Bafilomycin 1A Affects p62/SQSTM1 Autophagy Marker Protein Level and Autophagosome Puncta Formation Oppositely under Various Inflammatory Conditions in Cultured Rat Microglial Cells
Abstract
:1. Introduction
2. Results
2.1. BAF Inhibits Microglia Proliferation
2.2. Pro- and Anti-Inflammatory Drugs Affect Soluble Iba1 and p62/SQSTM1 Protein Contents Differently
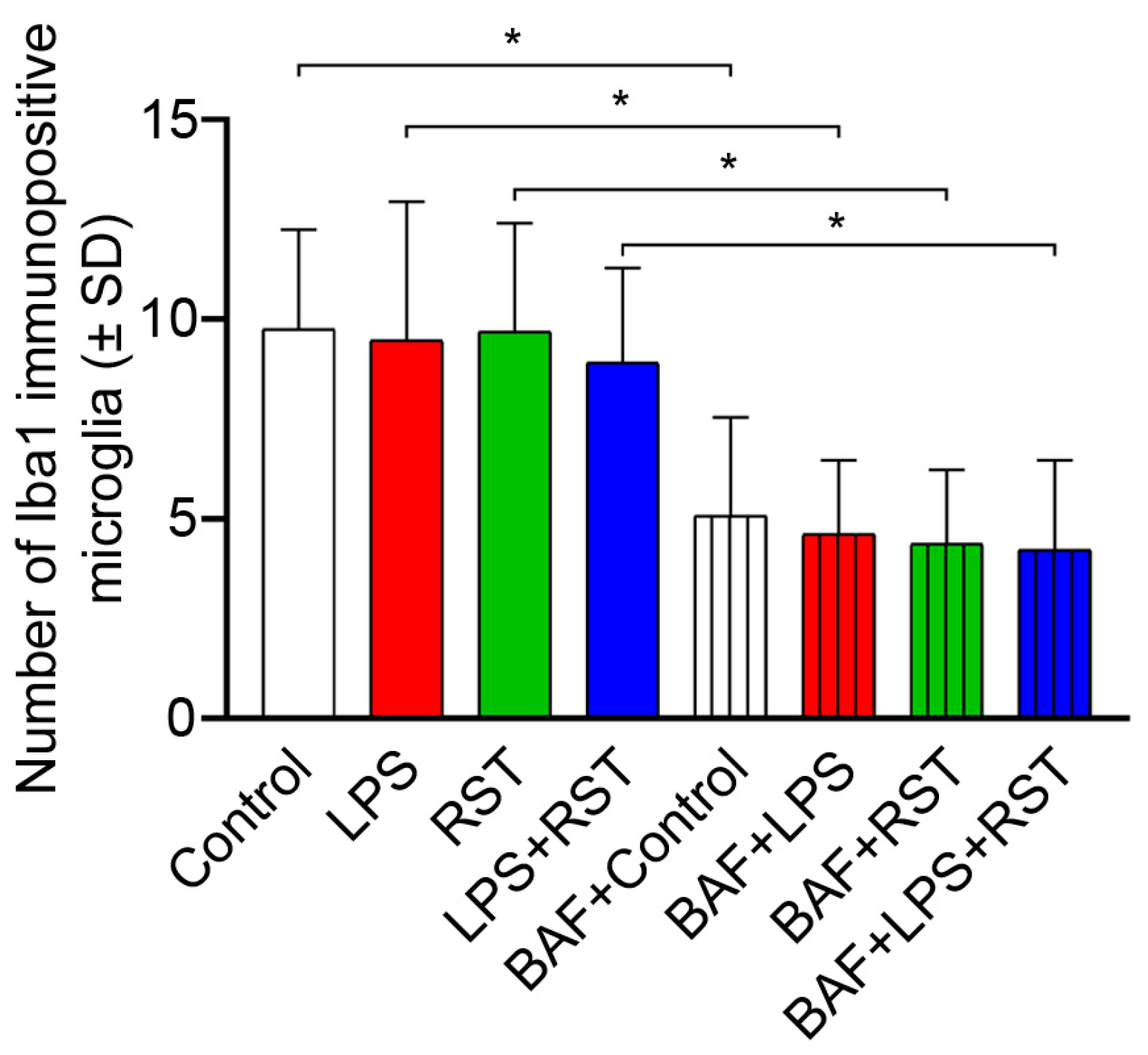
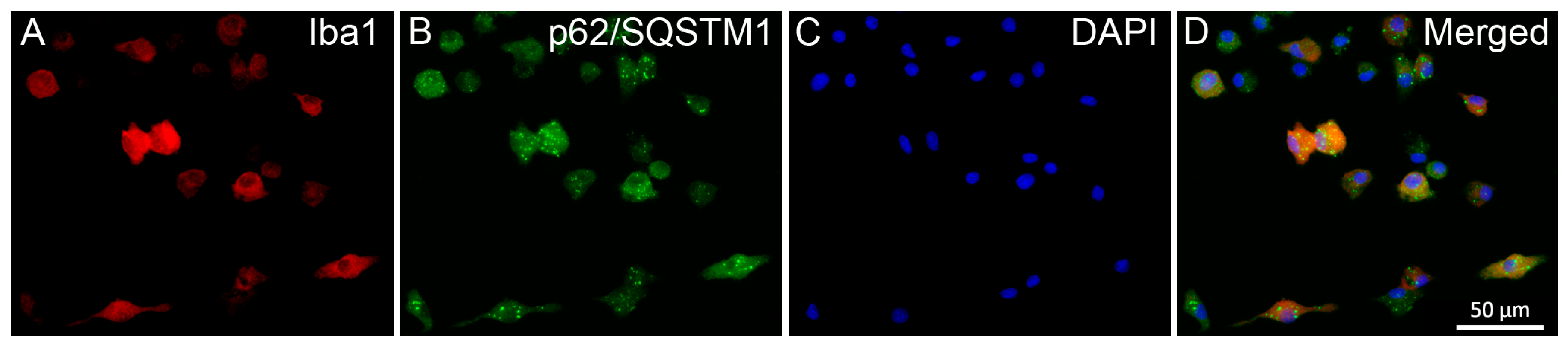
2.3. Pro- and Anti-Inflammatory Drugs Affect Iba1 and p62/SQSTM1 Immunocytochemical Signals Differently
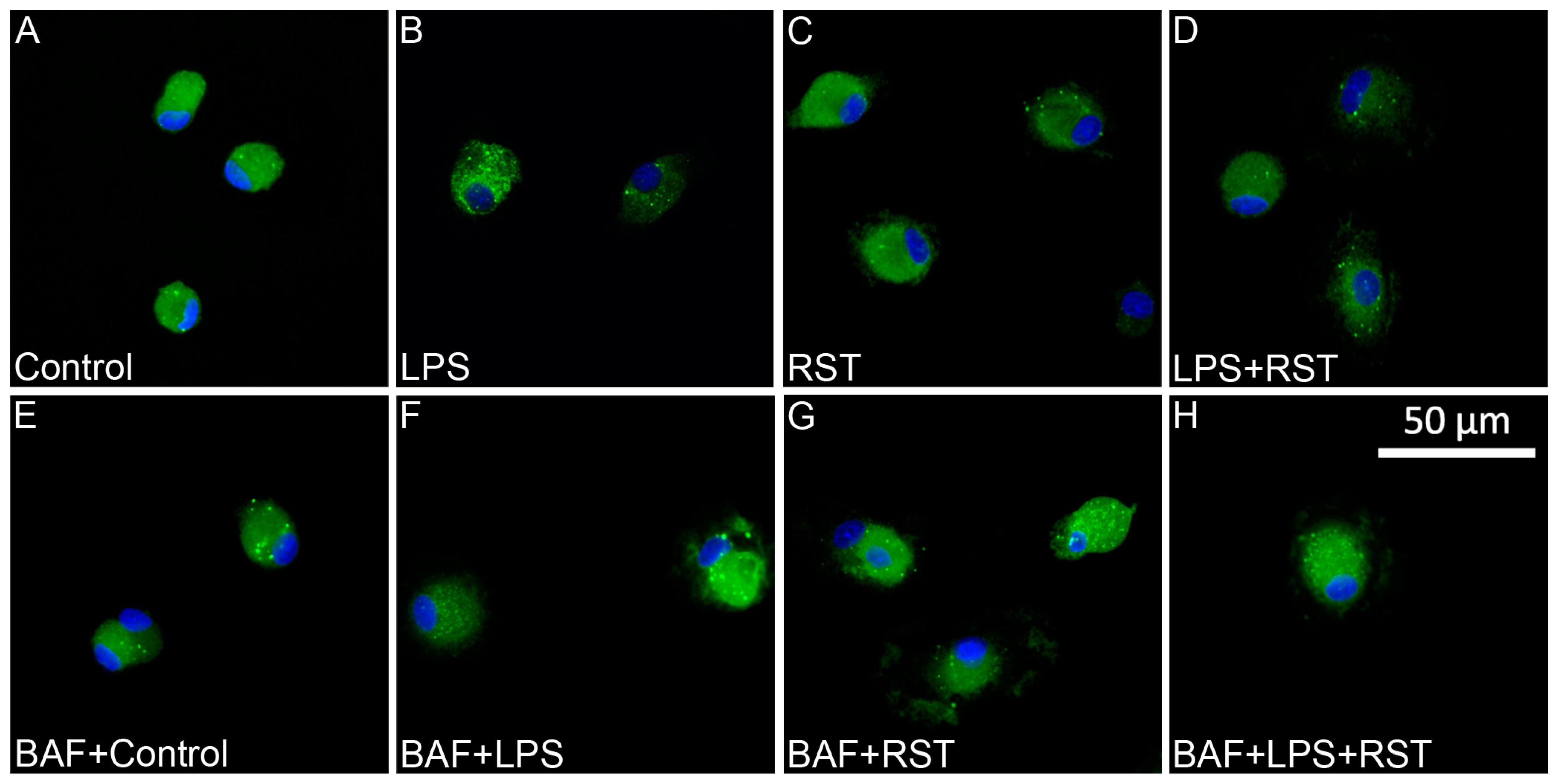
2.4. BAF Alters Autophagocytotic Puncta Formation and Affects Autophagic Flux Only in Cultures Treated with RST
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Reagents and Antibodies
4.3. Maintenance and Treatment of Cell Cultures
4.4. Determination of Microglia Cell Purity and Survival
4.5. Western Blot Analysis
4.6. Immunocytochemistry
4.7. Image Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANOVA | one-way analysis of variance |
| BAF | bafilomycin A1, ((3Z,5E,7R,8S,9S,11E,13E,15S,16R)-16-[(1S,2R,3S)-3-[(2R,4R,5S,6R)-2,4-dihydroxy-6-isopropyl-5-methyl-2-tetrahydropyranyl]-2-hydroxy-1-methylbutyl]-8-hydroxy-3,15-dimethoxy-5,7,9,11-tetramethyl-1-oxacyclohexadeca-3,5,11,13-tetraen-2-one) |
| DAPI | 2-[4-(aminoiminomethyl)phenyl]-1H-indole-6-carboximidamide hydrochloride |
| DIV | day(s) in vitro |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | fetal bovine serum |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase (EC 1.2.1.12) |
| Iba1 | ionized calcium-binding adaptor molecule 1 |
| LPS | lipopolysaccharide |
| p62 | ubiquitin-binding protein, 62 kDa (also known as sequestosome 1 (SQSTM1) |
| PBS | phosphate-buffered saline |
| rpm | revolutions per minute |
| RST | rosuvastatin, ((E)-7-[4-(4-Fluorophenyl)-6-isopropyl-2-[methyl(methylsulfonyl)amino]pyrimidin-5-yl]-(3R,5S)-3,5-dihydroxyhept-6-enoic acid) |
| RT | room temperature |
| SD | standard deviation |
| SDS | sodium dodecyl sulfate |
| SQSTM1 | autophagosome cargo protein sequestosome 1 (also known as ubiquitin-binding protein, 62 kDa (p62)) |
| subDIV | subcloned day(s) in vitro |
| TBS | Tris-buffered saline |
References
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef] [PubMed]
- Han, H.E.; Kim, T.K.; Son, H.J.; Park, W.J.; Han, P.L. Activation of autophagy pathway suppresses the expression of iNOS, IL6 and cell death of LPS-stimulated microglia cells. Biomol. Ther. 2013, 21, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Kata, D.; Földesi, I.; Feher, L.Z.; Hackler, L., Jr.; Puskas, L.G.; Gulya, K. Rosuvastatin enhances anti-inflammatory and inhibits pro-inflammatory functions in cultured microglial cells. Neuroscience 2016, 314, 47–63. [Google Scholar] [CrossRef]
- Jin, M.M.; Wang, F.; Qi, D.; Liu, W.W.; Gu, C.; Mao, C.J.; Yang, Y.P.; Zhao, Z.; Hu, L.F.; Liu, C.F. A critical role of autophagy in regulating microglia polarization in neurodegeneration. Front. Aging Neurosci. 2018, 10, 378. [Google Scholar] [CrossRef] [PubMed]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef]
- Komatsu, M.; Kageyama, S.; Ichimura, Y. p62/SQSTM1/A170: Physiology and pathology. Pharmacol. Res. 2012, 66, 457–462. [Google Scholar] [CrossRef]
- Itakura, E.; Mizushima, N. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell. Biol. 2011, 192, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Øvervatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Bjørkøy, G.; Lamark, T.; Pankiv, S.; Øvervatn, A.; Brech, A.; Johansen, T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009, 452, 181–197. [Google Scholar] [CrossRef]
- Sigmond, T.; Barna, J.; Tóth, M.L.; Takács-Vellai, K.; Pásti, G.; Kovács, A.L.; Vellai, T. Autophagy in Caenorhabditis elegans. Methods Enzymol. 2008, 451, 521–540. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Mansilla, A.; Menzies, F.M.; Rubinsztein, D.C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell 2009, 33, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Billes, V.; Komlós, M.; Hotzi, B.; Manzéger, A.; Tarnóci, A.; Papp, D.; Szikszai, F.; Szinyákovics, J.; Rácz, Á.; et al. The small molecule AUTEN-99 (autophagy enhancer-99) prevents the progression of neurodegenerative symptoms. Sci. Rep. 2017, 7, srep42014. [Google Scholar] [CrossRef] [PubMed]
- Rusten, T.; Stenmark, H. p62, an autophagy hero or culprit? Nat. Cell Biol. 2010, 12, 207–209. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Szinyákovics, J.; Billes, V.; Murányi, G.; Varga, V.B.; Bjelik, A.; Légrádi, Á.; Szabó, M.; Sándor, S.; Kubinyi, E.; et al. A conserved MTMR lipid phosphatase increasingly suppresses autophagy in brain neurons during aging. Sci. Rep. 2022, 12, 21817. [Google Scholar] [CrossRef] [PubMed]
- Billes, V.; Kovács, T.; Manzéger, A.; Lőrincz, P.; Szincsák, S.; Regős, Á.; Kulcsár, P.I.; Korcsmáros, T.; Lukácsovich, T.; Hoffmann, G.; et al. Developmentally regulated autophagy is required for eye formation in Drosophila. Autophagy 2018, 14, 1499–1519. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.G.; Zhou, X.G.; Qiao, G.; Yu, L.; Tang, Y.; Yan, L.; Qiu, W.Q.; Pan, R.; Yu, C.L.; Law, B.Y.; et al. Targeting microglial autophagic degradation in NLRP3 inflammasome-mediated neurodegenerative diseases. Ageing Res. Rev. 2021, 65, 101202. [Google Scholar] [CrossRef]
- Taylor, F.; Huffman, M.D.; Macedo, A.F.; Moore, T.H.; Burke, M.; Davey Smith, G.; Ward, K.; Ebrahim, S. Statins for the primary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2013, 1, CD004816. [Google Scholar] [CrossRef]
- Burg, J.S.; Espenshade, P.J. Regulation of HMG-CoA reductase in mammals and yeast. Prog. Lipid Res. 2011, 50, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Liu, Y.; Mai, H.; Zang, J.; Shen, L.; Zhang, Y.; Xu, A. Rosuvastatin reduces neuroinflammation in the hemorrhagic transformation after rt-PA treatment in a mouse model of experimental stroke. Front. Cell. Neurosci. 2018, 12, 225. [Google Scholar] [CrossRef]
- Chae, U.; Kim, H.S.; Lee, H.S.; Lee, S.R.; Lee, D.S. Drp1-dependent mitochondrial fission regulates p62-mediated autophagy in LPS-induced activated microglial cells. Biosci. Biotechnol. Biochem. 2019, 83, 409–416. [Google Scholar] [CrossRef]
- Bussi, C.; Peralta Ramos, J.M.; Arroyo, D.S.; Gaviglio, E.A.; Gallea, J.I.; Wang, J.M.; Celej, M.S.; Iribarren, P. Autophagy down regulates pro-inflammatory mediators in BV2 microglial cells and rescues both LPS and alpha-synuclein induced neuronal cell death. Sci. Rep. 2017, 7, 43153. [Google Scholar] [CrossRef]
- He, Y.; She, H.; Zhang, T.; Xu, H.; Cheng, L.; Yepes, M.; Zhao, Y.; Mao, Z. p38 MAPK inhibits autophagy and promotes microglial inflammatory responses by phosphorylating ULK1. J. Cell Biol. 2018, 217, 315–328. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Bowman, E.J.; Siebers, A.; Altendorf, K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 1988, 85, 7972–7976. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Hagenmaier, H.; Drautz, H.; Baumgartner, A.; Zähner, H. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. 1984, 37, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Redmann, M.; Benavides, G.A.; Berryhill, T.F.; Wani, W.Y.; Ouyang, X.; Johnson, M.S.; Ravi, S.; Barnes, S.; Darley-Usmar, V.M.; Zhang, J. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol. 2017, 11, 73–81. [Google Scholar] [CrossRef]
- Yoshimori, T.; Yamamoto, A.; Moriyama, Y.; Futai, M.; Tashiro, Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 1991, 266, 17707–17712. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Tagawa, Y.; Yoshimori, T.; Moriyama, Y.; Masaki, R.; Tashiro, Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell. Struct. Funct. 1998, 23, 33–42. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell. Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Klionsky, D.J. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell. Biol. 2007, 8, 931–937. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdalla, F.C.; Abeliovich, H.; Abraham, R.T.; Acevedo-Arozena, A.; Adeli, K.; Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-Ghiso, J.A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012, 8, 445–544. [Google Scholar] [CrossRef] [PubMed]
- Harry, G.J.; Kraft, A.D. Neuroinflammation and microglia: Considerations and approaches for neurotoxicity assessment. Expert Opin. Drug Metab. Toxicol. 2008, 10, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhu, M.; Che, X.; Wang, H.; Liang, X.J.; Wu, C.; Xue, X.; Yang, J. Lipopolysaccharide induces neuroinflammation in microglia by activating the MTOR pathway and downregulating Vps34 to inhibit autophagosome formation. J. Neuroinflamm. 2020, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222, Erratum in Autophagy 2016, 12, 443. [Google Scholar] [CrossRef]
- Lin, M.; Yu, H.; Xie, Q.; Xu, Z.; Shang, P. Role of microglia autophagy and mitophagy in age-related neurodegenerative diseases. Front. Aging Neurosci. 2023, 14, 1100133. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.-S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.-I.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Moscat, J.; Diaz-Meco, M.T. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009, 137, 1001–1004. [Google Scholar] [CrossRef] [PubMed]
- Bardag-Gorce, F.; Francis, T.; Nan, L.; Li, J.; He Lue, Y.; French, B.A.; French, S.W. Modifications in p62 occur due to proteasome inhibition in alcoholic liver disease. Life Sci. 2005, 77, 2594–2602. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Elazar, Z.; Seglen, P.O.; Rubinsztein, D.C. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 2008, 4, 849–850. [Google Scholar] [CrossRef]
- Chen, C.; Deng, M.; Sun, Q.; Loughran, P.; Billiar, T.R.; Scott, M.J. Lipopolysaccharide stimulates p62-dependent autophagy-like aggregate clearance in hepatocytes. Biomed. Res. Int. 2014, 2014, 267350. [Google Scholar] [CrossRef]
- Kim, S.-W.; Oh, S.-A.; Seol, S.-I.; Davaanyam, D.; Lee, J.-K. Cytosolic HMGB1 mediates LPS-induced autophagy in microglia by interacting with NOD2 and suppresses its proinflammatory function. Cells 2022, 11, 2410. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Ge, Q.; Li, R.; Lv, J.; Zhang, Y.; Feng, A.; Kelly, G.T.; Wang, H.; Wang, X.; Song, C.; et al. LPS restores protective immunity in macrophages against Mycobacterium tuberculosis via autophagy. Mol. Immunol. 2020, 124, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, L.; Chen, Y.; Shao, Q.; Qin, W. Bafilomycin A1 inhibits the growth and metastatic potential of the BEL-7402 liver cancer and HO-8910 ovarian cancer cell lines and induces alterations in their microRNA expression. Exp. Ther. Med. 2015, 10, 1829–1834. [Google Scholar] [CrossRef]
- Saris, N.E.; Andersson, M.A.; Mikkola, R.; Andersson, L.C.; Teplova, V.V.; Grigoriev, P.A.; Salkinoja-Salonen, M.S. Microbial toxin’s effect on mitochondrial survival by increasing K+ uptake. Toxicol. Ind. Health 2009, 25, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Xie, Y.; Xu, Y.; Zhou, H.; Xu, W.; Dong, Q. Bafilomycin A1 inhibits autophagy and induces apoptosis in MG63 osteosarcoma cells. Mol. Med. Rep. 2014, 10, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- Whitton, B.; Okamoto, H.; Packham, G.; Crabb, S.J. Vacuolar ATPase as a potential therapeutic target and mediator of treatment resistance in cancer. Cancer Med. 2018, 7, 3800–3811. [Google Scholar] [CrossRef]
- Wu, Y.C.; Wu, W.K.; Li, Y.; Yu, L.; Li, Z.J.; Wong, C.C.; Li, H.T.; Sung, J.J.; Cho, C.H. Inhibition of macroautophagy by bafilomycin A1 lowers proliferation and induces apoptosis in colon cancer cells. Biochem. Biophys. Res. Commun. 2009, 382, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Zabala, A.; Sierra-Torre, V.; Sierra, A. Assessing autophagy in microglia: A two-step model to determine autophagosome formation, degradation, and net turnover. Front. Immunol. 2021, 11, 620602, Erratum in Front. Immunol. 2021, 12, 724901. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Zhu, K.; Lin, Y.; An, L.; Huang, F.; Yao, Y.; Ren, L. Insulin secretion impairment induced by rosuvastatin partly though autophagy in INS-1E cells. Cell Biol. Int. 2020, 44, 127–136. [Google Scholar] [CrossRef]
- Han, F.; Xiao, Q.Q.; Peng, S.; Che, X.Y.; Jiang, L.S.; Shao, Q.; He, B. Atorvastatin ameliorates LPS-induced inflammatory response by autophagy via AKT/mTOR signaling pathway. J. Cell. Biochem. 2018, 119, 1604–1615. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, K.; Liu, P.; Zhang, X.; Dong, X.; Gao, J.; Liu, Q.; Barr, M.P.; Zhang, Q.; Hou, X.; et al. Bafilomycin A1 induces caspase-independent cell death in hepatocellular carcinoma cells via targeting of autophagy and MAPK pathways. Sci. Rep. 2016, 6, 37052. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Nam, H.; Kim, L.E.; Jeon, Y.; Min, H.; Ha, S.; Lee, Y.; Kim, S.Y.; Lee, S.J.; Kim, E.K.; et al. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy 2019, 15, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.Z.; Ke, P.; Xu, Z.Q.; Wei, W.; Cheng, M.H.; Han, B.Z.; Chen, X.W.; Su, D.F.; Liu, C. Autophagy plays an important role in anti-inflammatory mechanisms stimulated by alpha7 nicotinic acetylcholine receptor. Front. Immunol. 2017, 8, 553. [Google Scholar] [CrossRef] [PubMed]
- Billes, V.; Kovács, T.; Hotzi, B.; Manzéger, A.; Tagscherer, K.; Komlós, M.; Tarnóci, A.; Pádár, Z.; Erdős, A.; Bjelik, A.; et al. AUTEN-67 (Autophagy Enhancer-67) hampers the progression of neurodegenerative symptoms in a Drosophila model of Huntington’s Disease. J. Huntingt. Dis. 2016, 5, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Erie, C.; Lu, M.L.; Wei, J. Aberrant subcellular localization of SQSTM1/p62 contributes to increased vulnerability to proteotoxic stress recovery in Huntington’s disease. Mol. Cell. Neurosci. 2018, 88, 43–52. [Google Scholar] [CrossRef]
- Szabo, M.; Gulya, K. Development of the microglial phenotype in culture. Neuroscience 2013, 241, 280–295. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, M.; He, G.; Zhang, X.; Li, W.; Gao, Y.; Li, Z.; Wang, Z.; Zhang, C. Glyceraldehyde-3-phosphate dehydrogenase: A universal internal control for Western blots in prokaryotic and eukaryotic cells. Anal. Biochem. 2012, 423, 15–22. [Google Scholar] [CrossRef]
- Dulka, K.; Nacsa, K.; Lajkó, N.; Gulya, K. Quantitative morphometric and cell-type-specific population analysis of microglia-enriched cultures subcloned to high purity from newborn rat brains. IBRO Neurosci. Reports 2021, 10, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Singh, B.; Bhaskar, S. Methods for detection of autophagy in mammalian cells. Methods Mol. Biol. 2019, 2045, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Szabo, M.; Lajkó, N.; Dulka, K.; Barczánfalvi, G.; Lőrinczi, B.; Szatmári, I.; Mihály, A.; Vécsei, L.; Gulya, K. The kynurenic acid analog SZR104 induces cytomorphological changes associated with the anti-inflammatory phenotype in cultured microglia. Sci. Rep. 2023, 13, 11328. [Google Scholar] [CrossRef] [PubMed]



| Primary Antibody, Abbrev. Name | Primary Antibody, Full Name (Cat. No.) | Final Dilution | Company | Secondary Antibody, Full Name (Cat. No.) | Final Dilution | Company |
|---|---|---|---|---|---|---|
| Antibodies used in Western blot analyses | ||||||
| Iba1 | Rabbit anti-Iba1 polycl. ab. (019-19741) | 1/1000 | Abcam, Cambridge, UK | Anti-rabbit IgG, peroxidase conjug. (A-9169) | 1/1000 | Sigma, St. Louis, MO, USA |
| p62/SQSTM1 | Mouse anti- p62 monocl. ab. (ab56416) | 1/300 | Abcam, Cambridge, UK | Anti-mouse IgG, peroxidase conjug. (A-9044) | 1/1000 | Sigma, St. Louis, MO, USA |
| GAPDH | Mouse anti-GAPDH monocl. ab. (G8795) | 1/20,000 | Sigma, St. Louis, USA | Anti-mouse IgG, peroxidase conjug. (A-9044) | 1/1000 | Sigma, St. Louis, MO, USA |
| Antibodies used in fluorescent immunocytochemistry | ||||||
| Iba1 | Rabbit anti-Iba1 polycl. ab. (019-19741) | 1/1000 | Abcam, Cambridge, UK | Alexa Fluor 568 goat anti-rabbit (A-11011) | 1/1000 | Invitrogen, Carlsbad, CA, USA |
| p62/SQSTM1 | Mouse anti- p62 monocl. ab. (ab56416) | 1/1000 | Abcam, Cambridge, UK | Alexa Fluor 488 goat anti-mouse (A-11029) | 1/1000 | Invitrogen, Carlsbad, CA, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pesti, I.; Barczánfalvi, G.; Dulka, K.; Kata, D.; Farkas, E.; Gulya, K. Bafilomycin 1A Affects p62/SQSTM1 Autophagy Marker Protein Level and Autophagosome Puncta Formation Oppositely under Various Inflammatory Conditions in Cultured Rat Microglial Cells. Int. J. Mol. Sci. 2024, 25, 8265. https://doi.org/10.3390/ijms25158265
Pesti I, Barczánfalvi G, Dulka K, Kata D, Farkas E, Gulya K. Bafilomycin 1A Affects p62/SQSTM1 Autophagy Marker Protein Level and Autophagosome Puncta Formation Oppositely under Various Inflammatory Conditions in Cultured Rat Microglial Cells. International Journal of Molecular Sciences. 2024; 25(15):8265. https://doi.org/10.3390/ijms25158265
Chicago/Turabian StylePesti, István, Gábor Barczánfalvi, Karolina Dulka, Diana Kata, Eszter Farkas, and Karoly Gulya. 2024. "Bafilomycin 1A Affects p62/SQSTM1 Autophagy Marker Protein Level and Autophagosome Puncta Formation Oppositely under Various Inflammatory Conditions in Cultured Rat Microglial Cells" International Journal of Molecular Sciences 25, no. 15: 8265. https://doi.org/10.3390/ijms25158265
APA StylePesti, I., Barczánfalvi, G., Dulka, K., Kata, D., Farkas, E., & Gulya, K. (2024). Bafilomycin 1A Affects p62/SQSTM1 Autophagy Marker Protein Level and Autophagosome Puncta Formation Oppositely under Various Inflammatory Conditions in Cultured Rat Microglial Cells. International Journal of Molecular Sciences, 25(15), 8265. https://doi.org/10.3390/ijms25158265







