Exploring the PDZ, DUF, and LIM Domains of Pdlim5 in Dendrite Branching
Abstract
:1. Introduction
2. Results
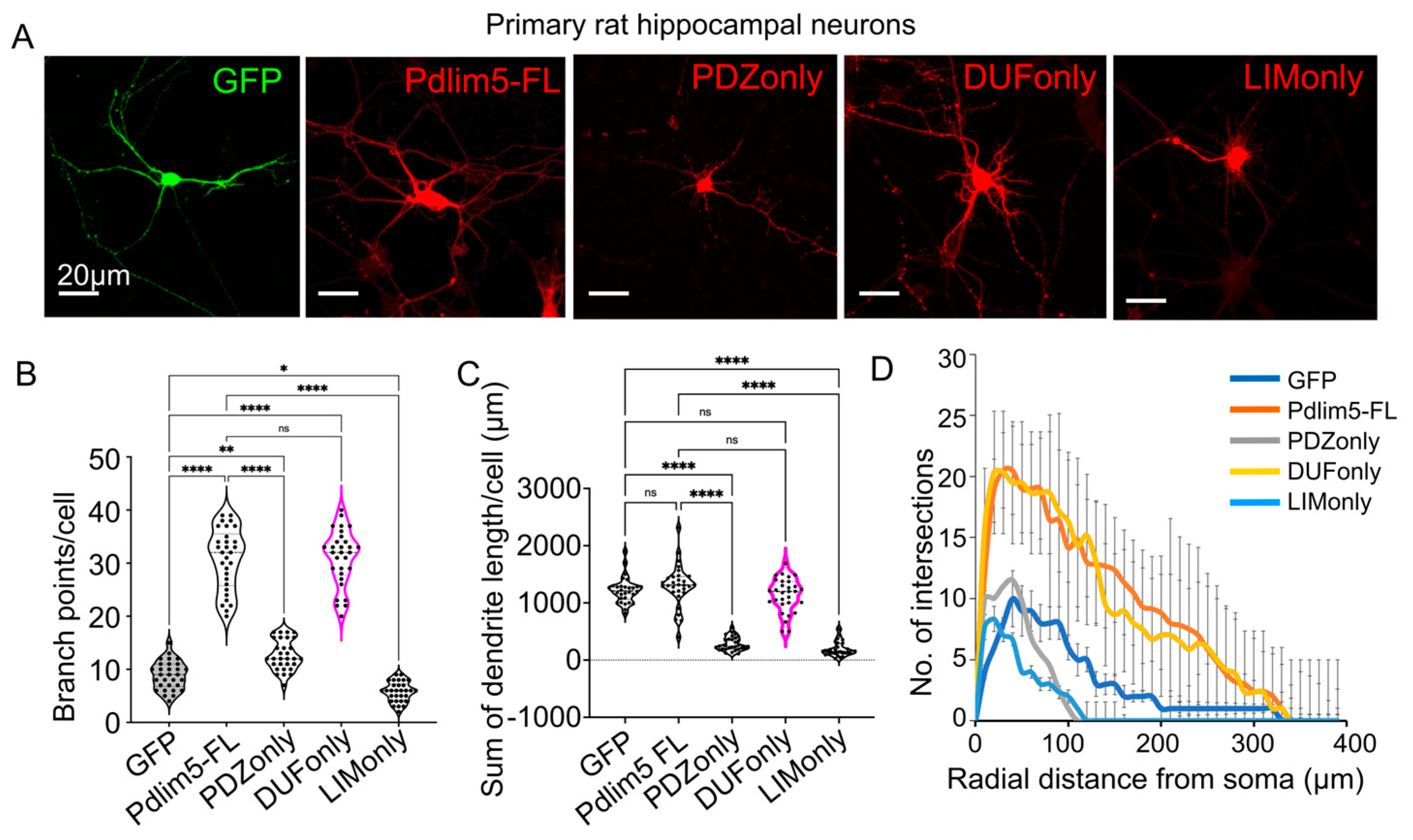
2.1. Impact of Expressing Isolated Pdlim5 Domains upon Dendrite Morphology
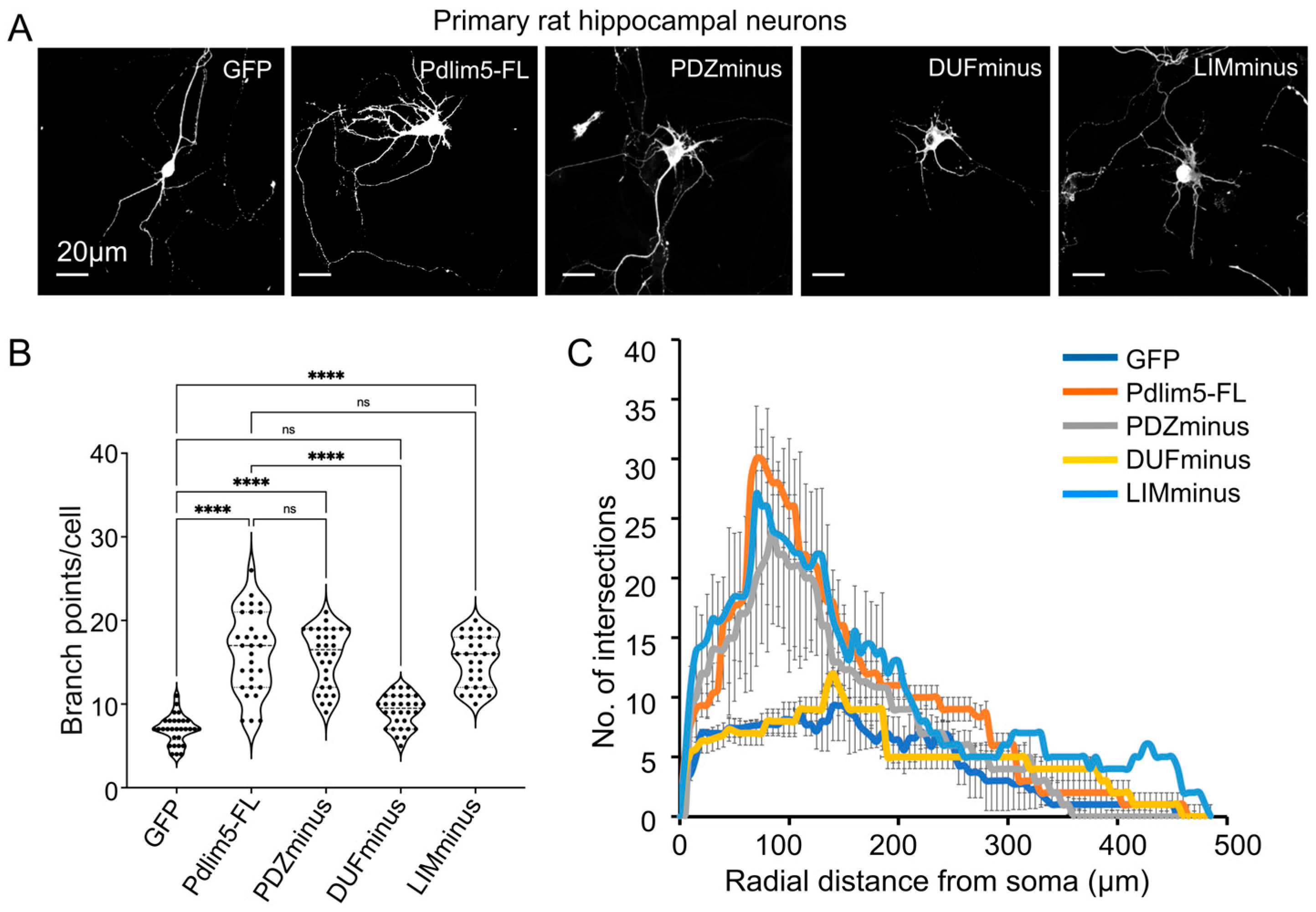
2.2. Domain Deletions of Pdlim5 Support the Importance of the DUF Domain in Dendrite Branching
2.3. Comparison of the DUF Domain between Species and a Potential Role for Phosphorylation at Residue Y251
2.4. Intra-Molecular Interactions of Pdlim5 May Regulate the Accessibility of the DUF Domain
3. Discussion
4. Materials and Methods
4.1. Neuronal Cultures and Transfection
4.2. HEK293 Cell Culture and Transfection
4.3. cDNA Constructs
4.4. Antibodies
4.5. Golgi Localization Assay (GLS)
4.6. Co-Immunoprecipitations and Immunoblots
4.7. Immunofluorescence Staining and Confocal Microscopy
4.8. Neuronal Morphological Analysis by IMARIS9.9
4.9. Protein Domain Array
4.10. AlphaFold Modeling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jan, Y.-N.; Jan, L.Y. Branching out: Mechanisms of dendritic arborization. Nat. Rev. Neurosci. 2010, 11, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Shen, K.; Bülow, H.E. Intrinsic and Extrinsic Mechanisms of Dendritic Morphogenesis. Annu. Rev. Physiol. 2015, 77, 271–300. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Kramer, D.A.; Chen, B.; Shen, K. A two-step actin polymerization mechanism drives dendrite branching. Neural Develop. 2021, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Stürner, T.; Ferreira Castro, A.; Philipps, M.; Cuntz, H.; Tavosanis, G. The branching code: A model of actin-driven dendrite arborization. Cell Rep. 2022, 39, 110746. [Google Scholar] [CrossRef] [PubMed]
- Healy, M.D.; Collins, B.M. The PDLIM family of actin-associated proteins and their emerging role in membrane trafficking. Biochem. Soc. Trans. 2023, 51, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qu, R.; Ouyang, J.; Zhong, S.; Dai, J. An Overview of the Cytoskeleton-Associated Role of PDLIM5. Front. Physiol. 2020, 11, 975. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, N.; Hoshijima, M.; Oyasu, M.; Saito, N.; Tanizawa, K.; Kuroda, S. ENH, Containing PDZ and LIM Domains, Heart/Skeletal Muscle-Specific Protein, Associates with Cytoskeletal Proteins through the PDZ Domain. Biochem. Biophys. Res. Commun. 2000, 272, 505–512. [Google Scholar] [CrossRef] [PubMed]
- te Velthuis, A.J.W.; Isogai, T.; Gerrits, L.; Bagowski, C.P. Insights into the Molecular Evolution of the PDZ/LIM Family and Identification of a Novel Conserved Protein Motif. PLoS ONE 2007, 2, e189. [Google Scholar] [CrossRef] [PubMed]
- Klaavuniemi, T.; Kelloniemi, A.; Ylänne, J. The ZASP-like Motif in Actinin-associated LIM Protein Is Required for Interaction with the α-Actinin Rod and for Targeting to the Muscle Z-line. J. Biol. Chem. 2004, 279, 26402–26410. [Google Scholar] [CrossRef]
- Wu, R.; Durick, K.; Songyang, Z.; Cantley, L.C.; Taylor, S.S.; Gill, G.N. Specificity of LIM Domain Interactions with Receptor Tyrosine Kinases. J. Biol. Chem. 1996, 271, 15934–15941. [Google Scholar] [CrossRef]
- Durick, K.; Wu, R.-Y.; Gill, G.N.; Taylor, S.S. Mitogenic Signaling by Ret/ptc2 Requires Association with Enigma via a LIM Domain. J. Biol. Chem. 1996, 271, 12691–12694. [Google Scholar] [CrossRef]
- Ahn, B.Y.; Saldanha-Gama, R.F.G.; Rahn, J.J.; Hao, X.; Zhang, J.; Dang, N.-H.; Alshehri, M.; Robbins, S.M.; Senger, D.L. Glioma invasion mediated by the p75 neurotrophin receptor (p75NTR/CD271) requires regulated interaction with PDLIM1. Oncogene 2016, 35, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-L.; Gou, C.-Y.; Wang, W.-H.; Li, Y.; Cui, Y.; Duan, J.-J.; Chen, Y. A novel effect of PDLIM5 in α7 nicotinic acetylcholine receptor upregulation and surface expression. Cell. Mol. Life Sci. 2022, 79, 64. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.W.; Nakagawa, T.; Licznerski, P.; Pawlak, V.; Kolleker, A.; Rozov, A.; Kim, J.; Dittgen, T.; Köhr, G.; Sheng, M.; et al. Actin/α-Actinin-Dependent Transport of AMPA Receptors in Dendritic Spines: Role of the PDZ-LIM Protein RIL. J. Neurosci. 2004, 24, 8584–8594. [Google Scholar] [CrossRef] [PubMed]
- Herrick, S.; Evers, D.M.; Lee, J.-Y.; Udagawa, N.; Pak, D.T.S. Postsynaptic PDLIM5/Enigma Homolog binds SPAR and causes dendritic spine shrinkage. Mol. Cell. Neurosci. 2010, 43, 188–200. [Google Scholar] [CrossRef]
- Baumert, R.; Ji, H.; Paulucci-Holthauzen, A.; Wolfe, A.; Sagum, C.; Hodgson, L.; Arikkath, J.; Chen, X.; Bedford, M.T.; Waxham, M.N.; et al. Novel phospho-switch function of delta-catenin in dendrite development. J. Cell Biol. 2020, 219, e201909166. [Google Scholar] [CrossRef]
- Donta, M.S.; Srivastava, Y.; Di Mauro, C.M.; Paulucci-Holthauzen, A.; Waxham, M.N.; McCrea, P.D. p120-catenin subfamily members have distinct as well as shared effects on dendrite morphology during neuron development in vitro. Front. Cell. Neurosci. 2023, 17, 1151249. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, Y.; Donta, M.; Mireles, L.L.; Paulucci-Holthauzen, A.; Waxham, M.N.; McCrea, P.D. Role of a Pdlim5:PalmD complex in directing dendrite morphology. Front. Cell. Neurosci. 2024, 18, 1315941. [Google Scholar] [CrossRef]
- Hansen, M.D.H.; Kwiatkowski, A.V. Chapter One—Control of Actin Dynamics by Allosteric Regulation of Actin Binding Proteins. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 303, pp. 1–25. [Google Scholar]
- Pérez-Mejías, G.; Velázquez-Cruz, A.; Guerra-Castellano, A.; Baños-Jaime, B.; Díaz-Quintana, A.; González-Arzola, K.; Ángel De la Rosa, M.; Díaz-Moreno, I. Exploring protein phosphorylation by combining computational approaches and biochemical methods. Comput. Struct. Biotechnol. J. 2020, 18, 1852–1863. [Google Scholar] [CrossRef]
- Martínez-Cerdeño, V. Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev. Neurobiol. 2017, 77, 393–404. [Google Scholar] [CrossRef]
- Kaufmann, W.E.; Moser, H.W. Dendritic Anomalies in Disorders Associated with Mental Retardation. Cereb. Cortex 2000, 10, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tsukamoto, O.; Nakano, A.; Kato, H.; Kioka, H.; Ito, N.; Higo, S.; Yamazaki, S.; Shintani, Y.; Matsuoka, K.; et al. Augmented AMPK activity inhibits cell migration by phosphorylating the novel substrate Pdlim5. Nat. Commun. 2015, 6, 6137. [Google Scholar] [CrossRef] [PubMed]
- Krcmery, J.; Camarata, T.; Kulisz, A.; Simon, H.-G. Nucleocytoplasmic functions of the PDZ-LIM protein family: New insights into organ development. BioEssays 2010, 32, 100–108. [Google Scholar] [CrossRef] [PubMed]
- te Velthuis, A.J.W.; Bagowski, C.P. PDZ and LIM Domain-Encoding Genes: Molecular Interactions and their Role in Development. Sci. World J. 2007, 7, 1470–1492. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Iwayama, Y.; Kakiuchi, C.; Iwamoto, K.; Yamada, K.; Minabe, Y.; Nakamura, K.; Mori, N.; Fujii, K.; Nanko, S.; et al. Gene expression and association analyses of LIM (PDLIM5) in bipolar disorder and schizophrenia. Mol. Psychiatry 2005, 10, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, Y.; Arai, M.; Niizato, K.; Iritani, S.; Noguchi, E.; Ohtsuki, T.; Koga, M.; Kato, T.; Itokawa, M.; Arinami, T. A Polymorphism in the PDLIM5 Gene Associated with Gene Expression and Schizophrenia. Biol. Psychiatry 2006, 59, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, Y.; Ishikawa, M.; Kaito, N.; Iijima, Y.; Tanabe, Y.; Ishiguro, H.; Arinami, T. Experimental Evidence for the Involvement of PDLIM5 in Mood Disorders in Hetero Knockout Mice. PLoS ONE 2013, 8, e59320. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, W.; Xiao, Z.; Wang, G.; Yin, S.; Zhu, F.; Wang, H.; Cheng, J.; Wang, X.; He, X.; et al. A Major Single Nucleotide Polymorphism of the PDLIM5 Gene Associated with Recurrent Major Depressive Disorder. J. Psychiatry Neurosci 2008, 33, 43–46. [Google Scholar] [PubMed]
- Chen, J.; Sagum, C.; Bedford, M.T. Protein domain microarrays as a platform to decipher signaling pathways and the histone code. Methods 2020, 184, 4–12. [Google Scholar] [CrossRef]
- Levy-Apter, E.; Finkelshtein, E.; Vemulapalli, V.; Li, S.S.-C.; Bedford, M.T.; Elson, A. Adaptor Protein GRB2 Promotes Src Tyrosine Kinase Activation and Podosomal Organization by Protein-tyrosine Phosphatase ϵ in Osteoclasts. J. Biol. Chem. 2014, 289, 36048–36058. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, Y.; Donta, M.; Mireles, L.L.; Paulucci-Holthauzen, A.; Shi, L.; Bedford, M.T.; Waxham, M.N.; McCrea, P.D. Exploring the PDZ, DUF, and LIM Domains of Pdlim5 in Dendrite Branching. Int. J. Mol. Sci. 2024, 25, 8326. https://doi.org/10.3390/ijms25158326
Srivastava Y, Donta M, Mireles LL, Paulucci-Holthauzen A, Shi L, Bedford MT, Waxham MN, McCrea PD. Exploring the PDZ, DUF, and LIM Domains of Pdlim5 in Dendrite Branching. International Journal of Molecular Sciences. 2024; 25(15):8326. https://doi.org/10.3390/ijms25158326
Chicago/Turabian StyleSrivastava, Yogesh, Maxsam Donta, Lydia L. Mireles, Adriana Paulucci-Holthauzen, Leilei Shi, Mark T. Bedford, M. Neal Waxham, and Pierre D. McCrea. 2024. "Exploring the PDZ, DUF, and LIM Domains of Pdlim5 in Dendrite Branching" International Journal of Molecular Sciences 25, no. 15: 8326. https://doi.org/10.3390/ijms25158326




