Insulin-Activated Signaling Pathway and GLUT4 Membrane Translocation in hiPSC-Derived Cardiomyocytes
Abstract
:1. Introduction
2. Results
2.1. HiPSC-CMs Express Key Components of the Insulin-Activated Pathway
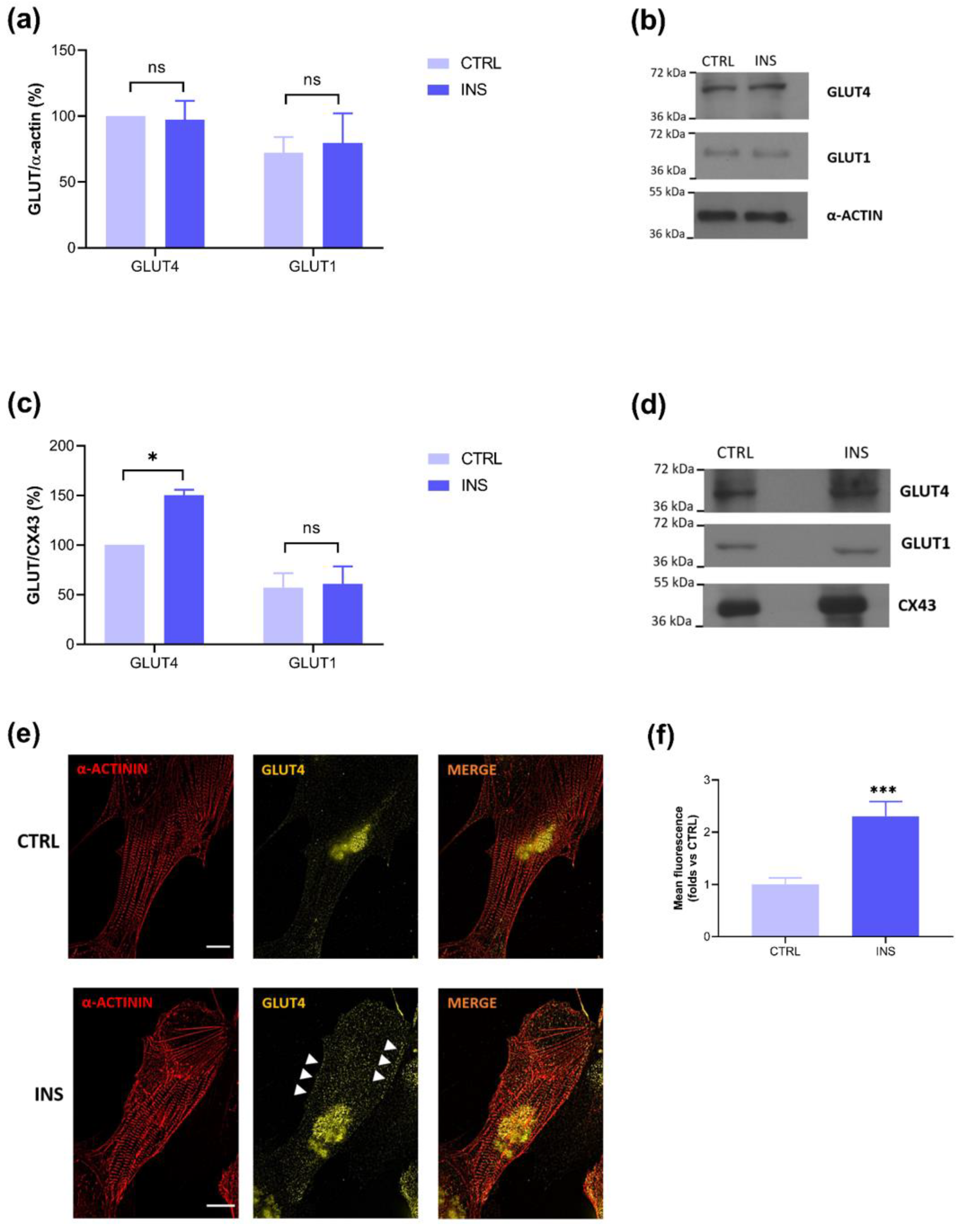
2.2. Insulin-Stimulated GLUT4 Membrane Translocation Is Visible Only on Membrane Lysates
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Line
4.3. Differentiation Protocol and Treatment
4.4. Total Lysates
4.5. Isolation of Membrane Lysates
4.6. Immunoblotting
4.7. Immunofluorescence
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitcheson, J.S.; Hancox, J.C.; Levi, A.J. Cultured Adult Cardiac Myocytes: Future Applications, Culture Methods, Morphological and Electrophysiological Properties. Cardiovasc. Res. 1998, 39, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Louch, W.E.; Sheehan, K.A.; Wolska, B.M. Methods in Cardiomyocyte Isolation, Culture, and Gene Transfer. J. Mol. Cell. Cardiol. 2011, 51, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Bistola, V.; Nikolopoulou, M.; Derventzi, A.; Kataki, A.; Sfyras, N.; Nikou, N.; Toutouza, M.; Toutouzas, P.; Stefanadis, C.; Konstadoulakis, M.M. Long-Term Primary Cultures of Human Adult Atrial Cardiac Myocytes: Cell Viability, Structural Properties and BNP Secretion in Vitro. Int. J. Cardiol. 2008, 131, 113–122. [Google Scholar] [CrossRef]
- Adegunsoye, A.; Gonzales, N.M.; Gilad, Y. Induced Pluripotent Stem Cells in Disease Biology and the Evidence for Their In Vitro Utility. Annu. Rev. Genet. 2023, 57, 341–360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wilson, G.F.; Soerens, A.G.; Koonce, C.H.; Yu, J.; Palecek, S.P.; Thomson, J.A.; Kamp, T.J. Functional Cardiomyocytes Derived From Human Induced Pluripotent Stem Cells. Circ. Res. 2009, 104, e30–e41. [Google Scholar] [CrossRef] [PubMed]
- Nijak, A.; Simons, E.; Vandendriessche, B.; Van de Sande, D.; Fransen, E.; Sieliwończyk, E.; Van Gucht, I.; Van Craenenbroeck, E.; Saenen, J.; Heidbuchel, H.; et al. Morpho-Functional Comparison of Differentiation Protocols to Create iPSC-Derived Cardiomyocytes. Biol. Open 2022, 11, bio059016. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, Y.; Kiskin, F.N.; Shen, M.; Zhang, J.Z. Recent Advances in Regulating the Proliferation or Maturation of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Res. Ther. 2023, 14, 228. [Google Scholar] [CrossRef]
- Ottaviani, D.; Ter Huurne, M.; Elliott, D.A.; Bellin, M.; Mummery, C.L. Maturing Differentiated Human Pluripotent Stem Cells in Vitro: Methods and Challenges. Development 2023, 150, dev201103. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Guo, L.; Fiene, S.J.; Anson, B.D.; Thomson, J.A.; Kamp, T.J.; Kolaja, K.L.; Swanson, B.J.; January, C.T. High Purity Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Electrophysiological Properties of Action Potentials and Ionic Currents. Am. J. Physiol.-Heart Circ. Physiol. 2011, 301, H2006–H2017. [Google Scholar] [CrossRef]
- Kamakura, T.; Makiyama, T.; Sasaki, K.; Yoshida, Y.; Wuriyanghai, Y.; Chen, J.; Hattori, T.; Ohno, S.; Kita, T.; Horie, M.; et al. Ultrastructural Maturation of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes in a Long-Term Culture. Circ. J. 2013, 77, 1307–1314. [Google Scholar] [CrossRef]
- Ebert, A.; Joshi, A.U.; Andorf, S.; Dai, Y.; Sampathkumar, S.; Chen, H.; Li, Y.; Garg, P.; Toischer, K.; Hasenfuss, G.; et al. Proteasome-Dependent Regulation of Distinct Metabolic States During Long-Term Culture of Human iPSC-Derived Cardiomyocytes. Circ. Res. 2019, 125, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, Y.; Yan, Y.; Terashvili, M.; Wells, C.; Horikoshi, H.; Fujita, S.; Bosnjak, Z.J.; Bai, X. Fatty Acid-Treated Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes Exhibit Adult Cardiomyocyte-Like Energy Metabolism Phenotypes. Cells 2019, 8, 1095. [Google Scholar] [CrossRef] [PubMed]
- Correia, C.; Koshkin, A.; Duarte, P.; Hu, D.; Teixeira, A.; Domian, I.; Serra, M.; Alves, P.M. Distinct Carbon Sources Affect Structural and Functional Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Sci. Rep. 2017, 7, 8590. [Google Scholar] [CrossRef]
- Kadari, A.; Mekala, S.; Wagner, N.; Malan, D.; Köth, J.; Doll, K.; Stappert, L.; Eckert, D.; Peitz, M.; Matthes, J.; et al. Robust Generation of Cardiomyocytes from Human iPS Cells Requires Precise Modulation of BMP and WNT Signaling. Stem Cell Rev. 2015, 11, 560–569. [Google Scholar] [CrossRef]
- Kieda, J.; Shakeri, A.; Landau, S.; Wang, E.Y.; Zhao, Y.; Lai, B.F.; Okhovatian, S.; Wang, Y.; Jiang, R.; Radisic, M. Advances in Cardiac Tissue Engineering and Heart-on-a-Chip. J. Biomed. Mater. Res. A 2023, 112, 492–511. [Google Scholar] [CrossRef]
- Dunn, K.K.; Reichardt, I.M.; Simmons, A.D.; Jin, G.; Floy, M.E.; Hoon, K.M.; Palecek, S.P. Coculture of Endothelial Cells with Human Pluripotent Stem Cell-Derived Cardiac Progenitors Reveals a Differentiation Stage-Specific Enhancement of Cardiomyocyte Maturation. Biotechnol. J. 2019, 14, e1800725. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ari, M.; Naor, S.; Zeevi-Levin, N.; Schick, R.; Ben Jehuda, R.; Reiter, I.; Raveh, A.; Grijnevitch, I.; Barak, O.; Rosen, M.R.; et al. Developmental Changes in Electrophysiological Characteristics of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Heart Rhythm. 2016, 13, 2379–2387. [Google Scholar] [CrossRef] [PubMed]
- Germanguz, I.; Sedan, O.; Zeevi-Levin, N.; Shtrichman, R.; Barak, E.; Ziskind, A.; Eliyahu, S.; Meiry, G.; Amit, M.; Itskovitz-Eldor, J.; et al. Molecular Characterization and Functional Properties of Cardiomyocytes Derived from Human Inducible Pluripotent Stem Cells. J. Cell Mol. Med. 2011, 15, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Emanuelli, G.; Zoccarato, A.; Reumiller, C.M.; Papadopoulos, A.; Chong, M.; Rebs, S.; Betteridge, K.; Beretta, M.; Streckfuss-Bömeke, K.; Shah, A.M. A Roadmap for the Characterization of Energy Metabolism in Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. J. Mol. Cell. Cardiol. 2022, 164, 136–147. [Google Scholar] [CrossRef]
- Dimasi, C.G.; Darby, J.R.T.; Morrison, J.L. A Change of Heart: Understanding the Mechanisms Regulating Cardiac Proliferation and Metabolism before and after Birth. J. Physiol. 2023, 601, 1319–1341. [Google Scholar] [CrossRef]
- Stein, J.M.; Mummery, C.L.; Bellin, M. Engineered Models of the Human Heart: Directions and Challenges. Stem Cell Rep. 2021, 16, 2049–2057. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.C.; Sicard, R.E. Lactate Metabolism of Isolated, Perfused Fetal, and Newborn Pig Hearts. Pediatr. Res. 1987, 22, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.C.; Whitman, V.; Musselman, J.; Schuler, G. Perinatal Changes in Mitochondrial Respiration of the Rabbit Heart. Biol. Neonate 2009, 42, 208–216. [Google Scholar] [CrossRef]
- Drake, R.R.; Louey, S.; Thornburg, K.L. Maturation of Lipid Metabolism in the Fetal and Newborn Sheep Heart. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2023, 325, R809–R819. [Google Scholar] [CrossRef] [PubMed]
- Szablewski, L. Glucose Transporters in Healthy Heart and in Cardiac Disease. Int. J. Cardiol. 2017, 230, 70–75. [Google Scholar] [CrossRef]
- Jia, G.; Hill, M.A.; Sowers, J.R. Diabetic Cardiomyopathy. Circ. Res. 2018, 122, 624–638. [Google Scholar] [CrossRef] [PubMed]
- Tohyama, S.; Hattori, F.; Sano, M.; Hishiki, T.; Nagahata, Y.; Matsuura, T.; Hashimoto, H.; Suzuki, T.; Yamashita, H.; Satoh, Y.; et al. Distinct Metabolic Flow Enables Large-Scale Purification of Mouse and Human Pluripotent Stem Cell-Derived Cardiomyocytes. Cell Stem Cell 2013, 12, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Gallo, M.P.; Femminò, S.; Antoniotti, S.; Querio, G.; Alloatti, G.; Levi, R. Catestatin Induces Glucose Uptake and GLUT4 Trafficking in Adult Rat Cardiomyocytes. Biomed. Res. Int. 2018, 2018, 2086109. [Google Scholar] [CrossRef] [PubMed]
- Steinbusch, L.K.M.; Schwenk, R.W.; Ouwens, D.M.; Diamant, M.; Glatz, J.F.C.; Luiken, J.J.F.P. Subcellular Trafficking of the Substrate Transporters GLUT4 and CD36 in Cardiomyocytes. Cell. Mol. Life Sci. 2011, 68, 2525–2538. [Google Scholar] [CrossRef]
- Montessuit, C.; Lerch, R. Regulation and Dysregulation of Glucose Transport in Cardiomyocytes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 848–856. [Google Scholar] [CrossRef]
- Bowman, P.R.T.; Smith, G.L.; Gould, G.W. GLUT4 Expression and Glucose Transport in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. PLoS ONE 2019, 14, e0217885. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.; Fajardo, G.; Ribeiro, A.J.S.; Kooiker, K.B.; Coronado, M.; Zhao, M.; Hu, D.-Q.; Reddy, S.; Kodo, K.; Sriram, K.; et al. Time-Dependent Evolution of Functional vs. Remodeling Signaling in Induced Pluripotent Stem Cell-Derived Cardiomyocytes and Induced Maturation with Biomechanical Stimulation. FASEB J. 2016, 30, 1464–1479. [Google Scholar] [CrossRef] [PubMed]
- Antoniotti, S.; Lovisolo, D.; Fiorio Pla, A.; Munaron, L. Expression and Functional Role of bTRPC1 Channels in Native Endothelial Cells. FEBS Lett. 2002, 510, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Pires, R.H.; Dau, T.H.; Manu, E.; Shree, N.; Otto, O. Switching in the Expression Pattern of Actin Isoforms Marks the Onset of Contractility and Distinct Mechanodynamic Behavior during Cardiomyocyte Differentiation. Physiol. Rep. 2022, 10, e15171. [Google Scholar] [CrossRef]
- Vozzi, C.; Dupont, E.; Coppen, S.R.; Yeh, H.I.; Severs, N.J. Chamber-Related Differences in Connexin Expression in the Human Heart. J. Mol. Cell. Cardiol. 1999, 31, 991–1003. [Google Scholar] [CrossRef]
- Granéli, C.; Hicks, R.; Brolén, G.; Synnergren, J.; Sartipy, P. Diabetic Cardiomyopathy Modelling Using Induced Pluripotent Stem Cell Derived Cardiomyocytes: Recent Advances and Emerging Models. Stem Cell Rev. 2019, 15, 13–22. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Querio, G.; Antoniotti, S.; Levi, R.; Fleischmann, B.K.; Gallo, M.P.; Malan, D. Insulin-Activated Signaling Pathway and GLUT4 Membrane Translocation in hiPSC-Derived Cardiomyocytes. Int. J. Mol. Sci. 2024, 25, 8197. https://doi.org/10.3390/ijms25158197
Querio G, Antoniotti S, Levi R, Fleischmann BK, Gallo MP, Malan D. Insulin-Activated Signaling Pathway and GLUT4 Membrane Translocation in hiPSC-Derived Cardiomyocytes. International Journal of Molecular Sciences. 2024; 25(15):8197. https://doi.org/10.3390/ijms25158197
Chicago/Turabian StyleQuerio, Giulia, Susanna Antoniotti, Renzo Levi, Bernd K. Fleischmann, Maria Pia Gallo, and Daniela Malan. 2024. "Insulin-Activated Signaling Pathway and GLUT4 Membrane Translocation in hiPSC-Derived Cardiomyocytes" International Journal of Molecular Sciences 25, no. 15: 8197. https://doi.org/10.3390/ijms25158197




