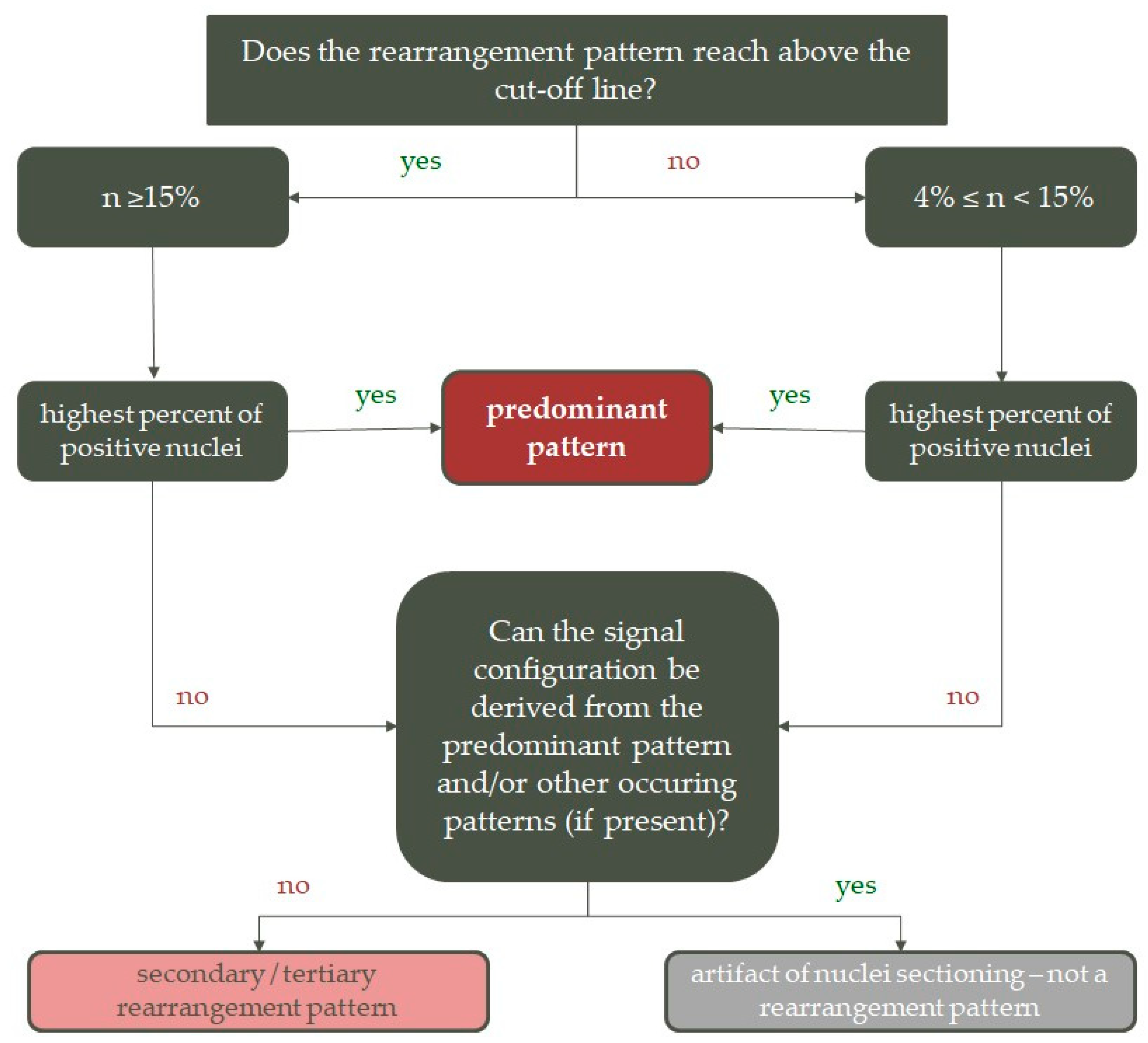
2.5.3. Statistical Analysis of Pattern Distribution
2.6. NGS Analysis
3. Discussion
4. Materials and Methods
4.1. Immunohistochemistry
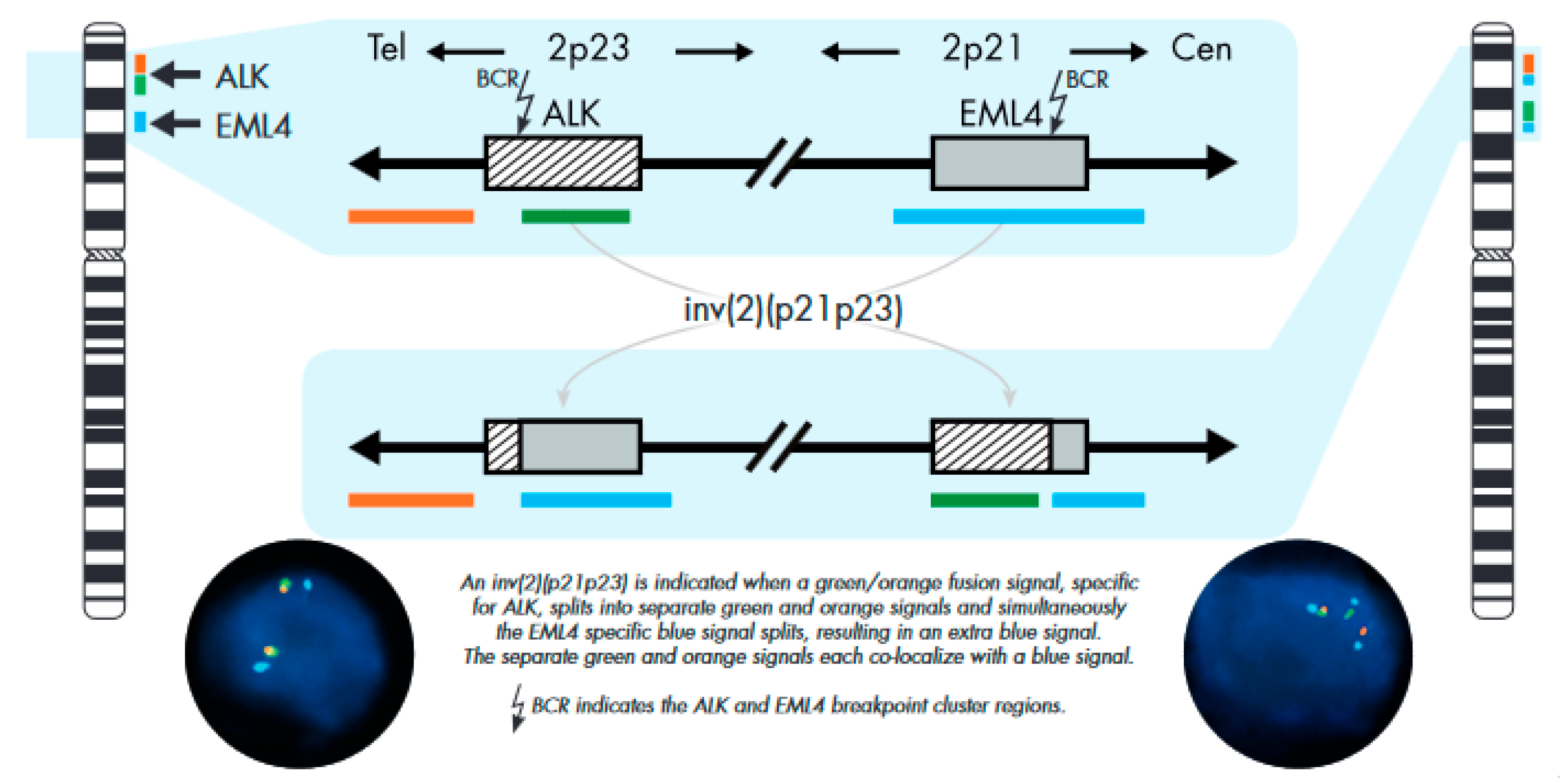
4.2. Fluorescence In Situ Hybridization
FISH Analysis
4.3. Next-Generation Sequencing
4.4. Comparison of IHC and FISH
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.W.; Kirstein, M.N.; Valentine, M.B.; Dittmer, K.G.; Shapiro, D.N.; Saltman, D.L.; Look, A.T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994, 263, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H.; Bartlett, C.H.; Mino-Kenudson, M.; Cui, J.; Iafrate, A.J. Crizotinib for the treatment of ALK-rearranged non-small cell lung cancer: A success story to usher in the second decade of molecular targeted therapy in oncology. Oncologist 2012, 17, 1351–1375. [Google Scholar] [CrossRef] [PubMed]
- Millett, R.L.; Elkon, J.M.; Tabbara, I.A. Directed therapies in anaplastic lymphoma kinase-rearranged non-small cell lung cancer. Anticancer Res. 2018, 38, 4969–4975. [Google Scholar] [CrossRef] [PubMed]
- Kwak, E.L.; Bang, Y.J.; Camidge, D.R.; Shaw, A.T.; Solomon, B.; Maki, R.G.; Ou, S.H.; Dezube, B.J.; Jänne, P.A.; Costa, D.B.; et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010, 363, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Camidge, D.R.; Bang, Y.J.; Kwak, E.L.; Iafrate, A.J.; Varella-Garcia, M.; Fox, S.B.; Riely, G.J.; Solomon, B.; Ou, S.H.; Kim, D.W.; et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: Updated results from a phase 1 study. Lancet Oncol. 2012, 13, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Kim, D.W.; Mehra, R.; Tan, D.S.; Felip, E.; Chow, L.Q.; Camidge, D.R.; Vansteenkiste, J.; Sharma, S.; De Pas, T.; et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 370, 1189–1197. [Google Scholar] [CrossRef]
- Peters, S.; Camidge, D.R.; Shaw, A.T.; Gadgeel, S.; Ahn, J.S.; Kim, D.W.; Ou, S.I.; Pérol, M.; Dziadziuszko, R.; Rosell, R.; et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 9, 829–838. [Google Scholar] [CrossRef]
- Gettinger, S.N.; Bazhenova, L.A.; Langer, C.J.; Salgia, R.; Gold, K.A.; Rosell, R.; Shaw, A.T.; Weiss, G.J.; Tugnait, M.; Narasimhan, N.I.; et al. Activity and safety of brigatinib in ALK-rearranged non-small-cell lung cancer and other malignancies: A single-arm, open-label, phase 1/2 trial. Lancet Oncol. 2016, 12, 1683–1696. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.; Infante, J.R.; Blumenschein, G.R.; Wakelee, H.A.; Arkenau, H.; Dukart, G.; Liang, C.; Harrow, K.; Gibbons, J.; Lovly, C.M.; et al. A phase I trial of X-396, a novel ALK inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 2014, 32, 8030. [Google Scholar] [CrossRef]
- Camidge, D.R.; Kono, S.A.; Flacco, A.; Tan, A.C.; Doebele, R.C.; Zhou, Q.; Crino, L.; Franklin, W.A.; Varella-Garcia, M. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin. Cancer Res. 2010, 16, 5581–5590. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Rekhtman, N.; Leighl, N.B.; Somerfield, M.R. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American society of clinical oncology endorsement of the college of american pathologists/international association for the study of lung cancer/association for molecular pathology guideline. J. Oncol. Pract. 2015, 2, 135–136. [Google Scholar] [CrossRef]
- Wynes, M.W.; Sholl, L.M.; Dietel, M.; Schuuring, E.; Tsao, M.S.; Yatabe, Y.; Tubbs, R.R.; Hirsch, F.R. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK and between evaluators. J. Thorac. Oncol. 2014, 9, 631–638. [Google Scholar] [CrossRef]
- Cabillic, F.; Gros, A.; Dugay, F.; Begueret, H.; Mesturoux, L.; Chiforeanu, D.C.; Dufrenot, L.; Jauffret, V.; Dachary, D.; Corre, R.; et al. Parallel FISH and immunohistochemical studies of ALK status in 3244 non-small-cell lung cancers reveal major discordances. J. Thorac. Oncol. 2014, 3, 295–306. [Google Scholar] [CrossRef]
- Ali, S.M.; Ou, S.I.; He, J.; Peled, N.; Chmielecki, J.; Pinder, M.C.; Palma, N.A.; Akerley, W.L.; Wang, K.; Molina, J.R.; et al. Identifying ALK rearrangements that are not detected by FISH with targeted next-generation sequencing of lung carcinoma. J. Clin. Oncol. 2014, 32, 8049a. [Google Scholar] [CrossRef]
- Lin, C.; Shi, X.; Yang, S.; Zhao, J.; He, Q.; Jin, Y.; Yu, X. Comparison of ALK detection by FISH, IHC and NGS to predict benefit from crizotinib in advanced non-small cell lung cancer. Lung Cancer 2019, 131, 62–68. [Google Scholar] [CrossRef]
- Savic, S.; Diebold, J.; Zimmermann, A.K.; Jochum, W.; Baschiera, B.; Grieshaber, S.; Tornillo, L.; Bisig, B.; Kerr, K.; Bubendorf, L. Screening for ALK in non-small cell lung carcinomas: 5A4 and D5F3 antibodies perform equally well, but combined use with FISH is recommended. Lung Cancer 2015, 2, 104–109. [Google Scholar] [CrossRef]
- Sholl, L.M.; Weremowicz, S.; Gray, S.W.; Wong, K.K.; Chirieac, L.R.; Lindeman, N.I.; Hornick, J.L. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J. Thorac. Oncol. 2013, 3, 322–328. [Google Scholar] [CrossRef]
- Sun, J.M.; Choi, Y.L.; Won, J.K.; Hirsch, F.R.; Ahn, J.S.; Ahn, M.; Park, K. A dramatic response to crizotinib in a non-small-cell lung cancer patient with IHC-positive and FISH-negative ALK. J. Thorac. Oncol. 2012, 7, e36–e38. [Google Scholar] [CrossRef] [PubMed]
- Peled, N.; Palmer, G.; Hirsch, F.R.; Wynes, M.W.; Ilouze, M.; Varella-Garcia, M.; Soussan-Gutman, L.; Otto, G.A.; Stephens, P.J.; Ross, J.S.; et al. Next-generation sequencing identifies and immunohistochemistry confirms a novel crizotinib-sensitive ALK rearrangement in a patient with metastatic non-small-cell lung cancer. J. Thorac. Oncol. 2012, 7, e14–e16. [Google Scholar] [CrossRef] [PubMed]
- Van der Wekken, A.J.; Pelgrim, R.; Hart, N.; Werner, N.; Mastik, M.F.; Hendriks, L.; van der Heijden, E.H.F.M.; Looijen-Salamon, M.; de Langen, A.J.; Staal-van den Brekel, J.; et al. Dichotomous ALK-IHC Is a Better Predictor for ALK Inhibition Outcome than Traditional ALK-FISH in Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Cabillic, F.; Hofman, P.; Ilie, M.; Peled, N.; Hochmair, M.; Dietel, M.; Von Laffert, M.; Gosney, J.R.; Lopez-Rios, F.; Erb, G.; et al. ALK IHC and FISH discordant results in patients with NSCLC and treatment response: For discussion of the question-to treat or not to treat? ESMO Open 2018, 3, e000419. [Google Scholar] [CrossRef] [PubMed]
- Scattone, A.; Catino, A.; Schirosi, L.; Caldarola, L.; Tommasi, S.; Lacalamita, R.; Montagna, E.S.; Galetta, D.; Serio, G.; Zito, F.A.; et al. Discordance between FISH, IHC, and NGS Analysis of ALK Status in Advanced Non-Small Cell Lung Cancer (NSCLC): A Brief Report of 7 Cases. Transl. Oncol. 2019, 12, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, E.; Lissenberg-Witte, B.I.; van den Heuvel, M.M.; Monkhorst, K.; Skov, B.G.; Sørensen, J.B.; Mellemgaard, A.; Dingemans, A.M.C.; Speel, E.J.M.; de Langen, A.J.; et al. ALK immunohistochemistry positive, FISH negative NSCLC is infrequent, but associated with impaired survival following treatment with crizotinib. Lung Cancer 2019, 138, 13–18. [Google Scholar] [CrossRef]
- Gao, X.; Sholl, L.M.; Nishino, M.; Heng, J.C.; Jänne, P.A.; Oxnard, G.R. Clinical Implications of Variant ALK FISH Rearrangement Patterns. J. Thorac. Oncol. 2015, 11, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- Clavé, S.; Rodon, N.; Pijuan, L.; Díaz, O.; Lorenzo, M.; Rocha, P.; Taus, Á.; Blanco, R.; Bosch-Barrera, J.; Reguart, N.; et al. Next-generation Sequencing for ALK and ROS1 Rearrangement Detection in Patients With Non-small-cell Lung Cancer: Implications of FISH-positive Patterns. Clin. Lung Cancer 2019, 20, e421–e429. [Google Scholar] [CrossRef] [PubMed]
- Dacic, S.; Villaruz, L.C.; Abberbock, S.; Mahaffey, A.; Incharoen, P.; Nikiforova, M.N. ALK FISH patterns and the detection of ALK fusions by next generation sequencing in lung adenocarcinoma. Oncotarget 2016, 7, 82943–82952. [Google Scholar] [CrossRef]
- Zito Marino, F.; Botti, G.; Aquino, G.; Ferrero, S.; Gaudioso, G.; Palleschi, A.; Rocco, D.; Salvi, R.; Micheli, M.C.; Micheli, P.; et al. Unproductive Effects of ALK Gene Amplification and Copy Number Gain in Non-Small-Cell Lung Cancer. ALK Gene Amplification and Copy Gain in NSCLC. Int. J. Mol. Sci. 2020, 21, 4927. [Google Scholar] [CrossRef] [PubMed]
- Peretti, U.; Ferrara, R.; Pilotto, S.; Kinspergher, S.; Caccese, M.; Santo, A.; Brunelli, M.; Caliò, A.; Carbognin, L.; Sperduti, I.; et al. ALK gene copy number gains in non-small-cell lung cancer: Prognostic impact and clinico-pathological correlations. Respir. Res. 2016, 17, 105. [Google Scholar] [CrossRef] [PubMed]
- Salido, M.; Pijuan, L.; Martínez-Avilés, L.; Galván, A.B.; Cañadas, I.; Rovira, A.; Zanui, M.; Martínez, A.; Longarón, R.; Sole, F.; et al. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Katayama, R.; Khan, T.M.; Benes, C.; Lifshits, E.; Ebi, H.; Rivera, V.M.; Shakespeare, W.C.; Iafrate, A.J.; Engelman, J.A.; Shaw, A.T. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc. Natl. Acad. Sci. USA 2011, 108, 7535–7540. [Google Scholar] [CrossRef] [PubMed]
- Katayama, R.; Shaw, A.T.; Khan, T.M.; Mino-Kenudson, M.; Solomon, B.J.; Halmos, B.; Jessop, N.A.; Wain, J.C.; Yeo, A.T.; Benes, C.; et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci. Transl. Med. 2012, 4, 120ra17. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, E.; Bubendorf, L.; Dietel, M.; Elmberger, G.; Kerr, K.; Lopez-Rios, F.; Moch, H.; Olszewski, W.; Pauwels, P.; Penault-Llorca, F.; et al. EML4-ALK testing in non-small cell carcinomas of the lung: A review with recommendations. Virchows Arch. 2012, 461, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Schildhaus, H.U.; Binot, E.; Wolf, J. Validation of a Simplified Approach to Detect ALK Translocations in Lung Cancer Samples by FISH. Mod. Pathol. 2016, 29, 482A. [Google Scholar]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M. Confidence Intervals. R package Version 0.1.2. 2022. Available online: https://CRAN.R-project.org/package=confintr (accessed on 22 February 2024).
- Murdoch, D. Formula-Driven Table Generation. R Package Version 0.9.22. 2023. Available online: https://dmurdoch.github.io/tables/ (accessed on 22 February 2024).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 22 February 2024).
- Zhu, H. kableExtra: Construct Complex Table with ‘kable’ andPipe Syntax. R Package Version 1.3.4. 2021. Available online: https://CRAN.R-project.org/package=kableExtra (accessed on 22 February 2024).
| Age | Total | Positive FISH | Negative FISH | Discordant Cases | |
|---|---|---|---|---|---|
| FISH−, IHC+ | FISH+, IHC− | ||||
| 20–29 | 6 | 2 | 4 | 1 | 0 |
| 20–29 men | 2 | 0 | 2 | 0 | 0 |
| 20–29 women | 4 | 2 | 2 | 1 | 0 |
| 30–39 | 28 | 9 | 19 | 0 | 0 |
| 30–39 men | 14 | 4 | 10 | 0 | 0 |
| 30–39 women | 14 | 5 | 9 | 0 | 0 |
| 40–49 | 133 | 33 | 100 | 1 | 2 |
| 40–49 men | 75 | 18 | 57 | 0 | 2 |
| 40–49 women | 58 | 15 | 43 | 1 | 0 |
| 50–59 | 487 | 55 | 432 | 1 | 7 |
| 50–59 men | 296 | 26 | 270 | 0 | 6 |
| 50–59 women | 191 | 29 | 162 | 1 | 1 |
| 60–69 | 1170 | 76 | 1094 | 6 | 18 |
| 60–69 men | 737 | 29 | 708 | 1 | 10 |
| 60–69 women | 433 | 47 | 386 | 5 | 8 |
| 70–79 | 743 | 41 | 702 | 7 | 6 |
| 70–79 men | 520 | 16 | 504 | 2 | 2 |
| 70–79 women | 223 | 25 | 198 | 5 | 4 |
| 80–89 | 114 | 6 | 108 | 1 | 0 |
| 80–89 men | 71 | 2 | 69 | 0 | 0 |
| 80–89 women | 43 | 4 | 39 | 1 | 0 |
| 90–99 | 2 | 0 | 2 | 0 | 0 |
| 90–99 men | 1 | 0 | 1 | 0 | 0 |
| 90–99 women | 1 | 0 | 1 | 0 | 0 |
| total | 2683 | 222 | 2461 | 17 | 33 |
| men | 1716 | 95 | 1621 | 3 | 20 |
| women | 967 | 127 | 840 | 14 | 13 |
| Histological Type | Total (n = 239) | Positive Discordant (n = 33) | Negative Discordant (n = 17) | Positive Concordant (n = 189) |
|---|---|---|---|---|
| Squamous cell carcinoma | 4 (1.67%) | 2 (6.06%) | 2 (11.76%) | 0 (0.00%) |
| Minimally invasive adenocarcinoma | 2 (0.83%) | 1 (3.03%) | 1 (5.88%) | 0 (0.00%) |
| Adenocarcinoma, NOS | 72 (30.12%) | 11 (33.33%) | 4 (23.53%) | 57 (30.16%) |
| Adenocarcinoma, NOS, mucin not detected | 6 (2.51%) | 1 (3.03%) | 0 (0.00%) | 5 (2.65%) |
| Adenocarcinoma, NOS, mucin producing | 8 (3.35%) | 0 (0.00%) | 0 (0.00%) | 8 (4.23%) |
| Lepidic predominant adenocarcinoma | 3 (1.26%) | 1 (3.03%) | 0 (0.00%) | 2 (1.06%) |
| Acinar predominant adenocarcinoma | 24 (10.04%) | 2 (6.06%) | 1 (5.88%) | 21 (11.11%) |
| Papillary predominant adenocarcinoma | 12 (5.02%) | 0 (0.00%) | 1 (5.88%) | 11 (5.82%) |
| Solid predominant adenocarcinoma | 29 (12.13%) | 4 (12.12%) | 4 (23.53%) | 21 (11.11%) |
| Micropapillary predominant adenocarcinoma | 6 (2.51%) | 0 (0.00%) | 0 (0.00%) | 6 (3.17%) |
| Signet ring cell adenocarcinoma | 4 (1.67%) | 0 (0.00%) | 0 (0.00%) | 4 (2.12%) |
| Mucinous adenocarcinoma | 9 (3.77%) | 1 (3.03%) | 0 (0.00%) | 8 (4.23%) |
| Clear cell adenocarcinoma | 1 (0.42%) | 1 (3.03%) | 0 (0.00%) | 0 (0.00%) |
| G3 adenocarcinoma, growth patterns not specified | 9 (3.77%) | 3 (9.09%) | 0 (0.00%) | 6 (3.17%) |
| G2 adenocarcinoma, growth patterns not specified | 1 (0.42%) | 0 (0.00%) | 0 (0.00%) | 1 (0.53%) |
| Pleomorphic carcinoma | 1 (0.42%) | 1 (3.03%) | 0 (0.00%) | 0 (0.00%) |
| Sarcomatoid carcinoma | 1 (0.42%) | 0 (0.00%) | 0 (0.00%) | 1 (0.53%) |
| Adenosquamous carcinoma | 3 (1.26%) | 1 (3.03%) | 0 (0.00%) | 2 (1.06%) |
| Combined NEC and adenocarcinoma | 2 (0.83%) | 1 (3.03%) | 0 (0.00%) | 1 (0.53%) |
| NSCLC, NOS | 3 (1.26%) | 0 (0.00%) | 0 (0.00%) | 3 (1.59%) |
| Metastatic adenocarcinoma | 26 (10.88%) | 3 (9.09%) | 2 (11.76%) | 21 (11.11%) |
| Data unavailable | 13 (5.44%) | 0 (0.00%) | 2 (11.76%) | 11 (5.82%) |
| Mucin production in one or more components | 41 (17.01%) | 2 (6.06%) | 1 (5.88%) | 38 (20.10%) |
| Mucin not detected | 11 (4.56%) | 3 (9.09%) | 2 (11.76%) | 6 (3.17%) |
| Not specified | 187 (77.59%) | 28 (84.85%) | 14 (82.35%) | 145 (76.72%) |
| Positive Discordant (n = 33) | Negative Discordant (n = 17) | Positive Concordant (n = 189) | |
|---|---|---|---|
| 3 or less ALK CNGs | 12 (36.36%) | 11 (64.70%) | 129 (68.25%) |
| 4–6 ALK CNGs | 8 (24.24%) | 2 (11.76%) | 40 (21.16%) |
| 7–9 ALK CNGs | 6 (18.18%) | 1 (5.88%) | 19 (10.05%) |
| 10+ ALK CNGs | 7 (21.21%) | 3 (17.65%) | 1 (0.53%) |
| Patterns <15% | At Least 1 Pattern ≥15% | Total | |
|---|---|---|---|
| Discordant samples (IHC−, FISH+) | 13 | 16 | 29 |
| 1 pattern identified | 5 | 7 | 12 |
| Inversion | 5 | 5 | 10 |
| Interstitial deletion | 0 | 0 | 0 |
| Translocation | 0 | 0 | 0 |
| 5′ deletion | 0 | 2 | 2 |
| 2 patterns identified | 7 | 7 | 14 |
| Interstitial deletion + inversion | 0 | 3 | 3 |
| 5′ deletion + inversion | 2 | 0 | 2 |
| 5′ deletion + translocation | 0 | 4 | 4 |
| Interstitial deletion + translocation | 1 | 0 | 1 |
| Translocation + inversion | 1 | 0 | 1 |
| Inversion + translocation | 3 | 0 | 3 |
| 3 patterns identified | 1 | 2 | 3 |
| 5′ deletion + translocation + inversion | 1 | 2 | 3 |
| Concordant samples (IHC+, FISH+) | 2 | 166 | 168 |
| 1 pattern identified | 2 | 135 | 137 |
| Inversion | 2 | 131 | 133 |
| Interstitial deletion | 0 | 3 | 3 |
| Translocation | 0 | 0 | 0 |
| 5′ deletion | 0 | 0 | 0 |
| 2 patterns identified | 0 | 30 | 30 |
| Interstitial deletion + inversion | 0 | 21 | 21 |
| 5′ deletion + interstitial deletion | 0 | 4 | 4 |
| Translocation + interstitial deletion | 0 | 1 | 1 |
| Interstitial deletion + translocation | 0 | 1 | 1 |
| Translocation + inversion | 0 | 3 | 3 |
| 3 patterns identified | 0 | 1 | 1 |
| 5′ deletion + translocation + inversion | 0 | 1 | 1 |
| Case No. | Gender | Age | ALK FISH Pattern | ALK Copy Number Gains | IHC | NGS |
|---|---|---|---|---|---|---|
| 1. | M | 69 | rearranged nuclei: 26% pattern: translocation + inversion | 16% 4–5 copies; 1% 9 copies | negative | IA: no variants reported; IB: TP53 p.P190L c.569C>T |
| 2. | M | 60 | rearranged nuclei: 68% pattern: inversion | 17% 4–10 copies; 8% >10 copies | negative | IA: no variants reported; IB: no variants reported |
| 3. | M | 53 | rearranged nuclei: 64% pattern: inversion | not detected | negative | IA: EML4, ALK fusion transcript; IB: MYC copy number gain (3 copies) |
| 4. | M | 70 | rearranged nuclei: 44% pattern: 5′ deletion + translocation | 11% 4–6 copies | negative | IA: no variants reported; IB: EGFR copy number gain (10 copies) MYC copy number gain (3 copies) TP53 p.I195T c.584T>C |
| 5. | M | 68 | rearranged nuclei: 30% pattern: translocation + interstitial deletion | 52% 4–10 copies | negative in limited material | IA: no variants reported; IB: MET copy number gain (8 copies) EGFR copy number gain (3 copies) BRAF copy number gain (5 copies) MYC copy number gain (3 copies) |
| 6. | F | 70 | adenocarcinoma component: rearranged nuclei: 26% pattern: inversion squamous cell carcinoma component: rearranged nuclei: 6% ALK negative | Adenocarcinoma component: 14% 4–7 copies Squamous cell carcinoma component: not detected | negative | IA: no variants reported; IB: no variants reported |
| 7. | M | 64 | rearranged nuclei: 18% pattern: 5′ deletion + inversion | not detected | negative | IA: no variants reported; IB: TP53 p.P152Rfs |
| 8. | F | 75 | rearranged nuclei: 17% pattern: 5′ deletion + translocation + inversion | 13% 4 copies; 12% 5–6 copies; 1% 7–8 copies; 5% more than 10 copies | negative | IA: no variants reported; IB: MYC copy number gain (22 copies) |
| 9. | M | 78 | rearranged nuclei: 60% pattern: 5′ deletion | 4% 4 copies; 4% 5 copies | negative | IA: no variants reported; IB: no variants reported |
| 10. | M | 63 | moderately differentiated component: rearranged nuclei: 4% ALK negative poorly differentiated component: rearranged nuclei: 16% pattern: translocation + inversion | Moderately differentiated component: 22% 4 copies; 18% 5–8 copies Poorly differentiated component: 10% 4 copies; 2% 5–6 copies | negative | IA: no variants reported; IB: no variants reported |
| 11. | F | 79 | rearranged nuclei: 4% ALK negative (38% positive in analysis with the break apart probe; population not found in further sections) | 4% 4 copies; 2% 7–8 copies | Negative, 100 positive nuclei | IA: no variants reported; IB: no variants reported |
| 12. | F | 77 | rearranged nuclei: 4% ALK negative (according to valid guidelines) abnormal signal configuration:
| not detected | positive | IA: EML4, ALK fusion transcript IB: no variants reported |
| 13. | F | 71 | ALK/EML4 probe not used rearranged nuclei: 2% using the ALK break apart probe
| not detected | positive | IA: CSFT3, ALK fusion transcript IB: no variants reported |
| 14. | F | 56 | rearranged nuclei: 50% pattern: inversion | not detected | positive | IA: EML4, ALK fusion transcript IB: no variants reported |
| 15. | F | 73 | rearranged nuclei: 48% pattern: interstitial deletion + inversion | not detected | positive | IA: EML4, ALK fusion transcript IB: EGFR copy number gain (3 copies) |
| 16. | M | 48 | rearranged nuclei: 62% pattern: 5′ deletion + interstitial deletion | not detected | positive | IA: HIP1, ALK fusion transcript IB: no variants reported |
| 17. | F | 64 | rearranged nuclei: 90% pattern: interstitial deletion | positive nuclei display up to 4 rearranged signals | positive | IA: EML4, ALK fusion transcript IB: no variants reported |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tobiášová, K.; Barthová, M.; Janáková, Ľ.; Lešková, K.; Farkašová, A.; Loderer, D.; Grendár, M.; Plank, L. Discordant ALK Status in Non-Small Cell Lung Carcinoma: A Detailed Reevaluation Comparing IHC, FISH, and NGS Analyses. Int. J. Mol. Sci. 2024, 25, 8168. https://doi.org/10.3390/ijms25158168
Tobiášová K, Barthová M, Janáková Ľ, Lešková K, Farkašová A, Loderer D, Grendár M, Plank L. Discordant ALK Status in Non-Small Cell Lung Carcinoma: A Detailed Reevaluation Comparing IHC, FISH, and NGS Analyses. International Journal of Molecular Sciences. 2024; 25(15):8168. https://doi.org/10.3390/ijms25158168
Chicago/Turabian StyleTobiášová, Katarína, Martina Barthová, Ľuboslava Janáková, Katarína Lešková, Anna Farkašová, Dušan Loderer, Marián Grendár, and Lukáš Plank. 2024. "Discordant ALK Status in Non-Small Cell Lung Carcinoma: A Detailed Reevaluation Comparing IHC, FISH, and NGS Analyses" International Journal of Molecular Sciences 25, no. 15: 8168. https://doi.org/10.3390/ijms25158168
APA StyleTobiášová, K., Barthová, M., Janáková, Ľ., Lešková, K., Farkašová, A., Loderer, D., Grendár, M., & Plank, L. (2024). Discordant ALK Status in Non-Small Cell Lung Carcinoma: A Detailed Reevaluation Comparing IHC, FISH, and NGS Analyses. International Journal of Molecular Sciences, 25(15), 8168. https://doi.org/10.3390/ijms25158168






