Genome-Wide Identification and Expression Analysis of SlNRAMP Genes in Tomato under Nutrient Deficiency and Cadmium Stress during Arbuscular Mycorrhizal Symbiosis
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of the SlNRAMP Family Genes in Tomato
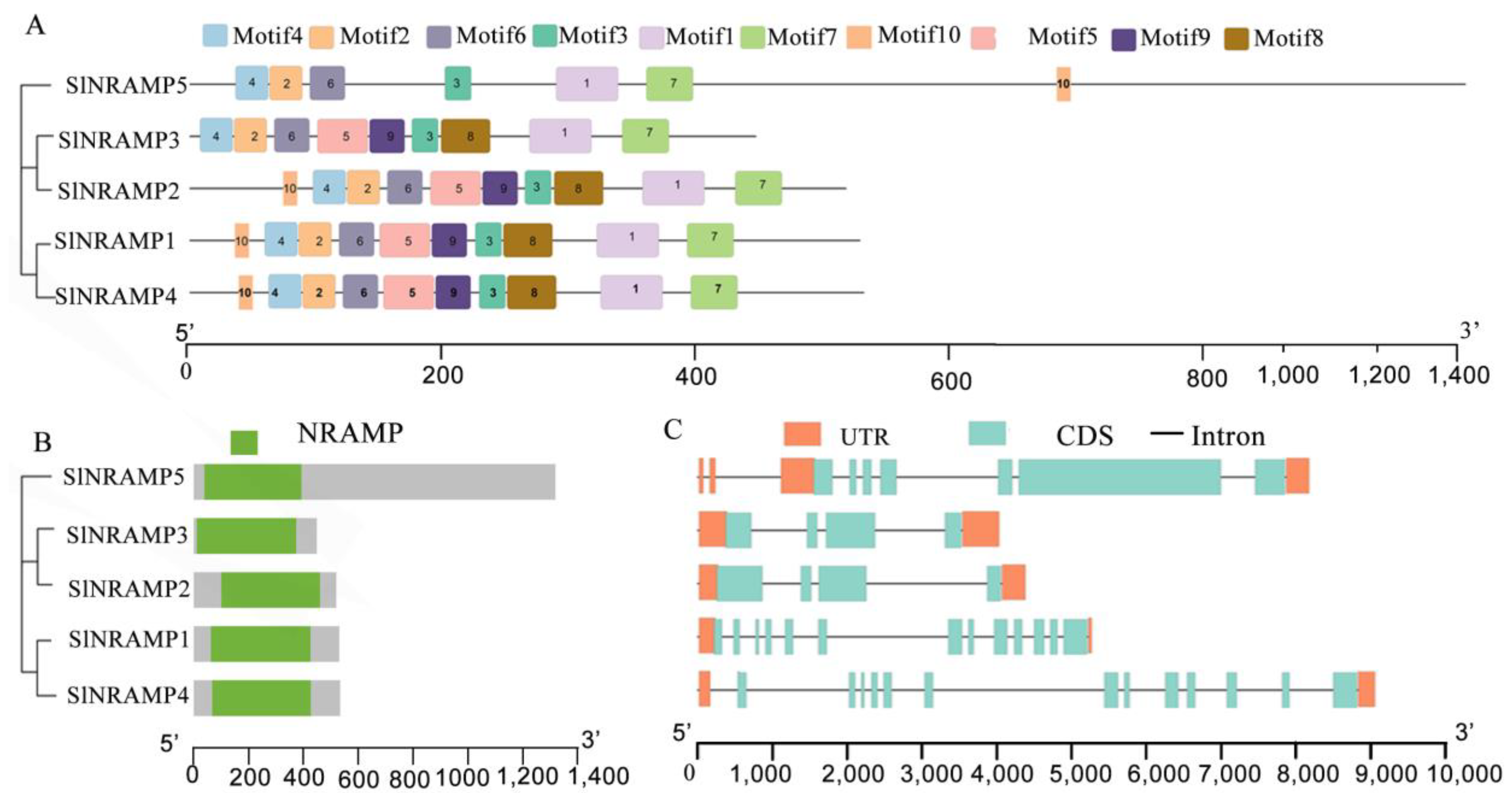
2.2. Domains, Conserved Motifs, and Models of the SlNRAMP Family Proteins
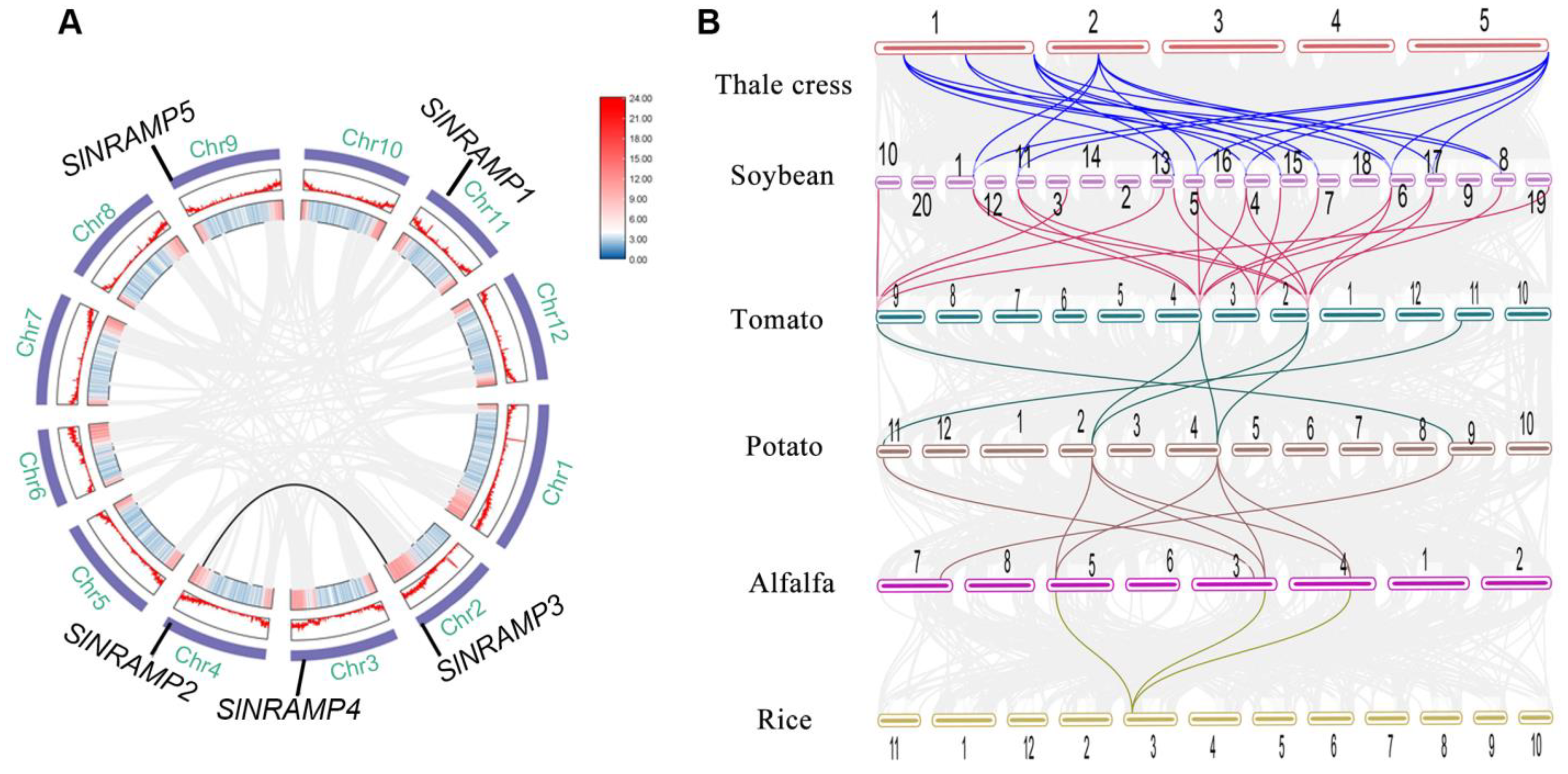
2.3. Chromosomal Organization and Duplication of the SlNRAMP Family Genes
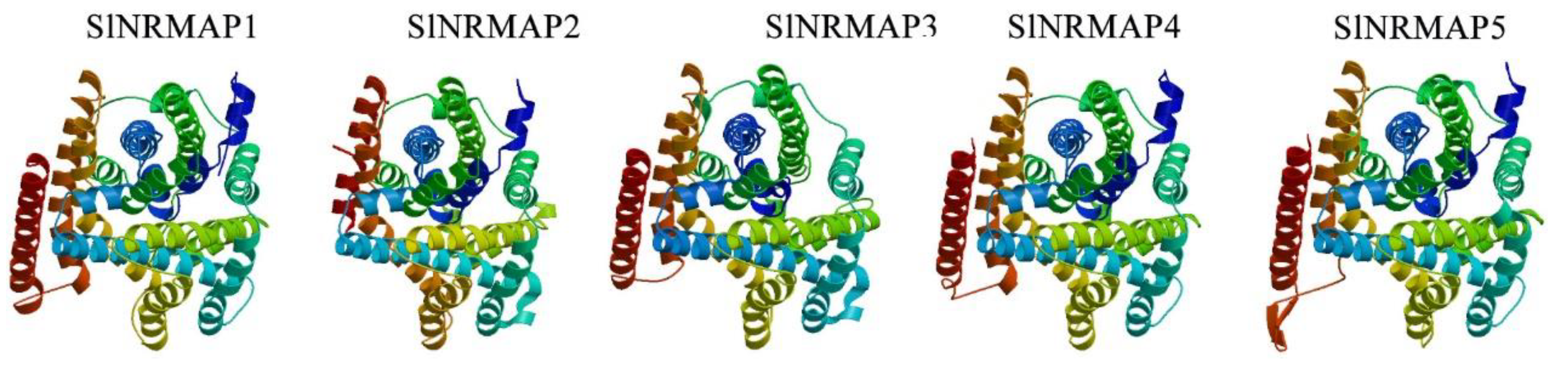
2.4. Multiple Sequence Alignment and 3D Model Predictions of the SlNRAMP Family Proteins
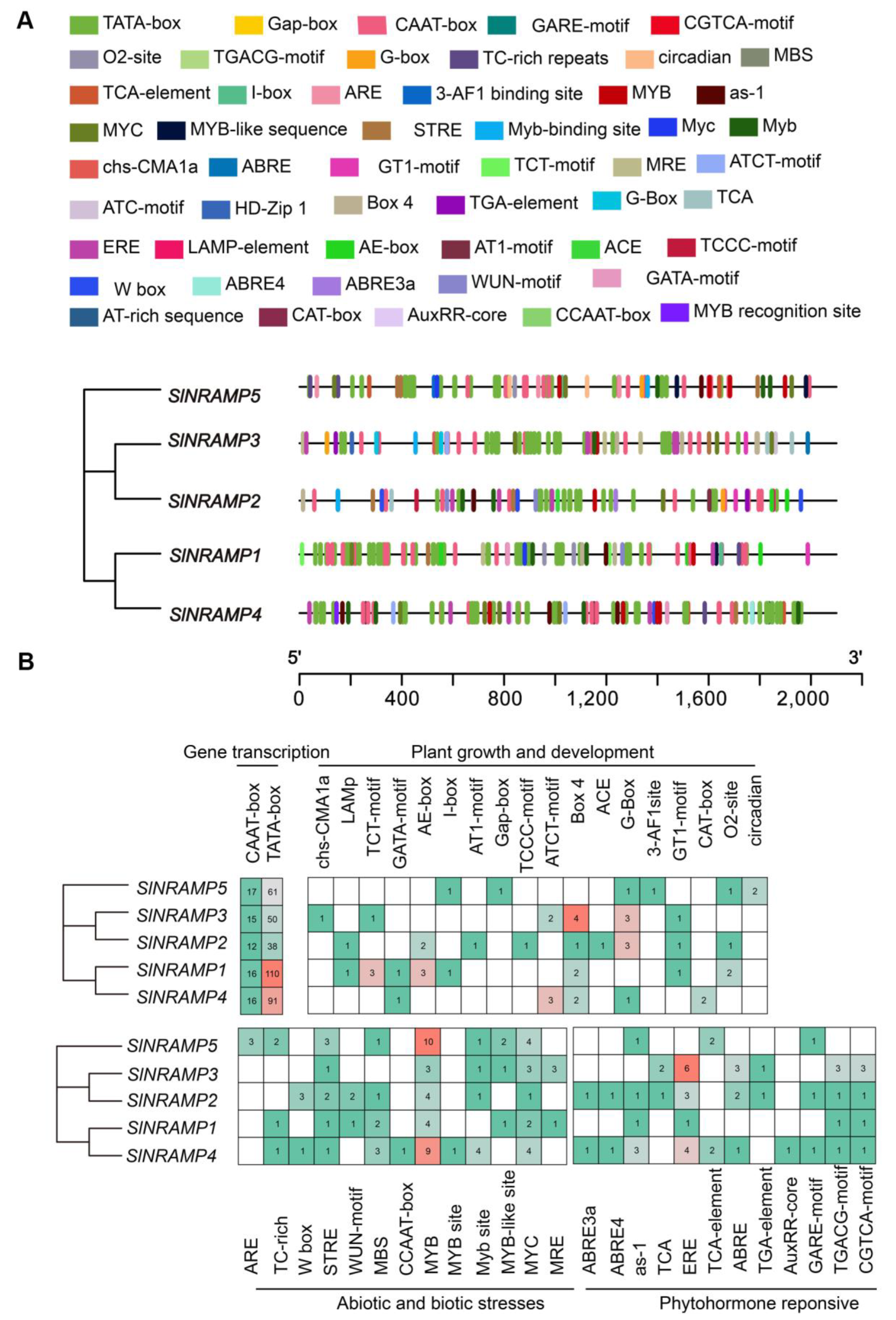
2.5. Cis-Element and Promoter Analysis of the SlNRAMP Family Genes
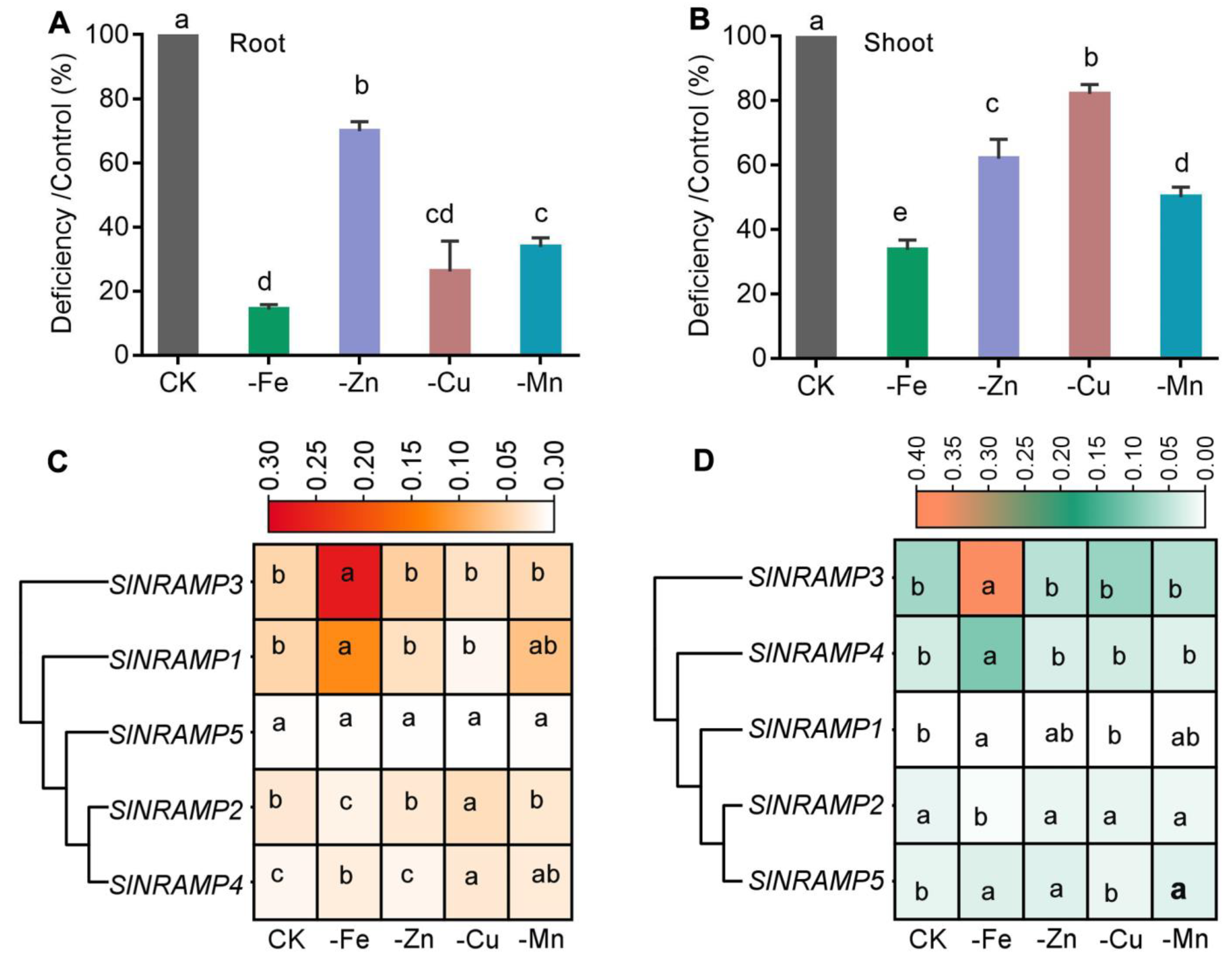
2.6. Expression Profiles of the SlNRAMP Family Genes in Response to Nutrient Deficiency in Tomato
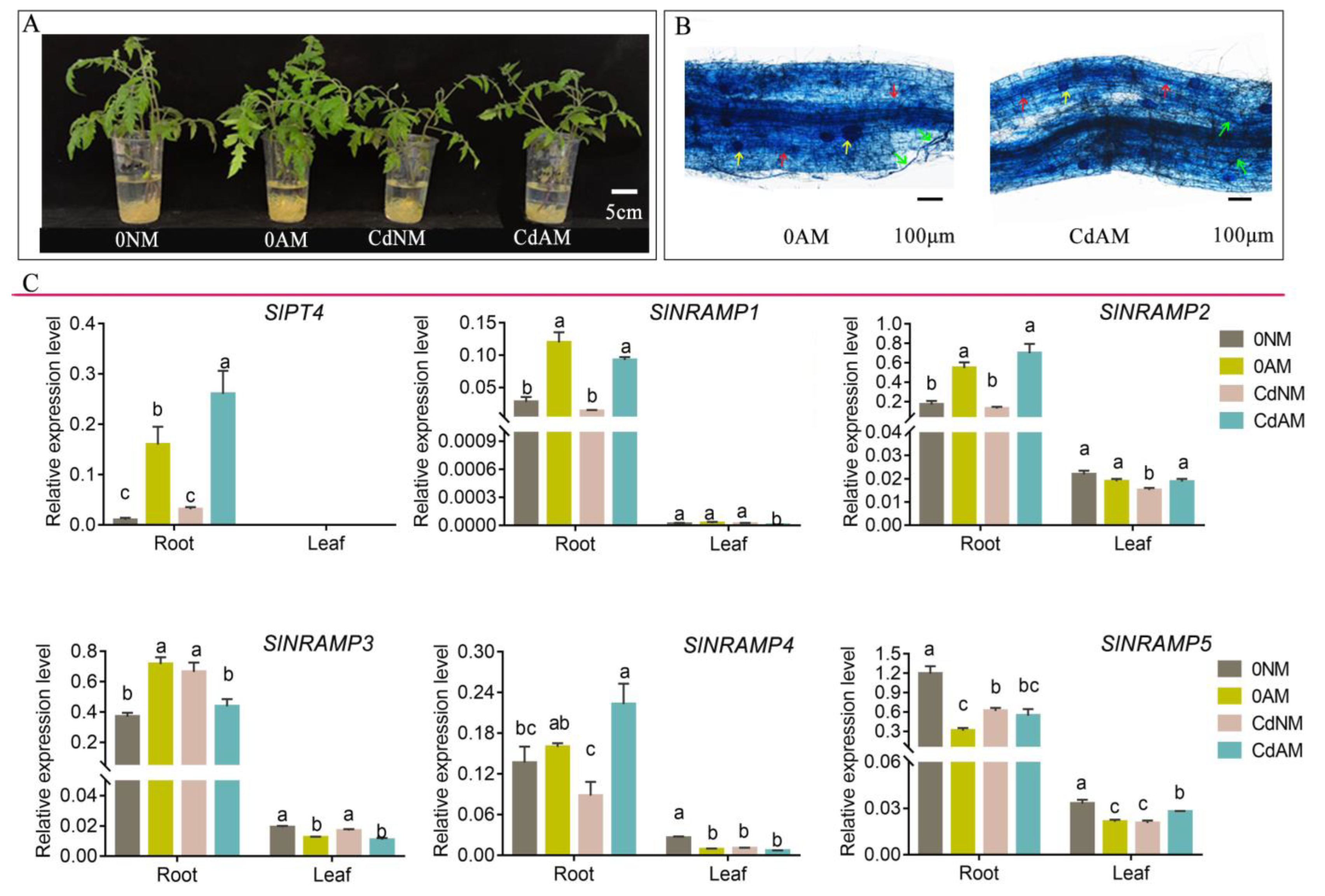
2.7. Expression Analysis of the SlNRAMP Family Genes in Response to AMF Colonization under Cd Stress
2.8. P and Cd Concentration in Tomato Root and Shoot Altered by AMF Colonization under Cd Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification and Sequence Analysis of the NRAMP Genes Family in Tomato
4.3. Physicochemical Properties and Structure Characteristics of SlNRAMP Proteins
4.4. Chromosomal Locations and Duplication of NRAMP Genes
4.5. Cis-Acting Regulatory Elements and MicroRNA Target Sites of NRAMP Genes
4.6. Phylogenetic Analysis
4.7. cDNA Preparation and qRT-PCR Analysis
4.8. Visualization of Mycorrhizal Fungal Structures
4.9. Plant Elements Concentration
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMF | Arbuscular mycorrhizal fungi |
| Cu | Copper |
| Cd | Cadmium |
| CDSs | Coding sequences |
| GMQE | Global model quality estimate |
| Fe | Iron |
| Mn | Manganese |
| MeJA | Methyl-jasmonate |
| NRAMP | Natural resistance-associated macrophage protein |
| PI | Isoelectric points |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| Zn | Zinc |
References
- Bozzi, A.T.; Gaudet, R. Molecular Mechanism of Nramp-Family Transition Metal Transport. J. Mol. Biol. 2021, 433, 166991. [Google Scholar] [CrossRef] [PubMed]
- Courville, P.; Chaloupka, R.; Cellier, M.F.M. Recent Progress in Structure-Function Analyses of Nramp Proton-Dependent Metal-Ion Transporters. Biochem. Cell Biol. 2006, 84, 960–978. [Google Scholar] [CrossRef] [PubMed]
- Nevo, Y.; Nelson, N. The NRAMP Family of Metal-Ion Transporters. Biochim. Biophys. Acta 2006, 1763, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Mani, A.; Sankaranarayanan, K. In Silico Analysis of Natural Resistance-Associated Macrophage Protein (NRAMP) Family of Transporters in Rice. Protein J. 2018, 37, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, G.; Yang, J.; Zhao, X.; Sun, Z.; Hou, H. Role of Nramp Transporter Genes of Spirodela Polyrhiza in Cadmium Accumulation. Ecotoxicol. Environ. Saf. 2021, 227, 112907. [Google Scholar] [CrossRef] [PubMed]
- Lanquar, V.; Ramos, M.S.; Lelièvre, F.; Barbier-Brygoo, H.; Krieger-Liszkay, A.; Krämer, U.; Thomine, S. Export of Vacuolar Manganese by AtNRAMP3 and AtNRAMP4 Is Required for Optimal Photosynthesis and Growth under Manganese Deficiency. Plant Physiol. 2010, 152, 1986–1999. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, G.; Kumar, R.; Prasad, B. Genome Wide Analysis and Identification of NRAMP Gene Family in Wheat (Triticum aestivum L.). Pharma Innov. J. 2022, 11, 499–504. [Google Scholar]
- Qin, L.; Han, P.; Chen, L.; Walk, T.C.; Li, Y.; Hu, X.; Xie, L.; Liao, H.; Liao, X. Genome-Wide Identification and Expression Analysis of NRAMP Family Genes in Soybean (Glycine Max L.). Front. Plant Sci. 2017, 8, 1436. [Google Scholar] [CrossRef]
- Tan, Z.; Li, J.; Guan, J.; Wang, C.; Zhang, Z.; Shi, G. Genome-Wide Identification and Expression Analysis Reveals Roles of the NRAMP Gene Family in Iron/Cadmium Interactions in Peanut. Int. J. Mol. Sci. 2023, 24, 1713. [Google Scholar] [CrossRef]
- Tian, W.; He, G.; Qin, L.; Li, D.; Meng, L.; Huang, Y.; He, T. Genome-Wide Analysis of the NRAMP Gene Family in Potato (Solanum tuberosum): Identification, Expression Analysis and Response to Five Heavy Metals Stress. Ecotoxicol. Environ. Saf. 2021, 208, 111661. [Google Scholar] [CrossRef]
- Ma, X.; Yang, H.; Bu, Y.; Zhang, Y.; Sun, N.; Wu, X.; Jing, Y. Genome-Wide Identification of the NRAMP Gene Family in Populus Trichocarpa and Their Function as Heavy Metal Transporters. Ecotoxicol. Environ. Saf. 2023, 261, 115110. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, M.; Song, H.; Zhao, J.; Shabala, S.; Tian, S.; Yang, X. A Novel Plasma Membrane-Based NRAMP Transporter Contributes to Cd and Zn Hyperaccumulation in Sedum Alfredii Hance. Environ. Exp. Bot. 2020, 176, 104121. [Google Scholar] [CrossRef]
- Curie, C.; Alonso, J.M.; Le Jean, M.; Ecker, J.R.; Briat, J.F. Involvement of NRAMP1 from Arabidopsis thaliana in Iron Transport. Biochem. J. 2000, 347 Pt 3, 749–755. [Google Scholar] [CrossRef]
- Lanquar, V.; Lelièvre, F.; Bolte, S.; Hamès, C.; Alcon, C.; Neumann, D.; Vansuyt, G.; Curie, C.; Schröder, A.; Krämer, U.; et al. Mobilization of Vacuolar Iron by AtNRAMP3 and AtNRAMP4 Is Essential for Seed Germination on Low Iron. EMBO J. 2005, 24, 4041–4051. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zheng, L.; Li, Y.; Zhou, X.; Li, J.; Gu, D.; Xu, E.; Lu, Y.; Chen, X.; et al. The Intracellular Transporter AtNRAMP6 Is Involved in Fe Homeostasis in Arabidopsis. Front. Plant Sci. 2019, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-D.; Xie, Y.; Zhang, H.; Zhang, S.; Zhao, F.-J. The Vacuolar Transporter OsNRAMP2 Mediates Fe Remobilization during Germination and Affects Cd Distribution to Rice Grain. Plant Soil 2022, 476, 79–95. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, W.; Dong, H.; Zhang, Y.; Lv, K.; Wang, D.; Lian, X. OsNRAMP3 Is a Vascular Bundles-Specific Manganese Transporter That Is Responsible for Manganese Distribution in Rice. PLoS ONE 2013, 8, e83990. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Senoura, T.; Sugimoto, K.; Ono, K.; Yano, M.; Ishikawa, S.; Arao, T.; et al. Characterizing the Role of Rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci. Rep. 2012, 2, 286. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 Is a Major Transporter Responsible for Manganese and Cadmium Uptake in Rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef]
- Zhang, Y.; He, T.; Tian, W.; Xia, Y.; He, Y.; Su, M.; He, G. The Expression of the StNRAMP2 Gene Determined the Accumulation of Cadmium in Different Tissues of Potato. Int. J. Mol. Sci. 2023, 24, 9322. [Google Scholar] [CrossRef]
- Guo, J.; Long, L.; Chen, A.; Dong, X.; Liu, Z.; Chen, L.; Wang, J.; Yuan, L. Tonoplast-Localized Transporter ZmNRAMP2 Confers Root-to-Shoot Translocation of Manganese in Maize. Plant Physiol. 2022, 190, 2601–2616. [Google Scholar] [CrossRef] [PubMed]
- Motaharpoor, Z.; Taheri, H.; Nadian, H. Rhizophagus Irregularis Modulates Cadmium Uptake, Metal Transporter, and Chelator Gene Expression in Medicago Sativa. Mycorrhiza 2019, 29, 389–395. [Google Scholar] [CrossRef]
- Gutjahr, C.; Parniske, M. Cell and Developmental Biology of Arbuscular Mycorrhiza Symbiosis. Annu. Rev. Cell Dev. Biol. 2013, 29, 593–617. [Google Scholar] [CrossRef] [PubMed]
- Varga, S.; Soulsbury, C.D. Arbuscular Mycorrhizal Fungi Change Host Plant DNA Methylation Systemically. Plant Biol. 2019, 21, 278–283. [Google Scholar] [CrossRef]
- Marschner, H.; Dell, B. Nutrient Uptake in Mycorrhizal Symbiosis. Plant Soil 1994, 159, 89–102. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Xie, K.; Tian, Y.; Yan, A.; Liu, J.; Huang, Y.; Wang, S.; Zhu, Y.; Chen, A.; et al. A Mycorrhiza-Specific H+-ATPase Is Essential for Arbuscule Development and Symbiotic Phosphate and Nitrogen Uptake. Plant Cell Environ. 2020, 43, 1069–1083. [Google Scholar] [CrossRef]
- Harrison, M.J. The Arbuscular Mycorrhizal Symbiosis. In Plant-Microbe Interactions; Stacey, G., Keen, N.T., Eds.; Springer: Boston, MA, USA, 1997; pp. 1–34. ISBN 978-1-4613-7758-0. [Google Scholar]
- Janeeshma, E.; Puthur, J.T. Direct and Indirect Influence of Arbuscular Mycorrhizae on Enhancing Metal Tolerance of Plants. Arch. Microbiol. 2020, 202, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Kamran, M.; Fang, Y.; Wang, Q.; Cao, H.; Yang, G.; Deng, L.; Wang, Y.; Zhou, Y.; Anastopoulos, I.; et al. Arbuscular Mycorrhizal Fungi-Induced Mitigation of Heavy Metal Phytotoxicity in Metal Contaminated Soils: A Critical Review. J. Hazard. Mater. 2021, 402, 123919. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, Y.; Chen, S.; Polle, A.; Rennenberg, H.; Luo, Z.-B. Physiological and Molecular Mechanisms of Heavy Metal Accumulation in Nonmycorrhizal versus Mycorrhizal Plants. Plant Cell Environ. 2019, 42, 1087–1103. [Google Scholar] [CrossRef]
- Fan, P.; Wu, L.; Wang, Q.; Wang, Y.; Luo, H.; Song, J.; Yang, M.; Yao, H.; Chen, S. Physiological and Molecular Mechanisms of Medicinal Plants in Response to Cadmium Stress: Current Status and Future Perspective. J. Hazard. Mater. 2023, 450, 131008. [Google Scholar] [CrossRef]
- Liang, J.; Wang, Z.; Ren, Y.; Jiang, Z.; Chen, H.; Hu, W.; Tang, M. The Alleviation Mechanisms of Cadmium Toxicity in Broussonetia Papyrifera by Arbuscular Mycorrhizal Symbiosis Varied with Different Levels of Cadmium Stress. J. Hazard. Mater. 2023, 459, 132076. [Google Scholar] [CrossRef]
- Gil-Cardeza, M.L.; Ferri, A.; Cornejo, P.; Gomez, E. Distribution of Chromium Species in a Cr-Polluted Soil: Presence of Cr(III) in Glomalin Related Protein Fraction. Sci. Total Environ. 2014, 493, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.W.; Wu, L.; Luo, N.; Mo, C.H.; Wong, M.H.; Li, H. Arbuscular Mycorrhizal Fungi and the Associated Bacterial Community Influence the Uptake of Cadmium in Rice. Geoderma 2019, 337, 749–757. [Google Scholar] [CrossRef]
- Cui, G.; Ai, S.; Chen, K.; Wang, X. Arbuscular Mycorrhiza Augments Cadmium Tolerance in Soybean by Altering Accumulation and Partitioning of Nutrient Elements, and Related Gene Expression. Ecotoxicol. Environ. Saf. 2019, 171, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Becerril, F.; van Tuinen, D.; Martin-Laurent, F.; Metwally, A.; Dietz, K.-J.; Gianinazzi, S.; Gianinazzi-Pearson, V. Molecular Changes in Pisum sativum L. Roots during Arbuscular Mycorrhiza Buffering of Cadmium Stress. Mycorrhiza 2005, 16, 51–60. [Google Scholar] [CrossRef]
- Jia, H.; Wang, X.; Wei, T.; Zhou, R.; Muhammad, H.; Hua, L.; Ren, X.; Guo, J.; Ding, Y. Accumulation and Fixation of Cd by Tomato Cell Wall Pectin under Cd Stress. Environ. Exp. Bot. 2019, 167, 103829. [Google Scholar] [CrossRef]
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.-P.; Lutts, S. Tomato Fruit Development and Metabolism. Front. Plant Sci. 2019, 10, 1554. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, J.; Zhao, L.; Zhou, R.; Yu, W.; Zhao, T. Application of Whole Genome Resequencing in Mapping of a Tomato Yellow Leaf Curl Virus Resistance Gene. Sci. Rep. 2018, 8, 9592. [Google Scholar] [CrossRef]
- Shreevastav, C.; Subedi, S.; Gajurel, S.; Basnet, P. A review on nutrient deficiency symptoms and effects on tomato plant. Food Agri Econ. Rev. 2022, 2, 34–36. [Google Scholar] [CrossRef]
- Lafuente, M.T.; Sampedro, R.; Vélez, D.; Romero, P. Deficient Copper Availability on Organoleptic and Nutritional Quality of Tomato Fruit. Plant Sci. 2023, 326, 111537. [Google Scholar] [CrossRef]
- Fei, C.; Zhang, S.; Sun, Z.; Ding, X. Assessment of Magnesium Deficiency in Greenhouse Tomato Crops Grown on Calcareous Soil. Soil Use Manag. 2024, 40, e12939. [Google Scholar] [CrossRef]
- Hashimoto, R.; Ueta, R.; Abe, C.; Osakabe, Y.; Osakabe, K. Efficient Multiplex Genome Editing Induces Precise, and Self-Ligated Type Mutations in Tomato Plants. Front. Plant Sci. 2018, 9, 916. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Hu, J.; Sun, S.; Xu, G. Conservation and Divergence of Both Phosphate- and Mycorrhiza-Regulated Physiological Responses and Expression Patterns of Phosphate Transporters in Solanaceous Species. New Phytol. 2007, 173, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Sun, C.; Liang, H.; Wang, Y.; Bian, X.; Dong, C.; Niu, X.; Yang, M.; Xu, G.; Chen, A.; et al. SlSPX1-SlPHR Complexes Mediate the Suppression of Arbuscular Mycorrhizal Symbiosis by Phosphate Repletion in Tomato. Plant Cell 2022, 34, 4045–4065. [Google Scholar] [CrossRef] [PubMed]
- Bereczky, Z.; Wang, H.-Y.; Schubert, V.; Ganal, M.; Bauer, P. Differential Regulation of Nramp and Irt Metal Transporter Genes in Wild Type and Iron Uptake Mutants of Tomato. J. Biol. Chem. 2003, 278, 24697–24704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, G.; Cao, N.; Yang, H.; Zhu, F. Genome-Wide Identification and Expression Analysis of NRAMP Transporter Genes in Cucumis sativus and Citrullus lanatus. Can. J. Plant Sci. 2021, 101, 377–392. [Google Scholar] [CrossRef]
- Ishida, J.K.; Caldas, D.G.G.; Oliveira, L.R.; Frederici, G.C.; Leite, L.M.P.; Mui, T.S. Genome-Wide Characterization of the NRAMP Gene Family in Phaseolus vulgaris Provides Insights into Functional Implications during Common Bean Development. Genet. Mol. Biol. 2018, 41, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Duan, Y.; Han, Z.; Shang, X.; Zhang, K.; Zou, Z.; Ma, Y.; Li, F.; Fang, W.; Zhu, X. Genome-Wide Identification and Expression Analysis of the NRAMP Family Genes in Tea Plant (Camellia sinensis). Plants 2021, 10, 1055. [Google Scholar] [CrossRef]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.M.; Sanders, D.; et al. Phylogenetic Relationships within Cation Transporter Families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef]
- Tang, L.; Mao, B.; Li, Y.; Lv, Q.; Zhang, L.; Chen, C.; He, H.; Wang, W.; Zeng, X.; Shao, Y.; et al. Knockout of OsNramp5 Using the CRISPR/Cas9 System Produces Low Cd-Accumulating Indica Rice without Compromising Yield. Sci. Rep. 2017, 7, 14438. [Google Scholar] [CrossRef]
- Xiaohua, H.; Mo, Y.; Ji, W.; Yang, X.; Xie, Z.; Huang, D.; Li, D.; Tian, L. The OsNramp4 Aluminum Transporter Is Involved in Cadmium Accumulation in Rice Grains. Reprod. Breed. 2022, 2, 125–132. [Google Scholar] [CrossRef]
- Cailliatte, R.; Lapeyre, B.; Briat, J.-F.; Mari, S.; Curie, C. The NRAMP6 Metal Transporter Contributes to Cadmium Toxicity. Biochem. J. 2009, 422, 217–228. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Zheng, L.; Wang, Y.; Chen, X.; Zhang, W. A Functional Study Identifying Critical Residues Involving Metal Transport Activity and Selectivity in Natural Resistance-Associated Macrophage Protein 3 in Arabidopsis Thaliana. Int. J. Mol. Sci. 2018, 19, 1430. [Google Scholar] [CrossRef]
- Guo, B.; Liang, Y.; Fu, Q.; Ding, N.; Liu, C.; Lin, Y.; Li, H.; Li, N. Cadmium Stabilization with Nursery Stocks through Transplantation: A New Approach to Phytoremediation. J. Hazard. Mater. 2012, 199–200, 233–239. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The Origins of Genomic Duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The Evolutionary Dynamics of Plant Duplicate Genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Wang, Y.; Eide, D.J.; Dunwell, J.M. Evolution, and Functional Analysis of Natural Resistance-Associated Macrophage Proteins (NRAMPs) from Theobroma cacao and Their Role in Cadmium Accumulation. Sci. Rep. 2018, 8, 14412. [Google Scholar] [CrossRef]
- Genome-Wide Identification and Expression Analysis of Metal Tolerance Protein (MTP) Gene Family in Soybean (Glycine Max) under Heavy Metal Stress | Molecular Biology Reports. Available online: https://link.springer.com/article/10.1007/s11033-022-08100-x (accessed on 25 December 2023).
- Chen, Y.; Zhao, X.; Li, G.; Kumar, S.; Sun, Z.; Li, Y.; Guo, W.; Yang, J.; Hou, H. Genome-Wide Identification of the Nramp Gene Family in Spirodela Polyrhiza and Expression Analysis under Cadmium Stress. Int. J. Mol. Sci. 2021, 22, 6414. [Google Scholar] [CrossRef] [PubMed]
- Ehrnstorfer, I.A.; Geertsma, E.R.; Pardon, E.; Steyaert, J.; Dutzler, R. Crystal Structure of a SLC11 (NRAMP) Transporter Reveals the Basis for Transition-Metal Ion Transport. Nat. Struct. Mol. Biol. 2014, 21, 990–996. [Google Scholar] [CrossRef]
- Manatschal, C.; Pujol-Giménez, J.; Poirier, M.; Reymond, J.-L.; Hediger, M.A.; Dutzler, R. Mechanistic Basis of the Inhibition of SLC11/NRAMP-Mediated Metal Ion Transport by Bis-Isothiourea Substituted Compounds. eLife 2019, 8, e51913. [Google Scholar] [CrossRef]
- Ehrnstorfer, I.A.; Manatschal, C.; Arnold, F.M.; Laederach, J.; Dutzler, R. Structural and Mechanistic Basis of Proton-Coupled Metal Ion Transport in the SLC11/NRAMP Family. Nat. Commun. 2017, 8, 14033. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.E.; Pittman, J.K.; Hall, J.L. Emerging Mechanisms for Heavy Metal Transport in Plants. Biochim. Biophys. Acta 2000, 1465, 104–126. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 Regulates Flavonoids Accumulation and Abiotic Stress Tolerance in Transgenic Arabidopsis Thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.-M.; Sánchez-Parra, B.; Ortiz-García, P.; Santamaría, M.E.; Díaz, I.; Pollmann, S. Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 9768. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yang, Y. Differential Expression of Rice Nramp Genes in Response to Pathogen Infection, Defense Signal Molecules and Metal Ions. Physiol. Mol. Plant Pathol. 2004, 65, 235–243. [Google Scholar] [CrossRef]
- Thomine, S.; Lelièvre, F.; Debarbieux, E.; Schroeder, J.I.; Barbier-Brygoo, H. AtNRAMP3, a Multispecific Vacuolar Metal Transporter Involved in Plant Responses to Iron Deficiency. Plant J. 2003, 34, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Wang, H.-Q.; Chen, J.; Chang, J.-D.; Zhao, F.-J. Molecular Mechanisms Underlying the Toxicity and Detoxification of Trace Metals and Metalloids in Plants. J. Integr. Plant Biol. 2023, 65, 570–593. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ding, L.; Zhou, M.; Chen, Z.; Ding, Y.; Zhu, C. Transcriptional Regulatory Network of Plant Cadmium Stress Response. Int. J. Mol. Sci. 2023, 24, 4378. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, C.; Zhu, J.; Zeng, J.; Kang, H.; Fan, X.; Sha, L.; Zhang, H.; Zhou, Y.; Wang, Y. Expression of TpNRAMP5, a Metal Transporter from Polish Wheat (Triticum Polonicum L.), Enhances the Accumulation of Cd, Co and Mn in Transgenic Arabidopsis Plants. Planta 2018, 247, 1395–1406. [Google Scholar] [CrossRef]
- Chang, J.-D.; Huang, S.; Yamaji, N.; Zhang, W.; Ma, J.F.; Zhao, F.-J. OsNRAMP1 Transporter Contributes to Cadmium and Manganese Uptake in Rice. Plant Cell Environ. 2020, 43, 2476–2491. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, X.W.; Wu, L.; Luo, N.; Huang, W.X.; Mo, C.H.; Wong, M.H. Effects of Arbuscular Mycorrhizal Fungi on Redox Homeostasis of Rice under Cd Stress. Plant Soil 2020, 455, 121–138. [Google Scholar] [CrossRef]
- Pan, J.; Cao, S.; Xu, G.; Rehman, M.; Li, X.; Luo, D.; Wang, C.; Fang, W.; Xiao, H.; Liao, C.; et al. Comprehensive Analysis Reveals the Underlying Mechanism of Arbuscular Mycorrhizal Fungi in Kenaf Cadmium Stress Alleviation. Chemosphere 2023, 314, 137566. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zheng, K.; Wang, Y.; Zhang, Y.; Lao, R.; Qin, Z.; Li, T.; Zhao, Z. Harnessing an Arbuscular Mycorrhizal Fungus to Improve the Adaptability of a Facultative Metallophytic Poplar (Populus yunnanensis) to Cadmium Stress: Physiological and Molecular Responses. J. Hazard. Mater. 2022, 424, 127430. [Google Scholar] [CrossRef]
- Liu, H.; Yuan, M.; Tan, S.; Yang, X.; Lan, Z.; Jiang, Q.; Ye, Z.; Jing, Y. Enhancement of Arbuscular Mycorrhizal Fungus (Glomus versiforme) on the Growth and Cd Uptake by Cd-Hyperaccumulator Solanum nigrum. Appl. Soil. Ecol. 2015, 89, 44–49. [Google Scholar] [CrossRef]
- Liu, M.; Sun, J.; Li, Y.; Xiao, Y. Nitrogen Fertilizer Enhances Growth and Nutrient Uptake of Medicago sativa Inoculated with Glomus tortuosum Grown in Cd-Contaminated Acidic Soil. Chemosphere 2017, 167, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Ouziad, F.; Hildebrandt, U.; Schmelzer, E.; Bothe, H. Differential Gene Expressions in Arbuscular Mycorrhizal-Colonized Tomato Grown under Heavy Metal Stress. J. Plant Physiol. 2005, 162, 634–649. [Google Scholar] [CrossRef] [PubMed]
- Kabir, A.H.; Debnath, T.; Das, U.; Prity, S.A.; Haque, A.; Rahman, M.M.; Parvez, M.S. Arbuscular Mycorrhizal Fungi Alleviate Fe-Deficiency Symptoms in Sunflower by Increasing Iron Uptake and Its Availability along with Antioxidant Defense. Plant Physiol. Biochem. 2020, 150, 254–262. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Tyerman, S.D.; Cavagnaro, T.R. The Dual Benefit of Arbuscular Mycorrhizal Fungi under Soil Zinc Deficiency and Toxicity: Linking Plant Physiology and Gene Expression. Plant Soil. 2017, 420, 375–388. [Google Scholar] [CrossRef]
- Cardini, A.; Pellegrino, E.; Declerck, S.; Calonne-Salmon, M.; Mazzolai, B.; Ercoli, L. Direct Transfer of Zinc between Plants Is Channelled by Common Mycorrhizal Network of Arbuscular Mycorrhizal Fungi and Evidenced by Changes in Expression of Zinc Transporter Genes in Fungus and Plant. Environ. Microbiol. 2021, 23, 5883–5900. [Google Scholar] [CrossRef]
- Liu, J.; Liu, J.; Chen, A.; Ji, M.; Chen, J.; Yang, X.; Gu, M.; Qu, H.; Xu, G. Analysis of Tomato Plasma Membrane H+-ATPase Gene Family Suggests a Mycorrhiza-Mediated Regulatory Mechanism Conserved in Diverse Plant Species. Mycorrhiza 2016, 26, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Tauriello, G.; Bienert, S.; Waterhouse, A.M.; Bertoni, M.; Bordoli, L.; Schwede, T.; Lepore, R. Modeling of Protein Tertiary and Quaternary Structures Based on Evolutionary Information. Methods Mol. Biol. 2019, 1851, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling Protein Quaternary Structure of Homo- and Hetero-Oligomers beyond Binary Interactions by Homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]


| Gene Name | SlNRAMP1 | SlNRAMP2 | SlNRAMP3 | SlNRAMP4 | SlNRAMP5 |
|---|---|---|---|---|---|
| Gene ID | Solyc11g018530.2.1 | Solyc04g078250.3.1 | Solyc02g092800.3.1 | Solyc03g116900.3.1 | Solyc09g007870.3.1 |
| Genomic location | 8,645,242–8,648,856 | 63,109,871–63,113,592 | 54,402,272–54,405,963 | 67,650,010–67,659,122 | 1,399,607–1,407,814 |
| Gene length (bp) | 1853 | 2128 | 2210 | 1984 | 4841 |
| CDS (bp) | 1593 | 1560 | 1347 | 1602 | 3951 |
| AA | 530 | 519 | 509 | 522 | 1316 |
| MW (kDa) | 57.78 | 57.15 | 56.15 | 56.71 | 142.67 |
| PI | 7.15 | 5.23 | 5.61 | 8.85 | 6.03 |
| Instability | 31.99 | 48.13 | 29.73 | 35.21 | 40.57 |
| Aliphatic index | 124.55 | 111.43 | 119.35 | 121.26 | 88.76 |
| Gravy | 0.6 | 0.357 | 0.581 | 0.509 | −0.013 |
| TN | 12 | 9 | 11 | 12 | 11 |
| SL | PM | V | V | V, PM | PM |
| Treatments | Cd Concentration (mg/g) | P Concentration (mg/g) | ||
|---|---|---|---|---|
| Root | Shoot | Root | Shoot | |
| 0 NM | 0.004 ± 0 c | 0.001 ± 0 c | 1.446 ± 0.068 bc | 0.755 ± 0.006 b |
| 0 AM | 0.003 ± 0.001 c | 0.001 ± 0 c | 2.288 ± 0.074 a | 1.129 ± 0.075 a |
| 100 NM | 3.473 ± 0.530 a | 0.118 ± 0.013 a | 1.199 ± 0.046 c | 0.782 ± 0.015 b |
| 100 AM | 2.073 ± 0.236 b | 0.073 ± 0.007 b | 1.544 ± 0.121 b | 0.977 ± 0.061 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Bao, X.; Qiu, G.; Li, H.; Wang, Y.; Chen, X.; Fu, Q.; Guo, B. Genome-Wide Identification and Expression Analysis of SlNRAMP Genes in Tomato under Nutrient Deficiency and Cadmium Stress during Arbuscular Mycorrhizal Symbiosis. Int. J. Mol. Sci. 2024, 25, 8269. https://doi.org/10.3390/ijms25158269
Liu J, Bao X, Qiu G, Li H, Wang Y, Chen X, Fu Q, Guo B. Genome-Wide Identification and Expression Analysis of SlNRAMP Genes in Tomato under Nutrient Deficiency and Cadmium Stress during Arbuscular Mycorrhizal Symbiosis. International Journal of Molecular Sciences. 2024; 25(15):8269. https://doi.org/10.3390/ijms25158269
Chicago/Turabian StyleLiu, Junli, Xiaoqi Bao, Gaoyang Qiu, Hua Li, Yuan Wang, Xiaodong Chen, Qinglin Fu, and Bin Guo. 2024. "Genome-Wide Identification and Expression Analysis of SlNRAMP Genes in Tomato under Nutrient Deficiency and Cadmium Stress during Arbuscular Mycorrhizal Symbiosis" International Journal of Molecular Sciences 25, no. 15: 8269. https://doi.org/10.3390/ijms25158269





