The Increase in the Peroxidase Activity of the Cytochrome C with Substitutions in the Universal Binding Site Is Associated with Changes in the Ability to Interact with External Ligands
Abstract
:1. Introduction
2. Results
2.1. Peroxidase Activity of Mutant CytC Forms
2.2. Effect of Mutations in UBS on Cyanide Binding to CytC
2.2.1. Acceleration of Cyanide Binding to Oxidized Forms of Mutant CytC as an Indicator of Changes in the Strength of Interaction between Heme Iron and S(Met80)
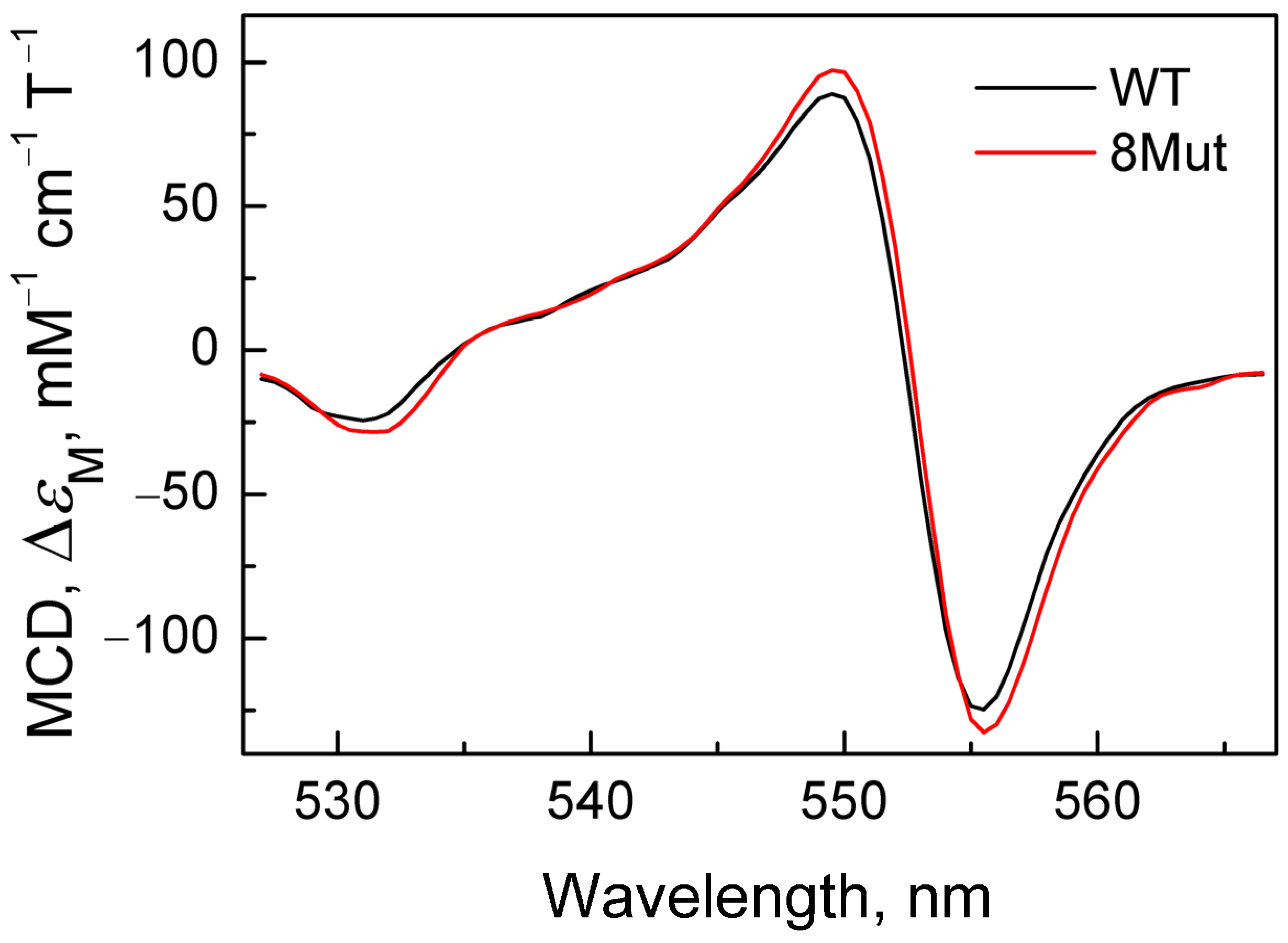
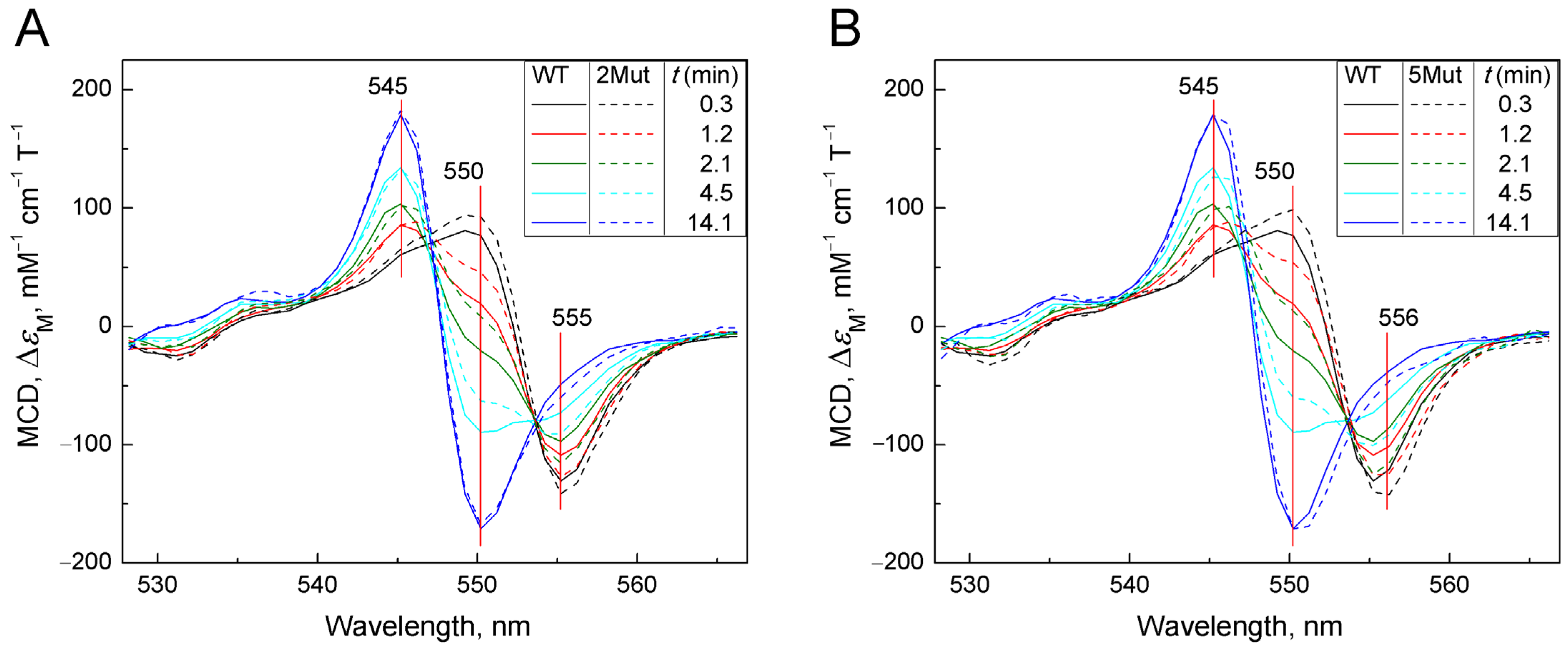
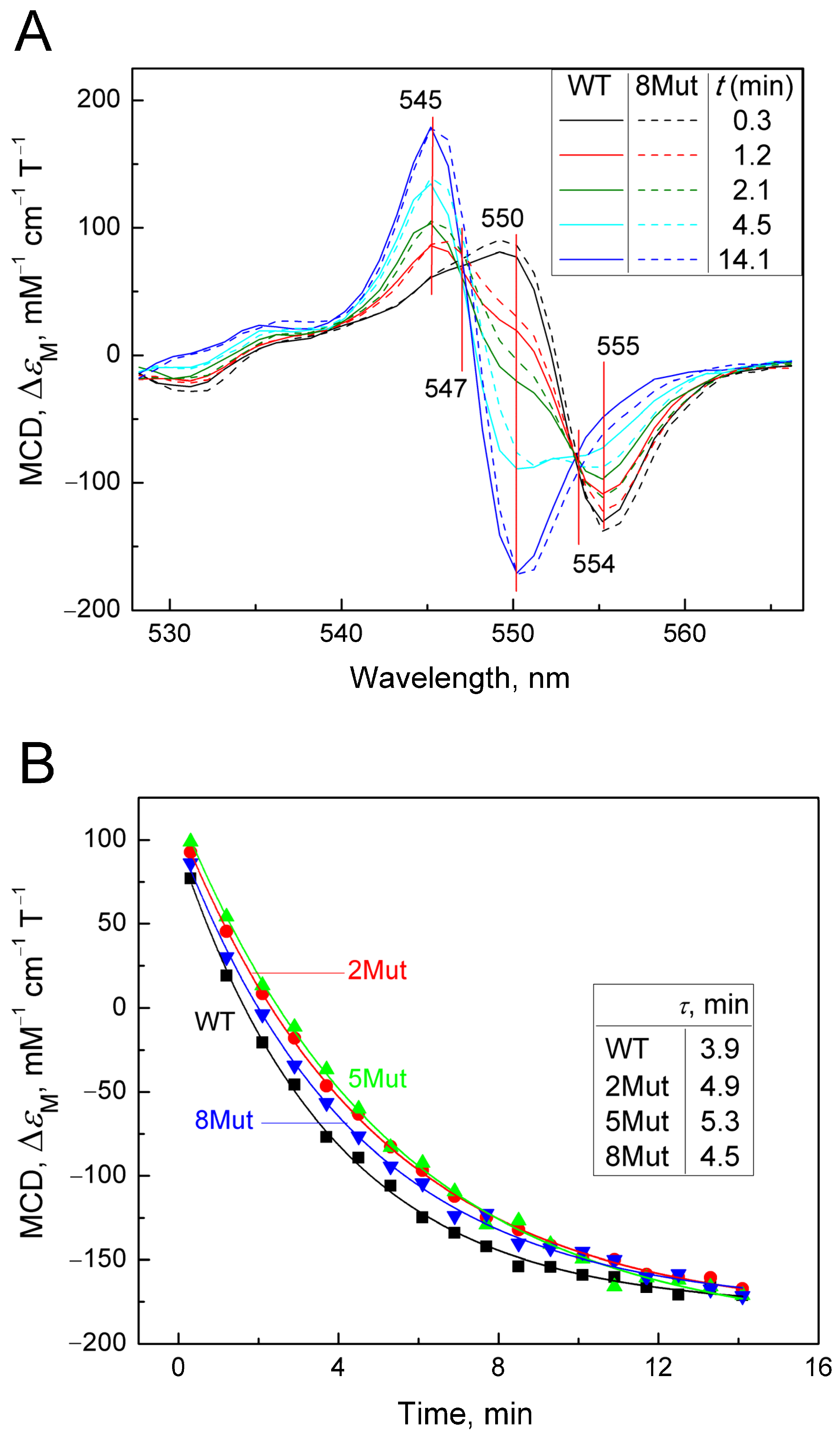
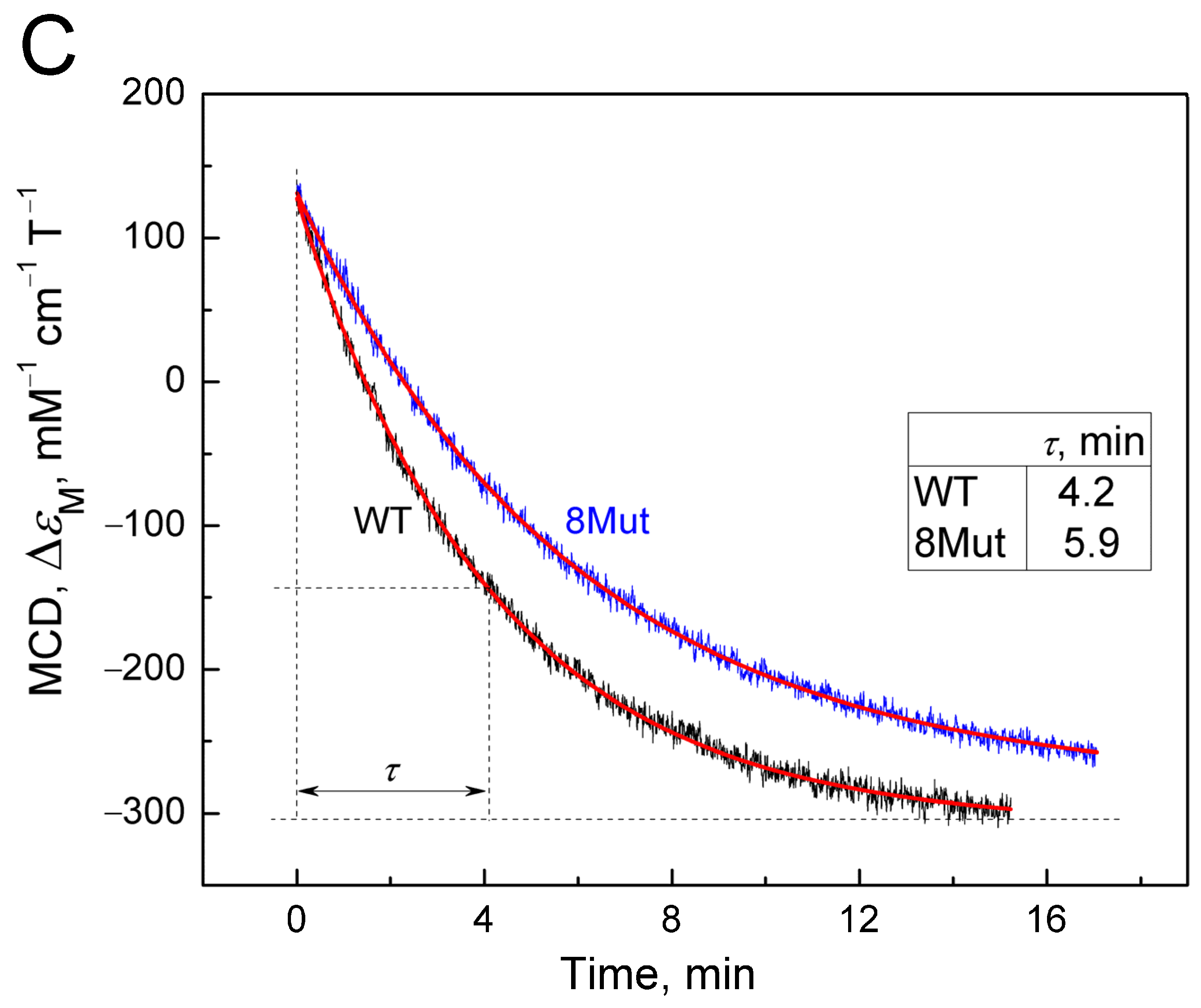
2.2.2. Deceleration of the Decay of the Cyanide Complex with the Reduced form of CytC, Caused by Mutations in UBS, Recorded by the MCD Method
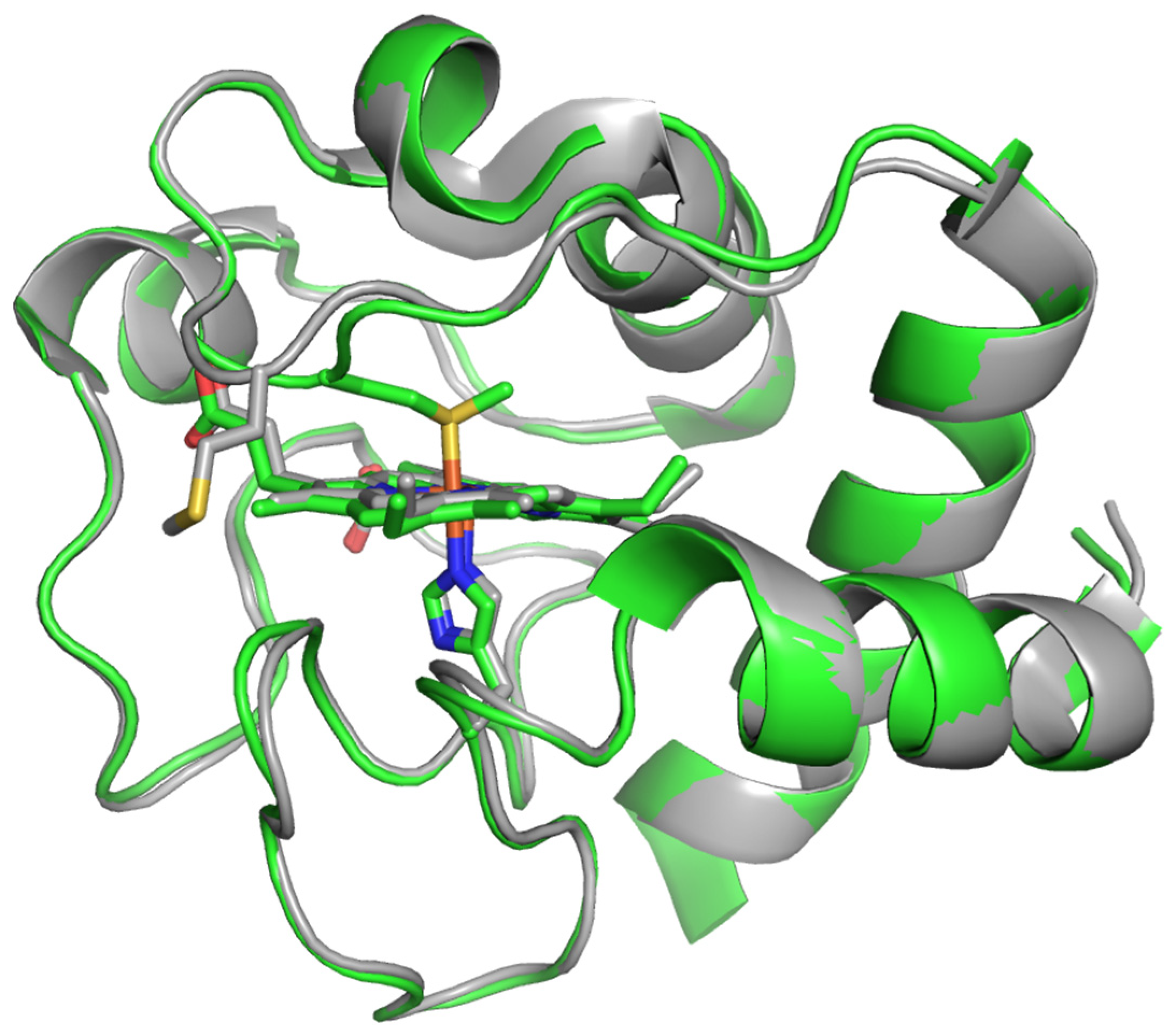
2.3. Molecular Modeling of the Structure of CytC WT and Its Mutant Variants 2Mut, 5Mut, and 8Mut
3. Discussion
4. Materials and Methods
4.1. Mutant Genes Construction
4.2. Expression of the Mutant Genes of CytC, Protein Isolation, and Purification
4.3. Spectrophotometric Assays
4.4. MCD Spectroscopy
4.5. Molecular Dynamics
4.6. Isoelectric Surfaces
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
Appendix A
References
- Muenzner, J.; Pletneva, E.V. Structural transformations of cytochrome c upon interaction with cardiolipin. Chem. Phys. Lipids 2014, 179, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer-Stenner, R. Relating the multi-functionality of cytochrome c to membrane binding and structural conversion. Biophys. Rev. 2018, 10, 1151–1185. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Tyurin, V.A.; Jiang, J.; Tyurina, Y.Y.; Ritov, V.B.; Amoscato, A.A.; Osipov, A.N.; Belikova, N.A.; Kapralov, A.A.; Kini, V.; et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005, 1, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Bayir, H.A.; Belikova, N.A.; Kapralov, O.; Tyurina, V.A.; Jiang, J.; Stoyanovsky, D.A.; Wipf, P.; Kochanek, P.M.; Greenberger, J.S.; et al. Cytochrome c/cardiolipin relations in mitochondria: A kiss of death. Free Radic. Biol. Med. 2009, 46, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, L.; Erba, F.; Santucci, R.; Sinibaldi, F. Cytochrome c interaction with cardiolipin plays a key role in cell apoptosis: Implications for human diseases. Symmetry 2022, 14, 767. [Google Scholar] [CrossRef]
- Pandey, V.P.; Awasthi, M.; Singh, S.; Tiwari, S. A comprehensive review on function and application of plant peroxidases. Biochem. Anal. Biochem. 2017, 6, 308. [Google Scholar] [CrossRef]
- Vygodina, T.V.; Konstantinov, A.A. Peroxidase activity of mitochondrial cytochrome c oxidase. Biochemistry 2007, 72, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Kinnunen, P.K.J.; Kaarniranta, K.; Mahalka, A.K. Protein-oxidized phospholipid interactions in cellular signaling for cell death: From biophysics to clinical correlations. Biochim. Biophys. Acta 2012, 1818, 2446–2455. [Google Scholar] [CrossRef]
- Gorbenko, G.P.; Molotkovsky, J.G.; Paavo, K.; Kinnunen, J. Cytochrome C interaction with cardiolipin/phosphatidylcholine model membranes: Effect of cardiolipin protonation. Biophys. J. 2006, 90, 4093–4103. [Google Scholar] [CrossRef]
- Alvarez-Paggi, D.; Hannibal, L.; Castro, M.A.; Oviedo-Rouco, S.; Demicheli, V.; Tortora, V.; Tomasina, F.; Radi, R.; Murgida, D.H. Multifunctional cytochrome c: Learning new tricks from an old dog. Chem. Rev. 2017, 117, 13382–13460. [Google Scholar] [CrossRef]
- Fiorucci, L.; Sinibaldi, F.; Chimenti, M.; Perricone, R.; Santucci, R. Cytochrome c as a Clinical Biomarker in Diseases Characterized by Cell Apoptosis. In Cytochrome c: Roles and Therapeutic Implications; Arias, N., Ed.; Nova Science Publishers: New York, NY, USA, 2019; pp. 119–147. [Google Scholar]
- Lopes, J.; Marques-da-Silva, D.; Videira, P.A.; Samhan-Arias, A.K.; Lagoa, R. Cardiolipin Membranes Promote Cytochrome c Transformation of Polycyclic Aromatic Hydrocarbons and Their In Vivo Metabolites. Molecules 2024, 29, 1129. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Niibayashi, R.; Koubori, S.; Moriyama, I.; Miyoshi, H. Molecular mechanisms for the induction of peroxidase activity of the cytochrome c−cardiolipin complex. Biochemistry 2011, 50, 8383–8391. [Google Scholar] [CrossRef] [PubMed]
- Belikova, N.A.; Vladimirov, Y.A.; Osipov, A.N.; Kapralov, A.A.; Tyurin, V.A.; Potapovich, M.V.; Basova, L.V.; Peterson, J.; Kurnikov, I.V.; Kagan, V.E. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry 2006, 45, 4998–5009. [Google Scholar] [CrossRef] [PubMed]
- Kawai, C.; Prado, F.M.; Nunes, G.L.C.; Di Mascio, P.; Carmona-Ribeiro, A.M.; Nantes, I.L. pH-dependent interaction of cytochrome c with mitochondrial mimetic membranes: The role of an array of positively charged amino acids. J. Biol. Chem. 2005, 280, 34709–34717. [Google Scholar] [CrossRef]
- Pepelina, T.Y.; Chertkova, R.V.; Ostroverkhova, T.V.; Dolgikh, D.A.; Kirpichnikov, M.P.; Grivennikova, V.G.; Vinogradov, A.D. Site-directed mutagenesis of cytochrome c: Reactions with respiratory chain components and superoxide radical. Biochemistry 2009, 74, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Chertkova, R.V.; Oleynikov, I.P.; Pakhomov, A.A.; Sudakov, R.V.; Orlov, V.N.; Semenova, M.A.; Arutyunyan, A.M.; Ptushenko, V.V.; Kirpichnikov, M.P.; Dolgikh, D.A.; et al. Mutant cytochrome c as a potential detector of superoxide generation: Effect of mutations on the function and properties. Cells 2023, 12, 2316. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Jeong, M.S.; Choi, S.Y.; Kang, J.H. Peroxidase activity of cytochrome c. Bull. Korean Chem. Soc. 2004, 25, 1889–1892. [Google Scholar] [CrossRef]
- Wang, Z.; Matsuo, T.; Nagao, S.; Hirota, S. Peroxidase activity enhancement of horse cytochrome c by dimerization. Org. Biomol. Chem. 2011, 9, 4766–4769. [Google Scholar] [CrossRef]
- McClelland, L.J.; Mou, T.-C.; Jeakins-Cooley, M.E.; Sprang, S.R.; Bowler, B.E. Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity. Proc. Nat. Acad. Sci. USA 2014, 111, 6648–6653. [Google Scholar] [CrossRef]
- Yin, V.; Shaw, G.S.; Konermann, L. Cytochrome c as a peroxidase: Activation of the precatalytic native state by H2O2-induced covalent modifications. J. Am. Chem. Soc. 2017, 139, 15701–15709. [Google Scholar] [CrossRef]
- Chertkova, R.V.; Firsov, A.M.; Brazhe, N.A.; Nikelshparg, E.I.; Bochkova, Z.V.; Bryntseva, T.V.; Semenova, M.A.; Baizhumanov, A.A.; Kotova, E.A.; Kirpichnikov, M.P.; et al. Multiple mutations in the non-ordered red Ω-loop enhance the membrane-permeabilizing and peroxidase-like activity of cytochrome c. Biomolecules 2022, 12, 665. [Google Scholar] [CrossRef] [PubMed]
- Antonini, E.; Brunori, M. Hemoglobin. Annu. Rev. Biochem. 1970, 39, 977–1042. [Google Scholar] [CrossRef] [PubMed]
- Sharonov, Y.A. Room-temperature and low-temperature magnetic circular dichroism of hemoproteins and related compounds. Sov. Sci. Rev. D Physicochem. Biol. 1991, 10, 1–118. [Google Scholar]
- Arutyunyan, A.M.; Sharonov, Y.A. Fine structure of the magnetooptical rotatory dispersion curves and the surroundings of heme in ferrocytochrome c and its model compounds. Mol. Biol. 1974, 7, 478–484. [Google Scholar]
- George, P.; Schejter, A. The Reactivity of Ferrocytochrome c with Iron-binding Ligands. J. Biol. Chem. 1964, 239, 1504–1508. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Schejter, A.; Ryan, M.D.; Blizzard, E.R.; Zhang, C.; Margoliash, E.; Feinberg, B.A. The redox couple of the cytochrome c cyanide complex: The contribution of heme iron ligation to the structural stability, chemical reactivity, and physiological behavior of horse cytochrome c. Protein Sci. 2006, 15, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Varhac, R.; Tomaskova, N.; Fabian, M.; Sedlak, E. Kinetics of cyanide binding as a probe of local stability/flexibility of cytochrome c. Biophys. Chem. 2009, 144, 21–26. [Google Scholar] [CrossRef]
- Nold, S.M.; Lei, H.; Mou, T.-C.; Bowler, B.E. Effect of a K72A mutation on the structure, stability, dynamics, and peroxidase activity of human cytochrome c. Biochemistry 2017, 56, 3358–3368. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, B.P.; Volkenstein, M.V.; Sharonov, Y.A. Investigation of modified cytochrome c with myoglobin-like properties by the magnetooptic rotation method. Mol. Biol. 1969, 3, 406–412. [Google Scholar]
- Chertkova, R.V.; Firsov, A.M.; Kotova, E.A.; Gusev, I.D.; Dolgikh, D.A.; Kirpichnikov, M.P.; Antonenko, Y.N. Lysine 72 substitutions differently affect lipid membrane permeabilizing and proapoptotic activities of horse heart cytochrome c. Biochem. Biophys. Res. Commun. 2021, 548, 74–77. [Google Scholar] [CrossRef]
- Belousov, V.V.; Fradkov, A.F.; Lukyanov, K.A.; Staroverov, D.B.; Shakhbazov, K.S.; Terskikh, A.V.; Lukyanov, S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 2006, 3, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Lyublinskaya, O.; Antunes, F. Measuring intracellular concentration of hydrogen peroxide with the use of genetically encoded H2O2 biosensor HyPer. Redox Biol. 2019, 24, 101200. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Sharma, H.; Singh, N. Hydrogen peroxide induces apoptosis in HeLa cells through mitochondrial pathway. Mitochondrion 2007, 7, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Antonenko, Y.N.; Avetisyan, A.V.; Bakeeva, L.E.; Chernyak, B.V.; Chertkov, V.A.; Domnina, L.V.; Ivanova, O.Y.; Izyumov, D.S.; Khailova, L.S.; Klishin, S.S.; et al. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 1. Cationic plastoquinone derivatives: Synthesis and in vitro studies. Biochemistry 2008, 73, 1273–1287. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.J.; Millett, F. Use of specific lysine modifications to identify the site of reaction between cytochrome c and ferricyanide. J. Biol. Chem. 1981, 256, 1611–1615. [Google Scholar] [CrossRef] [PubMed]
- Konstantinov, A.A.; Vygodina, T.; Capitanio, N.; Papa, S. Ferrocyanide-peroxidase activity of cytochrome c oxidase. Biochim. Biophys. Acta 1998, 1363, 11–23. [Google Scholar] [CrossRef]
- George, P.; Lyster, R.L.J. Crevice structures in hemoprotein reactions. Proc. Natl. Acad. Sci. USA 1958, 44, 1013–1029. [Google Scholar] [CrossRef] [PubMed]
- Bergmayer, H.U.; Gawehn, K.; Grassl, M. Methoden der Enzymatischen Analyze; Bergmayer, H.U., Ed.; Verlag Chemie: Weinheim, Germany, 1970; p. 440. [Google Scholar]
- Margoliash, E.; Frohwirt, N. Spectrum of horse-heart cytochrome c. Biochem. J. 1959, 71, 570–572. [Google Scholar] [CrossRef]
- Pepelina, T.Y.; Chertkova, R.V.; Dolgikh, D.A.; Kirpichnikov, M.P. The role of individual lysine residues of horse cytochrome c in the formation of reactive complexes with components of the respiratory chain. Russ. J. Bioorg. Chem. 2010, 36, 90–96. [Google Scholar] [CrossRef]
- Pakhomov, A.A.; Frolova, A.Y.; Tabakmakher, V.M.; Chugunov, A.O.; Efremov, R.G.; Martynov, V.I. Impact of external amino acids on fluorescent protein chromophore biosynthesis revealed by molecular dynamics and mutagenesis studies. J. Photochem. Photobiol. 2020, 206, 111853. [Google Scholar] [CrossRef] [PubMed]
- Goga, N.; Rzepiela, A.J.; de Vries, A.H.; Marrink, S.J.; Berendsen, H.J.C. Efficient algorithms for Langevin and DPD dynamics. J. Chem. Theory Comput. 2012, 8, 3637–3649. [Google Scholar] [CrossRef] [PubMed]
- Khruschev, S.S.; Abaturova, A.M.; Diakonova, A.N.; Ustinin, D.M.; Zlenko, D.V.; Fedorov, V.A.; Kovalenko, I.B.; Riznichenko, G.Y.; Rubin, A.B. Multi-particle Brownian dynamics software ProKSim for protein-protein interactions modeling. Comput. Res. Model. 2013, 5, 47–64. [Google Scholar] [CrossRef]
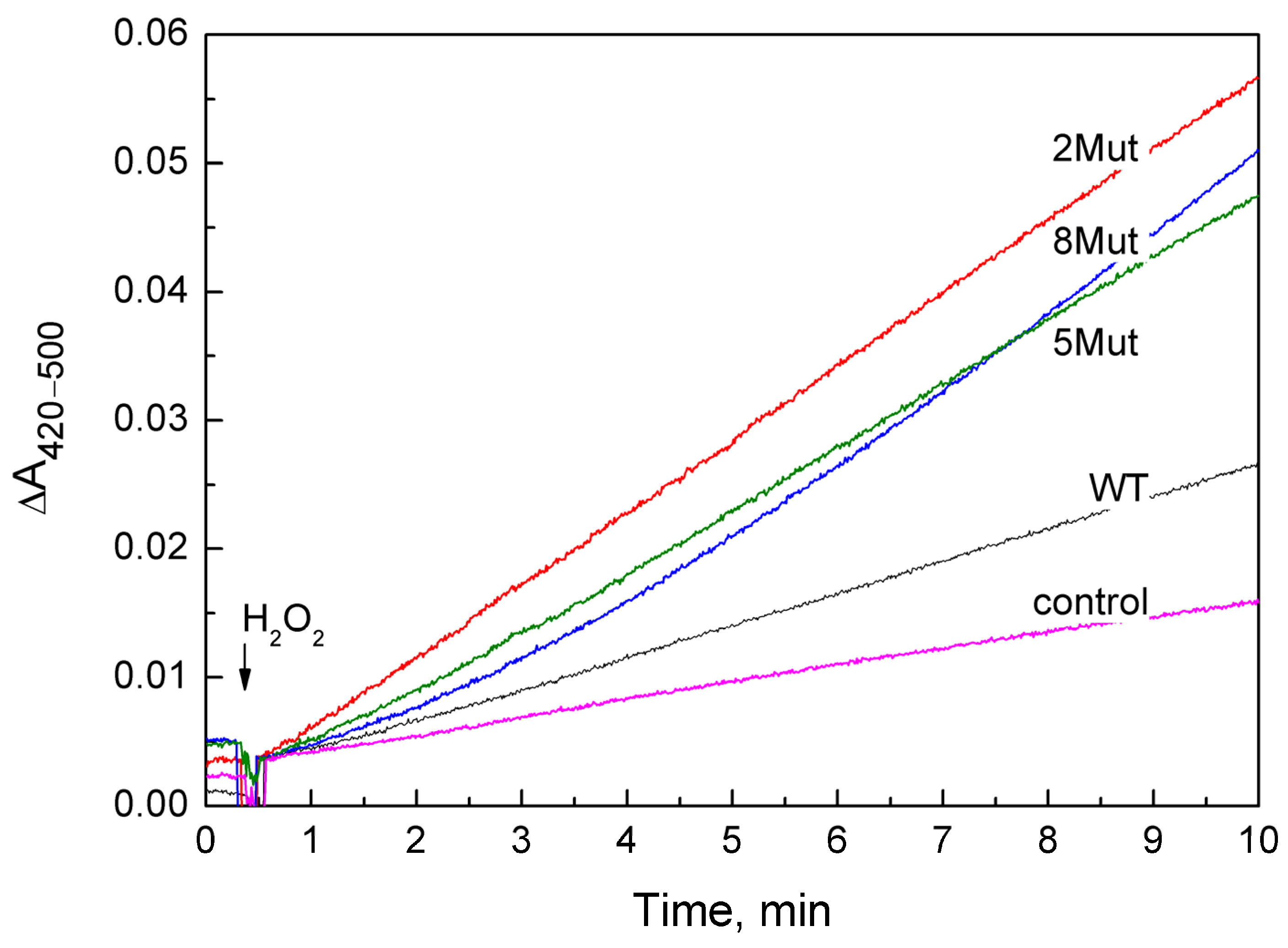
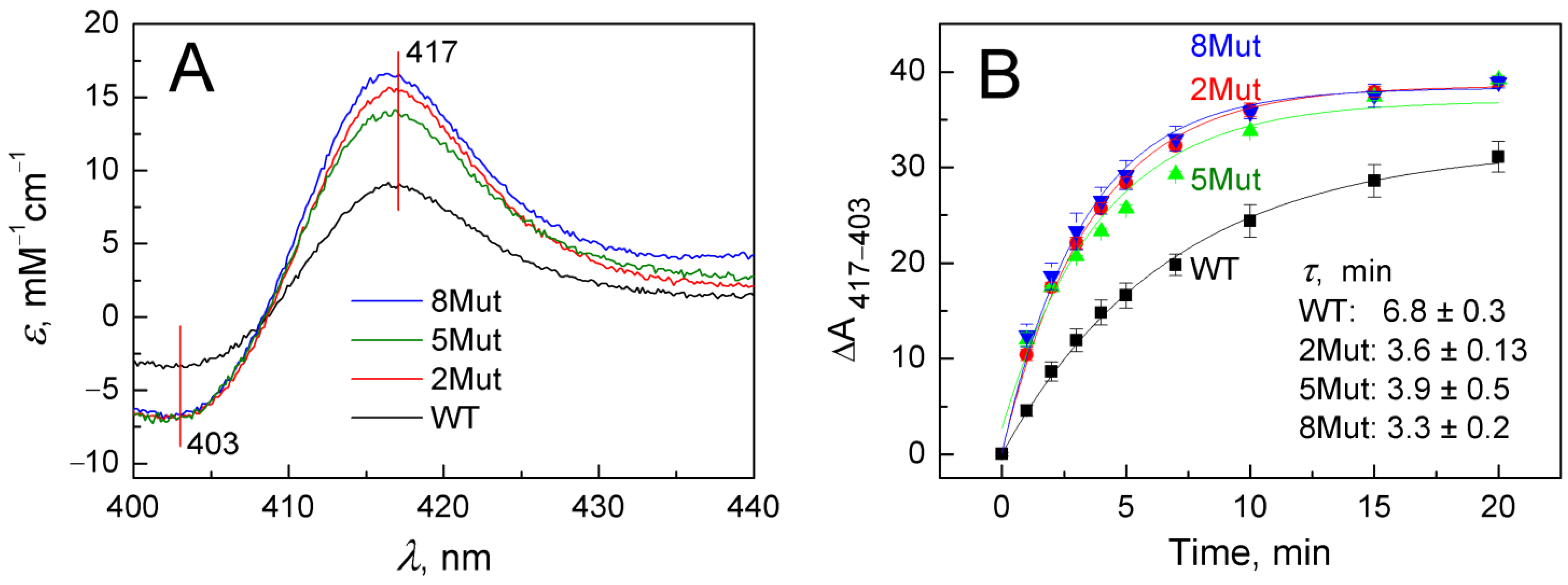



| Type of CytC | WT | 2Mut | 5Mut | 8Mut | Control without CytC |
|---|---|---|---|---|---|
| µM ferri/min | 1.7 ± 0.1 | 5.6 ± 0.03 | 5.0 ± 0.5 | 6.6 ± 0.2 | 0.77 ± 0.11 |
| (TNs-control)/[c] | 0.31 min−1 | 1.61 | 1.41 | 1.95 |
| Time, min | 0.3 | 1.2 | 2.1 | 2.9 | 3.7 | 4.5 | 5.3 | 6.1 | 6.9 |
| Mut8-CN | 97 | 86.5 | 65.1 | 52 | 44 | 32 | 28.7 | 23 | 19 |
| WT-CN | 95 | 75 | 59 | 48.5 | 38 | 30.6 | 26 | 20 | 16 |
| Time, min | 7.7 | 8.5 | 9.3 | 10.1 | 10.9 | 11.7 | 12.5 | 13.3 | 14.1 |
| Mut8-CN | 18 | 11.9 | 10.7 | 9.4 | 9.4 | 6.8 | 5 | 1.5 | 0.5 |
| WT-CN | 13 | 10 | 9.6 | 7.0 | 3.9 | 3.9 | 3 | 0.5 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chertkova, R.V.; Oleynikov, I.P.; Pakhomov, A.A.; Sudakov, R.V.; Semenova, M.A.; Arutyunyan, A.M.; Ptushenko, V.V.; Kirpichnikov, M.P.; Dolgikh, D.A.; Vygodina, T.V. The Increase in the Peroxidase Activity of the Cytochrome C with Substitutions in the Universal Binding Site Is Associated with Changes in the Ability to Interact with External Ligands. Int. J. Mol. Sci. 2024, 25, 8237. https://doi.org/10.3390/ijms25158237
Chertkova RV, Oleynikov IP, Pakhomov AA, Sudakov RV, Semenova MA, Arutyunyan AM, Ptushenko VV, Kirpichnikov MP, Dolgikh DA, Vygodina TV. The Increase in the Peroxidase Activity of the Cytochrome C with Substitutions in the Universal Binding Site Is Associated with Changes in the Ability to Interact with External Ligands. International Journal of Molecular Sciences. 2024; 25(15):8237. https://doi.org/10.3390/ijms25158237
Chicago/Turabian StyleChertkova, Rita V., Ilya P. Oleynikov, Alexey A. Pakhomov, Roman V. Sudakov, Marina A. Semenova, Alexander M. Arutyunyan, Vasily V. Ptushenko, Mikhail P. Kirpichnikov, Dmitry A. Dolgikh, and Tatiana V. Vygodina. 2024. "The Increase in the Peroxidase Activity of the Cytochrome C with Substitutions in the Universal Binding Site Is Associated with Changes in the Ability to Interact with External Ligands" International Journal of Molecular Sciences 25, no. 15: 8237. https://doi.org/10.3390/ijms25158237







