Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective
Abstract
:1. Introduction
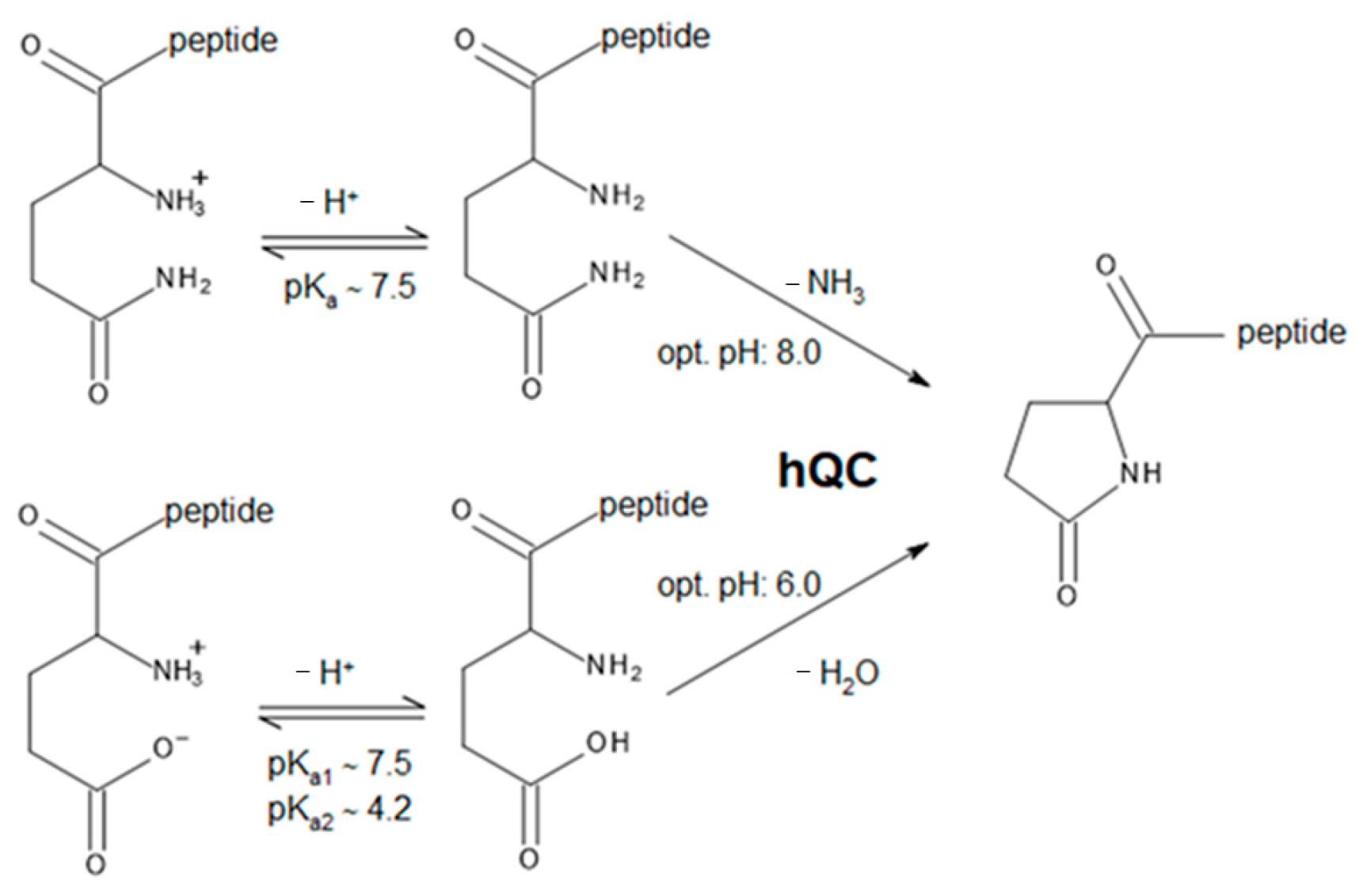
2. Results and Discussion
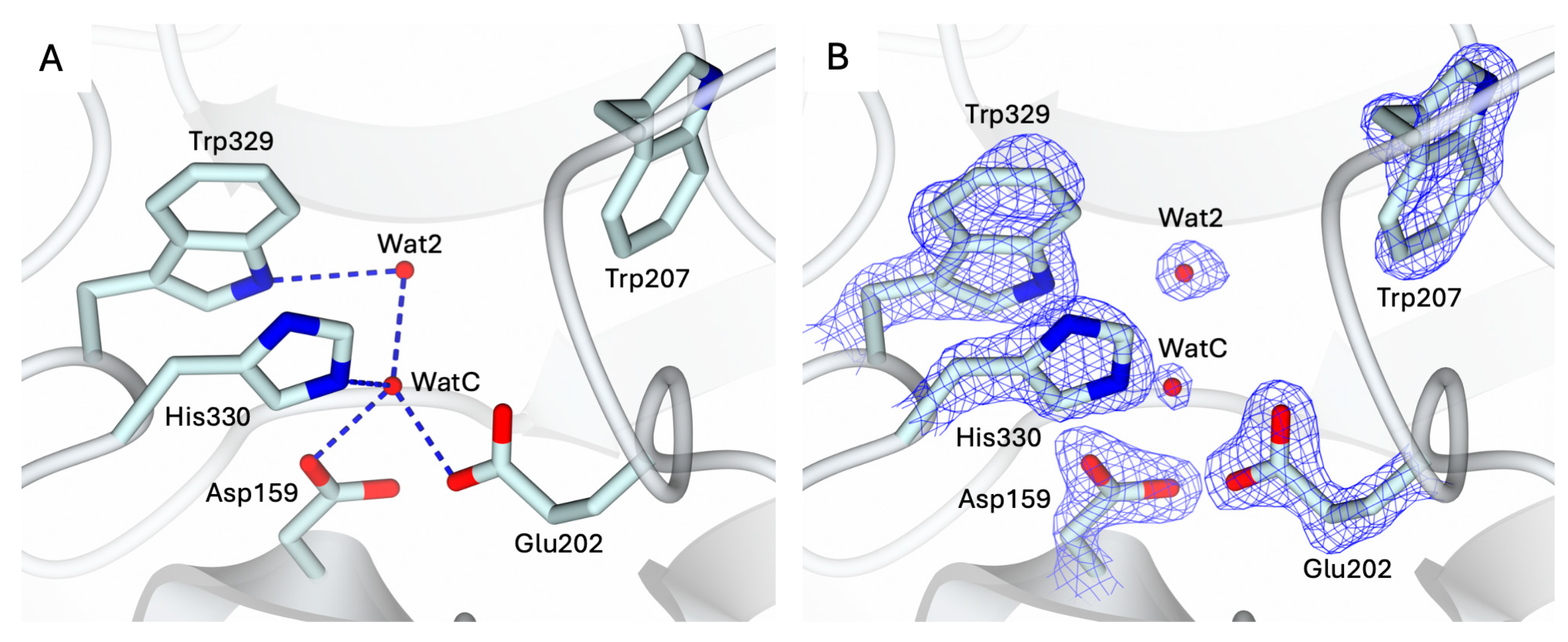
2.1. Crystal Structure of the Apo-State of hQC-2X
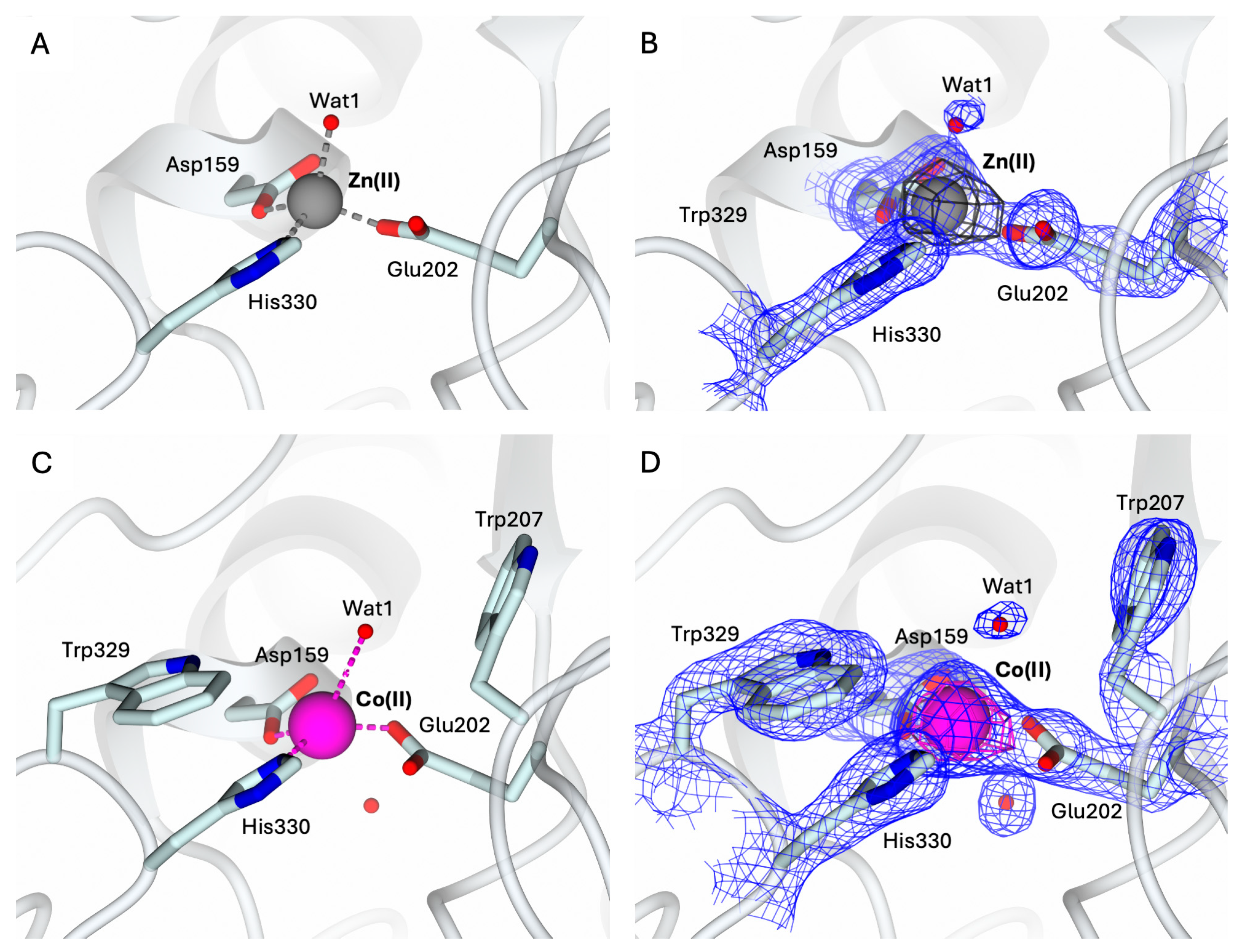
2.2. Structural Characterization of the Holo-States of hQC-2X with Zn(II) and Co(II) Metal Ions
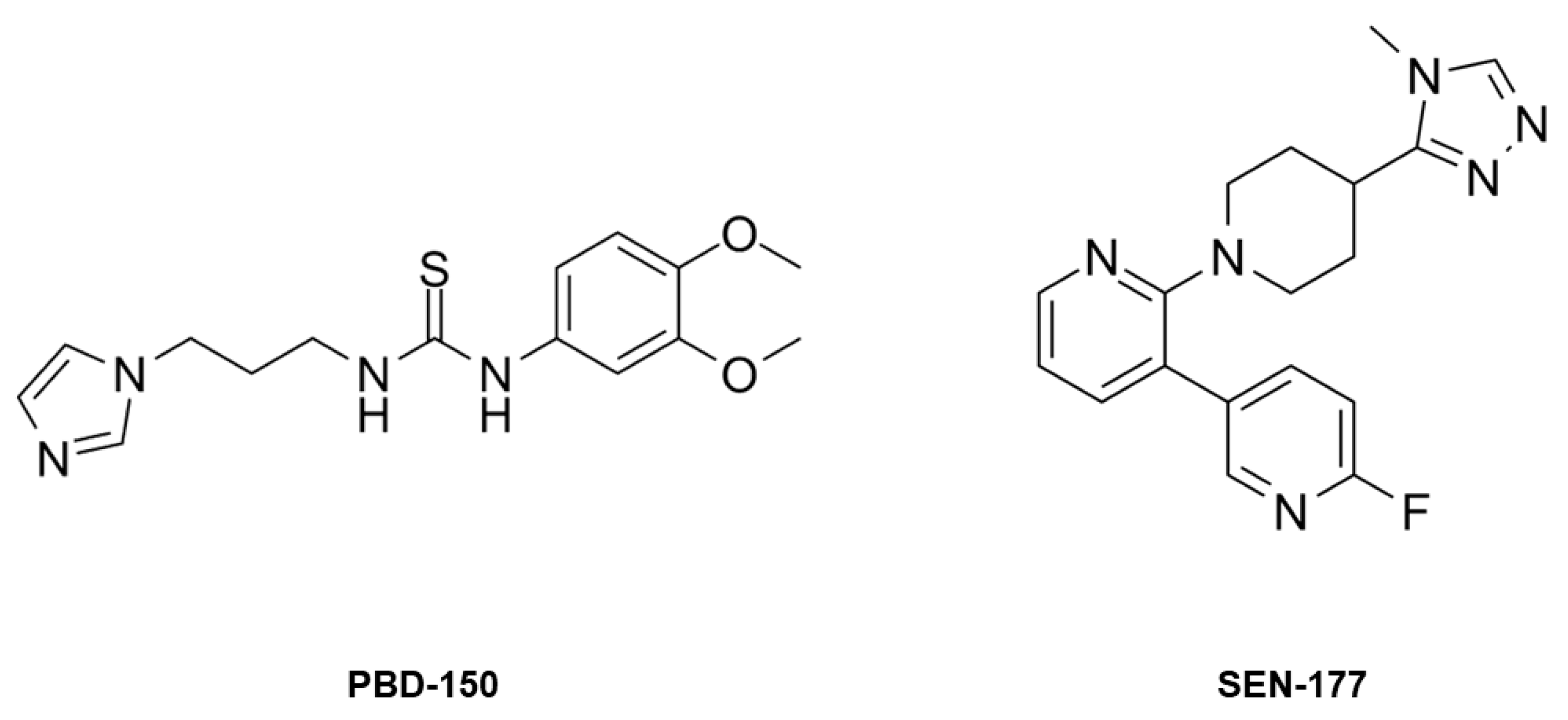
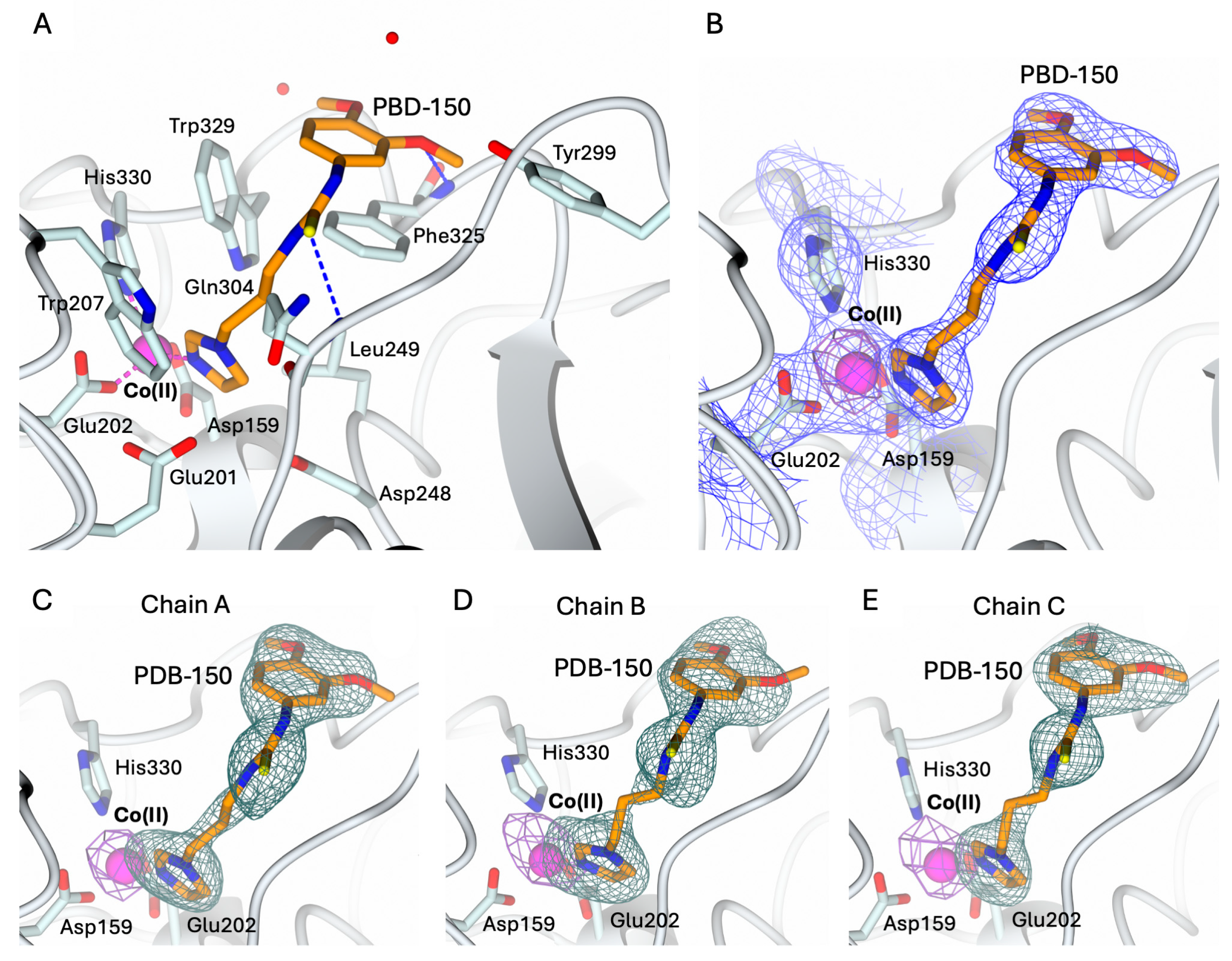
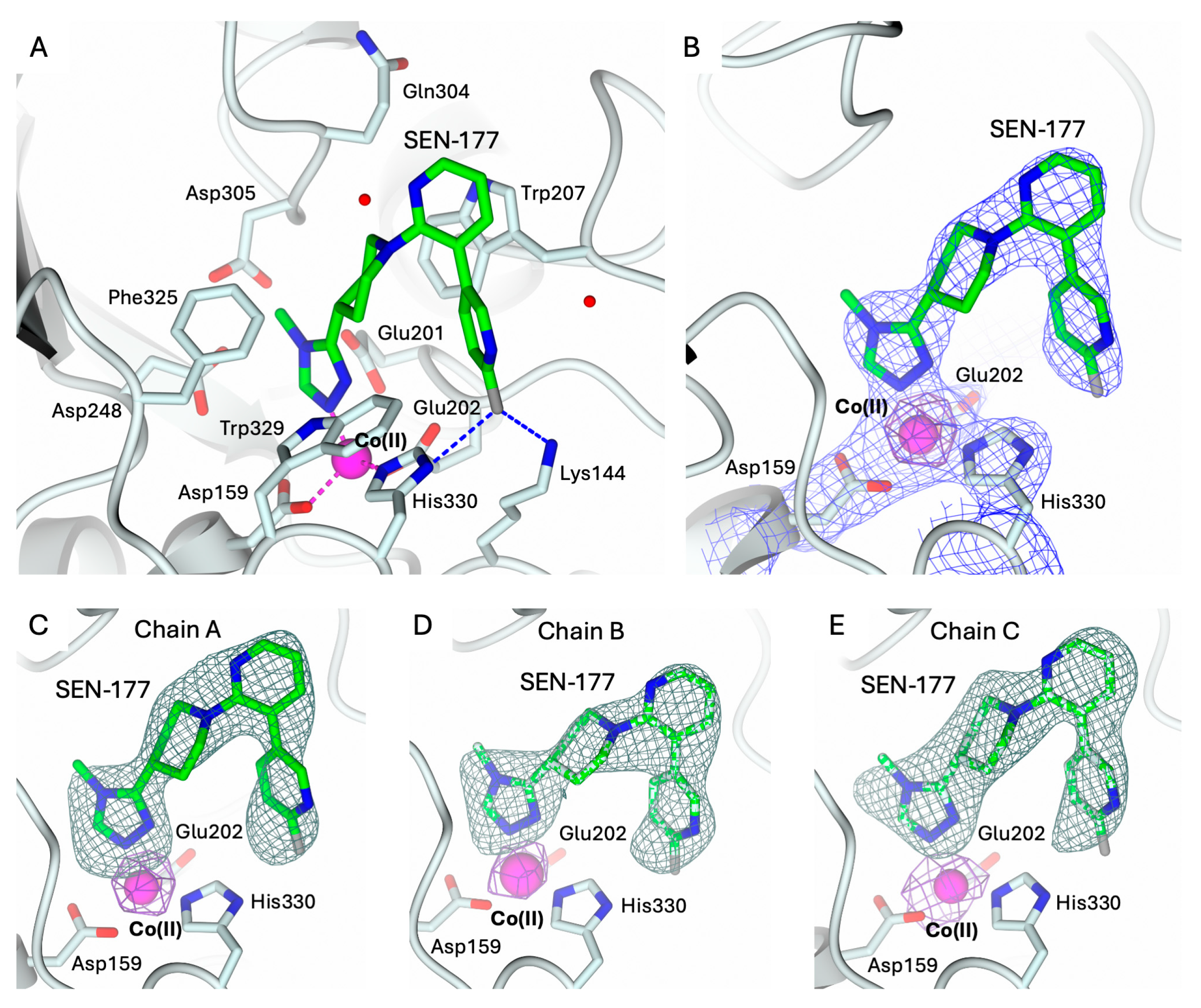
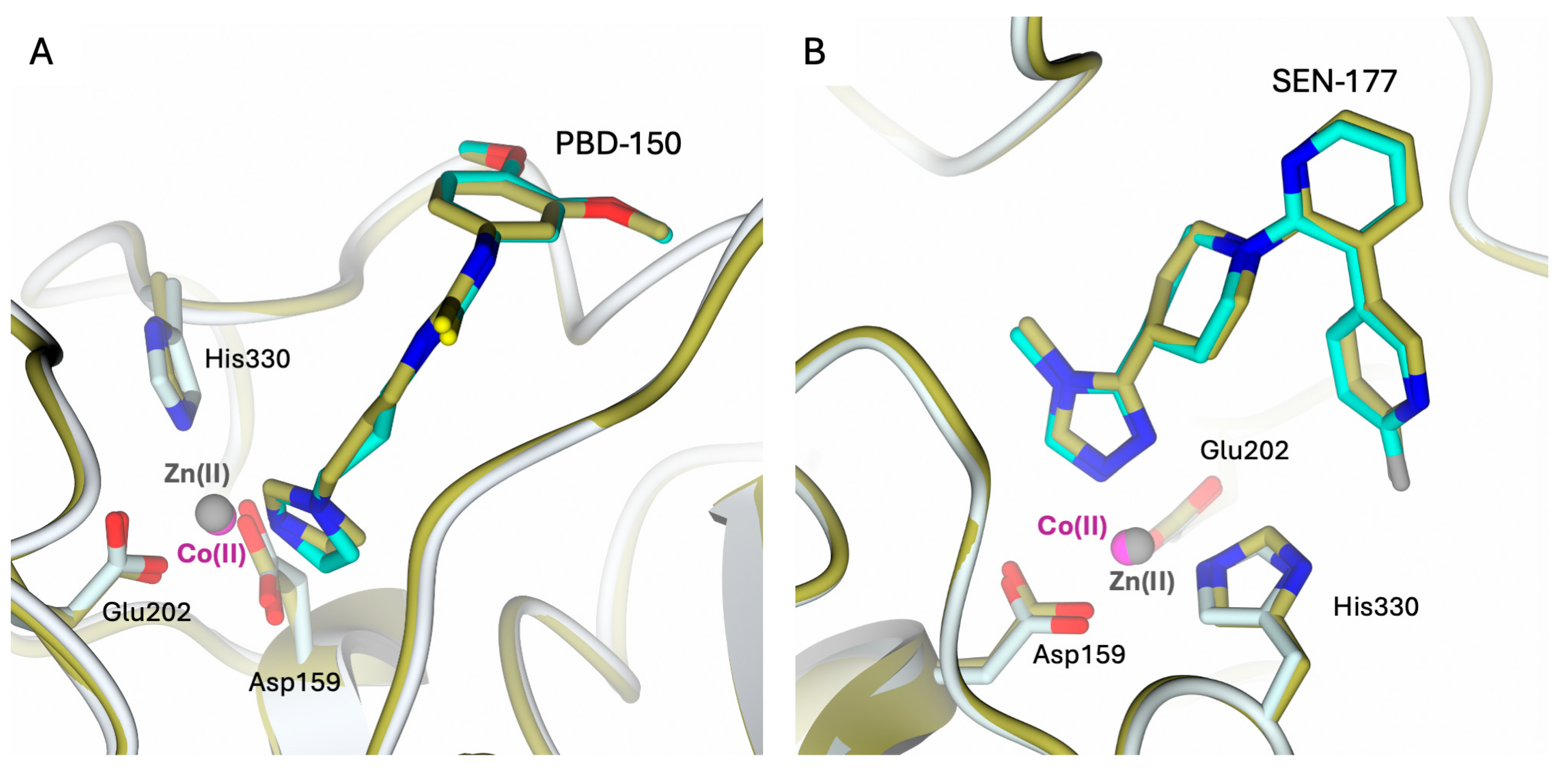
2.3. Structural Characterization of hQC-2X–Co(II) in Complex with PBD-150 and SEN-177
2.4. Comparison of hQC with Bacterial di-zinc Aminopeptidases
3. Conclusions
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Preparation of Apo-hQC-2X and Co(II)-hQC-2X
4.3. Crystallization
4.4. Data Collection, Structure Solution, and Refinement
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schilling, S.; Cynis, H.; von Bohlen, A.; Hoffmann, T.; Wermann, M.; Heiser, U.; Buchholz, M.; Zunkel, K.; Demuth, H.-U. Isolation, Catalytic Properties, and Competitive Inhibitors of the Zinc-Dependent Murine Glutaminyl Cyclase. Biochemistry 2005, 44, 13415–13424. [Google Scholar] [CrossRef]
- Seifert, F.; Schulz, K.; Koch, B.; Manhart, S.; Demuth, H.-U.; Schilling, S. Glutaminyl Cyclases Display Significant Catalytic Proficiency for Glutamyl Substrates. Biochemistry 2009, 48, 11831–11833. [Google Scholar] [CrossRef]
- Coimbra, J.R.M.; Moreira, P.I.; Santos, A.E.; Salvador, J.A.R. Therapeutic Potential of Glutaminyl Cyclases: Current Status and Emerging Trends. Drug Discov. Today 2023, 28, 103644. [Google Scholar] [CrossRef]
- Shih, Y.-P.; Chou, C.-C.; Chen, Y.-L.; Huang, K.-F.; Wang, A.H.-J. Linked Production of Pyroglutamate-Modified Proteins via Self-Cleavage of Fusion Tags with TEV Protease and Autonomous N-Terminal Cyclization with Glutaminyl Cyclase In Vivo. PLoS ONE 2014, 9, e94812. [Google Scholar] [CrossRef]
- Schilling, S.; Hoffmann, T.; Manhart, S.; Hoffmann, M.; Demuth, H.-U. Glutaminyl Cyclases Unfold Glutamyl Cyclase Activity under Mild Acid Conditions. FEBS Lett. 2004, 563, 191–196. [Google Scholar] [CrossRef]
- Messer, M. Enzymatic Cyclization of L-Glutamine and L-Glutaminyl Peptides. Nature 1963, 197, 1299. [Google Scholar] [CrossRef]
- Messer, M.; Ottesen, M. Isolation and Properties of Glutamine Cyclotransferase of Dried Papaya Latex. Biochim. Biophys. Acta BBA-Spec. Sect. Enzymol. Subj. 1964, 92, 409–411. [Google Scholar] [CrossRef]
- Schilling, S.; Wasternack, C.; Demuth, H.-U. Glutaminyl Cyclases from Animals and Plants: A Case of Functionally Convergent Protein Evolution. Biol. Chem. 2008, 389, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Taudte, N.; Linnert, M.; Rahfeld, J.-U.; Piechotta, A.; Ramsbeck, D.; Buchholz, M.; Kolenko, P.; Parthier, C.; Houston, J.A.; Veillard, F.; et al. Mammalian-like Type II Glutaminyl Cyclases in Porphyromonas gingivalis and Other Oral Pathogenic Bacteria as Targets for Treatment of Periodontitis. J. Biol. Chem. 2021, 296, 100263. [Google Scholar] [CrossRef]
- Cynis, H.; Rahfeld, J.-U.; Stephan, A.; Kehlen, A.; Koch, B.; Wermann, M.; Demuth, H.-U.; Schilling, S. Isolation of an Isoenzyme of Human Glutaminyl Cyclase: Retention in the Golgi Complex Suggests Involvement in the Protein Maturation Machinery. J. Mol. Biol. 2008, 379, 966–980. [Google Scholar] [CrossRef]
- Stephan, A.; Wermann, M.; von Bohlen, A.; Koch, B.; Cynis, H.; Demuth, H.-U.; Schilling, S. Mammalian Glutaminyl Cyclases and Their Isoenzymes Have Identical Enzymatic Characteristics. FEBS J. 2009, 276, 6522–6536. [Google Scholar] [CrossRef]
- Busby, W.H.; Quackenbush, G.E.; Humm, J.; Youngblood, W.W.; Kizer, J.S. An Enzyme(s) That Converts Glutaminyl-Peptides into Pyroglutamyl-Peptides. Presence in Pituitary, Brain, Adrenal Medulla, and Lymphocytes. J. Biol. Chem. 1987, 262, 8532–8536. [Google Scholar] [CrossRef]
- Hoffmann, T.; Meyer, A.; Heiser, U.; Kurat, S.; Böhme, L.; Kleinschmidt, M.; Bühring, K.-U.; Hutter-Paier, B.; Farcher, M.; Demuth, H.-U.; et al. Glutaminyl Cyclase Inhibitor PQ912 Improves Cognition in Mouse Models of Alzheimer’s Disease-Studies on Relation to Effective Target Occupancy. J. Pharmacol. Exp. Ther. 2017, 362, 119–130. [Google Scholar] [CrossRef]
- Schilling, S.; Zeitschel, U.; Hoffmann, T.; Heiser, U.; Francke, M.; Kehlen, A.; Holzer, M.; Hutter-Paier, B.; Prokesch, M.; Windisch, M.; et al. Glutaminyl Cyclase Inhibition Attenuates Pyroglutamate Aβ and Alzheimer’s Disease–like Pathology. Nat. Med. 2008, 14, 1106–1111. [Google Scholar] [CrossRef]
- Jawhar, S.; Wirths, O.; Schilling, S.; Graubner, S.; Demuth, H.-U.; Bayer, T.A. Overexpression of Glutaminyl Cyclase, the Enzyme Responsible for Pyroglutamate Aβ Formation, Induces Behavioral Deficits, and Glutaminyl Cyclase Knock-out Rescues the Behavioral Phenotype in 5XFAD Mice. J. Biol. Chem. 2011, 286, 4454–4460. [Google Scholar] [CrossRef]
- Ross, C.A.; Wood, J.D.; Schilling, G.; Peters, M.F.; Nucifora, F.C.; Cooper, J.K.; Sharp, A.H.; Margolis, R.L.; Borchelt, D.R. Polyglutamine Pathogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 1005–1011. [Google Scholar] [CrossRef]
- Murphy, R.M. Peptide Aggregation in Neurodegenerative Disease. Annu. Rev. Biomed. Eng. 2002, 4, 155–174. [Google Scholar] [CrossRef]
- Jimenez-Sanchez, M.; Lam, W.; Hannus, M.; Sönnichsen, B.; Imarisio, S.; Fleming, A.; Tarditi, A.; Menzies, F.; Dami, T.E.; Xu, C.; et al. siRNA Screen Identifies QPCT as a Druggable Target for Huntington’s Disease. Nat. Chem. Biol. 2015, 11, 347–354. [Google Scholar] [CrossRef]
- Xiao, G.; Fan, Q.; Wang, X.; Zhou, B. Huntington Disease Arises from a Combinatory Toxicity of Polyglutamine and Copper Binding. Proc. Natl. Acad. Sci USA 2013, 110, 14995–15000. [Google Scholar] [CrossRef]
- Hartlage-Rübsamen, M.; Morawski, M.; Waniek, A.; Jäger, C.; Zeitschel, U.; Koch, B.; Cynis, H.; Schilling, S.; Schliebs, R.; Demuth, H.-U.; et al. Glutaminyl Cyclase Contributes to the Formation of Focal and Diffuse Pyroglutamate (pGlu)-Aβ Deposits in Hippocampus via Distinct Cellular Mechanisms. Acta Neuropathol. 2011, 121, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Wirths, O.; Hillmann, A.; Pradier, L.; Härtig, W.; Bayer, T.A. Oligomeric Pyroglutamate Amyloid-β Is Present in Microglia and a Subfraction of Vessels in Patients with Alzheimer’s Disease: Implications for Immunotherapy. J. Alzheimer’s Dis. 2013, 35, 741–749. [Google Scholar] [CrossRef]
- Wu, G.; Miller, R.A.; Connolly, B.; Marcus, J.; Renger, J.; Savage, M.J. Pyroglutamate-Modified Amyloid-β Protein Demonstrates Similar Properties in an Alzheimer’s Disease Familial Mutant Knock-in Mouse and Alzheimer’s Disease Brain. Neurodegener. Dis. 2014, 14, 53–66. [Google Scholar] [CrossRef]
- Harigaya, Y.; Saido, T.C.; Eckman, C.B.; Prada, C.M.; Shoji, M.; Younkin, S.G. Amyloid Beta Protein Starting Pyroglutamate at Position 3 Is a Major Component of the Amyloid Deposits in the Alzheimer’s Disease Brain. Biochem. Biophys. Res. Commun. 2000, 276, 422–427. [Google Scholar] [CrossRef]
- Schilling, S.; Lauber, T.; Schaupp, M.; Manhart, S.; Scheel, E.; Böhm, G.; Demuth, H.-U. On the Seeding and Oligomerization of pGlu-Amyloid Peptides (In Vitro). Biochemistry 2006, 45, 12393–12399. [Google Scholar] [CrossRef]
- Nussbaum, J.M.; Schilling, S.; Cynis, H.; Silva, A.; Swanson, E.; Wangsanut, T.; Tayler, K.; Wiltgen, B.; Hatami, A.; Rönicke, R.; et al. Prion-like Behaviour and Tau-Dependent Cytotoxicity of Pyroglutamylated Amyloid-β. Nature 2012, 485, 651–655. [Google Scholar] [CrossRef]
- Becker, A.; Kohlmann, S.; Alexandru, A.; Jagla, W.; Canneva, F.; Bäuscher, C.; Cynis, H.; Sedlmeier, R.; Graubner, S.; Schilling, S.; et al. Glutaminyl Cyclase-Mediated Toxicity of Pyroglutamate-Beta Amyloid Induces Striatal Neurodegeneration. BMC Neurosci. 2013, 14, 108. [Google Scholar] [CrossRef]
- Gunn, A.P.; Masters, C.L.; Cherny, R.A. Pyroglutamate-Aβ: Role in the Natural History of Alzheimer’s Disease. Int. J. Biochem. Cell Biol. 2010, 42, 1915–1918. [Google Scholar] [CrossRef]
- Soto, C.; Estrada, L.D. Protein Misfolding and Neurodegeneration. Arch. Neurol. 2008, 65, 184–189. [Google Scholar] [CrossRef]
- Buchholz, M.; Hamann, A.; Aust, S.; Brandt, W.; Böhme, L.; Hoffmann, T.; Schilling, S.; Demuth, H.-U.; Heiser, U. Inhibitors for Human Glutaminyl Cyclase by Structure Based Design and Bioisosteric Replacement. J. Med. Chem. 2009, 52, 7069–7080. [Google Scholar] [CrossRef]
- Li, M.; Dong, Y.; Yu, X.; Zou, Y.; Zheng, Y.; Bu, X.; Quan, J.; He, Z.; Wu, H. Inhibitory Effect of Flavonoids on Human Glutaminyl Cyclase. Bioorg. Med. Chem. 2016, 24, 2280–2286. [Google Scholar] [CrossRef]
- Mason, S.L.; Barker, R.A. Novel Targets for Huntington’s Disease: Future Prospects. Degener. Neurol. Neuromuscul. Dis. 2016, 6, 25–36. [Google Scholar] [CrossRef]
- Pozzi, C.; Di Pisa, F.; Benvenuti, M.; Mangani, S. The Structure of the Human Glutaminyl Cyclase–SEN177 Complex Indicates Routes for Developing New Potent Inhibitors as Possible Agents for the Treatment of Neurological Disorders. J. Biol. Inorg Chem. 2018, 23, 1219–1226. [Google Scholar] [CrossRef]
- Ruiz-Carrillo, D.; Koch, B.; Parthier, C.; Wermann, M.; Dambe, T.; Buchholz, M.; Ludwig, H.-H.; Heiser, U.; Rahfeld, J.-U.; Stubbs, M.T.; et al. Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center. Biochemistry 2011, 50, 6280–6288. [Google Scholar] [CrossRef]
- Schilling, S.; Niestroj, A.J.; Rahfeld, J.-U.; Hoffmann, T.; Wermann, M.; Zunkel, K.; Wasternack, C.; Demuth, H.-U. Identification of Human Glutaminyl Cyclase as a Metalloenzyme. Potent Inhibition by Imidazole Derivatives and Heterocyclic Chelators. J. Biol. Chem. 2003, 278, 49773–49779. [Google Scholar] [CrossRef]
- Huang, K.-F.; Liu, Y.-L.; Cheng, W.-J.; Ko, T.-P.; Wang, A.H.-J. Crystal Structures of Human Glutaminyl Cyclase, an Enzyme Responsible for Protein N-Terminal Pyroglutamate Formation. Proc. Natl. Acad. Sci. USA 2005, 102, 13117–13122. [Google Scholar] [CrossRef]
- DiPisa, F.; Pozzi, C.; Benvenuti, M.; Andreini, M.; Marconi, G.; Mangani, S. The Soluble Y115E-Y117E Variant of Human Glutaminyl Cyclase Is a Valid Target for X-ray and NMR Screening of Inhibitors against Alzheimer Disease. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 986–992. [Google Scholar] [CrossRef]
- Booth, R.E.; Lovell, S.C.; Misquitta, S.A.; Bateman, R.C. Human Glutaminyl Cyclase and Bacterial Zinc Aminopeptidase Share a Common Fold and Active Site. BMC Biol. 2004, 2, 2. [Google Scholar] [CrossRef]
- Huang, K.-F.; Liaw, S.-S.; Huang, W.-L.; Chia, C.-Y.; Lo, Y.-C.; Chen, Y.-L.; Wang, A.H.-J. Structures of Human Golgi-Resident Glutaminyl Cyclase and Its Complexes with Inhibitors Reveal a Large Loop Movement upon Inhibitor Binding. J. Biol. Chem. 2011, 286, 12439–12449. [Google Scholar] [CrossRef]
- Hoang, V.-H.; Tran, P.-T.; Cui, M.; Ngo, V.T.H.; Ann, J.; Park, J.; Lee, J.; Choi, K.; Cho, H.; Kim, H.; et al. Discovery of Potent Human Glutaminyl Cyclase Inhibitors as Anti-Alzheimer’s Agents Based on Rational Design. J. Med. Chem. 2017, 60, 2573–2590. [Google Scholar] [CrossRef]
- Benvenuti, M.; Mangani, S. Crystallization of Soluble Proteins in Vapor Diffusion for X-ray Crystallography. Nat. Protoc. 2007, 2, 1633–1651. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P. Scaling and Assessment of Data Quality. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 72–82. [Google Scholar] [CrossRef]
- Evans, P.R. An Introduction to Data Reduction: Space-Group Determination, Scaling and Intensity Statistics. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 282–292. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular Replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Read, R.J.; Schierbeek, A.J. A Phased Translation Function. J. Appl. Cryst. 1988, 21, 490–495. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Potterton, L.; McNicholas, S.; Krissinel, E.; Gruber, J.; Cowtan, K.; Emsley, P.; Murshudov, G.N.; Cohen, S.; Perrakis, A.; Noble, M. Developments in the CCP4 Molecular-Graphics Project. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2288–2294. [Google Scholar] [CrossRef]


| Apo-hQC-2X | hQC-2X–Co(II) | hQC-2X–Co(II)–SEN177 | hQC-2X–Co(II)–PBD-150 | |
|---|---|---|---|---|
| PDB ID | 9FXG | 9FXH | 9FXI | 9FXJ |
| Data Collection Statistics | ||||
| Diffraction source | I04 (DLS) | I04 (DLS) | I04 (DLS) | I04 (DLS) |
| Wavelength (Å) | 0.9795 | 0.9537 | 1.5896 | 1.5896 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Detector | Eiger2 XE 16M | Eiger2 XE 16M | Eiger2 XE 16M | Eiger2 XE 16M |
| Crystal detector distance (mm) | 221.1 | 307.4 | 175.3 | 175.2 |
| Exposure time per image (s) | 0.08 | 0.02 | 0.03 | 0.03 |
| Δφ (°) | 0.2 | 0.1 | 0.5 | 0.5 |
| Space group | C2 | C2 | C2 | C2 |
| No. of subunits in ASU | 3 | 3 | 3 | 3 |
| a, b, c (Å) | 86.28, 149.31, 95.71 | 86.18, 149.27, 96.01 | 86.43, 149.74, 95.65 | 86.39, 149.63, 96.16 |
| β (°) | 96.96 | 96.87 | 97.10 | 96.82 |
| Resolution range (Å) | 58.70–1.96 (2.00–1.96) | 61.76–2.30 (2.38–2.30) | 74.87–3.06 (3.27–3.06) | 74.81–3.06 (3.27–3.06) |
| Total no. of reflections | 224,450 (11,905) | 273,314 (24,880) | 81,001 (15,548) | 87,233 (16,860) |
| No. of unique reflections | 80,418 (4597) | 50,664 (4761) | 19,865 (3889) | 20,036 (3926) |
| Completeness (%) | 93.8 (94.5) | 95.2 (97.0) | 92.1 (94.7) | 92.5 (95.0) |
| Redundancy | 2.8 (2.6) | 5.4 (5.2) | 4.1 (4.0) | 4.4 (4.3) |
| 〈I/σ(I)〉 | 6.2 (2.7) | 7.0 (2.2) | 4.9 (2.0) | 5.1 (2.1) |
| Rmerge | 0.102 (0.286) | 0.157 (0.621) | 0.210 (0.536) | 0.227 (0.531) |
| Rmeas | 0.136 (0.389) | 0.194 (0.772) | 0.265 (0.676) | 0.289 (0.676) |
| Rpim | 0.090 (0.263) | 0.112 (0.452) | 0.129 (0.333) | 0.139 (0.329) |
| Overall B factor from Wilson plot (Å2) | 10.0 | 19.9 | 23.6 | 20.0 |
| Refinements Statistics | ||||
| Resolution range (Å) | 58.77–1.96 (2.01–1.96) | 61.83–2.30 (2.36–2.30) | 74.98–3.06 (3.14–3.06) | 74.93–3.06 (3.14–3.06) |
| Completeness (%) | 93.5 (94.7) | 94.9 (97.0) | 92.0 (94.7) | 92.3 (95.3) |
| No. of reflections, working set | 76,430 (5715) | 48,122 (3613) | 18,848 (1015) | 18,993 (1043) |
| No. of reflections, test set | 3973 (317) | 2540 (182) | 1501 (73) | 1517 (79) |
| Final Rcryst | 0.208 (0.248) | 0.205 (0.319) | 0.174 (0.278) | 0.171 (0.241) |
| Final Rfree | 0.257 (0.308) | 0.257 (0.363) | 0.228 (0.319) | 0.202 (0.258) |
| No. of non-H atoms | ||||
| Protein | 7721 | 7758 | 7731 | 7765 |
| Co(II) ion | - | 3 | 3 | 3 |
| Sulfate ion | 20 | 15 | 15 | 15 |
| Ligand | - | - | 75 | 66 |
| Other | 36 (ethylene glycol) | 6 (glycerol) | - | - |
| Water | 2063 | 1070 | 278 | 305 |
| Total | 9840 | 8852 | 8102 | 8154 |
| R.m.s. deviation bonds (Å) | 0.008 | 0.006 | 0.004 | 0.005 |
| Angles (°) | 1.543 | 1.382 | 1.191 | 1.253 |
| Average B factors (Å2) | 22.5 | 29.1 | 30.7 | 26.4 |
| Estimate error on coordinates based on R value (Å) | 0.22 | 0.40 | 0.49 | 0.49 |
| Ramachandran plot Most favored (%) Allowed (%) | 96.6% 3.4% | 94.6% 5.4% | 93.0% 7.0% | |
| 93.5% | ||||
| 6.5% | ||||
| RSCC ligand chain A, B, C | - | - | 0.98, 0.98, 0.98 | 0.98, 0.98, 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tassone, G.; Pozzi, C.; Mangani, S. Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective. Int. J. Mol. Sci. 2024, 25, 8279. https://doi.org/10.3390/ijms25158279
Tassone G, Pozzi C, Mangani S. Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective. International Journal of Molecular Sciences. 2024; 25(15):8279. https://doi.org/10.3390/ijms25158279
Chicago/Turabian StyleTassone, Giusy, Cecilia Pozzi, and Stefano Mangani. 2024. "Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective" International Journal of Molecular Sciences 25, no. 15: 8279. https://doi.org/10.3390/ijms25158279






