Research Progress on the Role of M6A in Regulating Economic Traits in Livestock
Abstract
:1. Introduction
2. Basic Regulatory Mechanisms of m6A
3. Function of m6A Modifications in Animal Growth and Development
3.1. Regulation of m6A Modifications in Muscle Development
3.2. Regulation of m6A Modifications in Adipogenesis
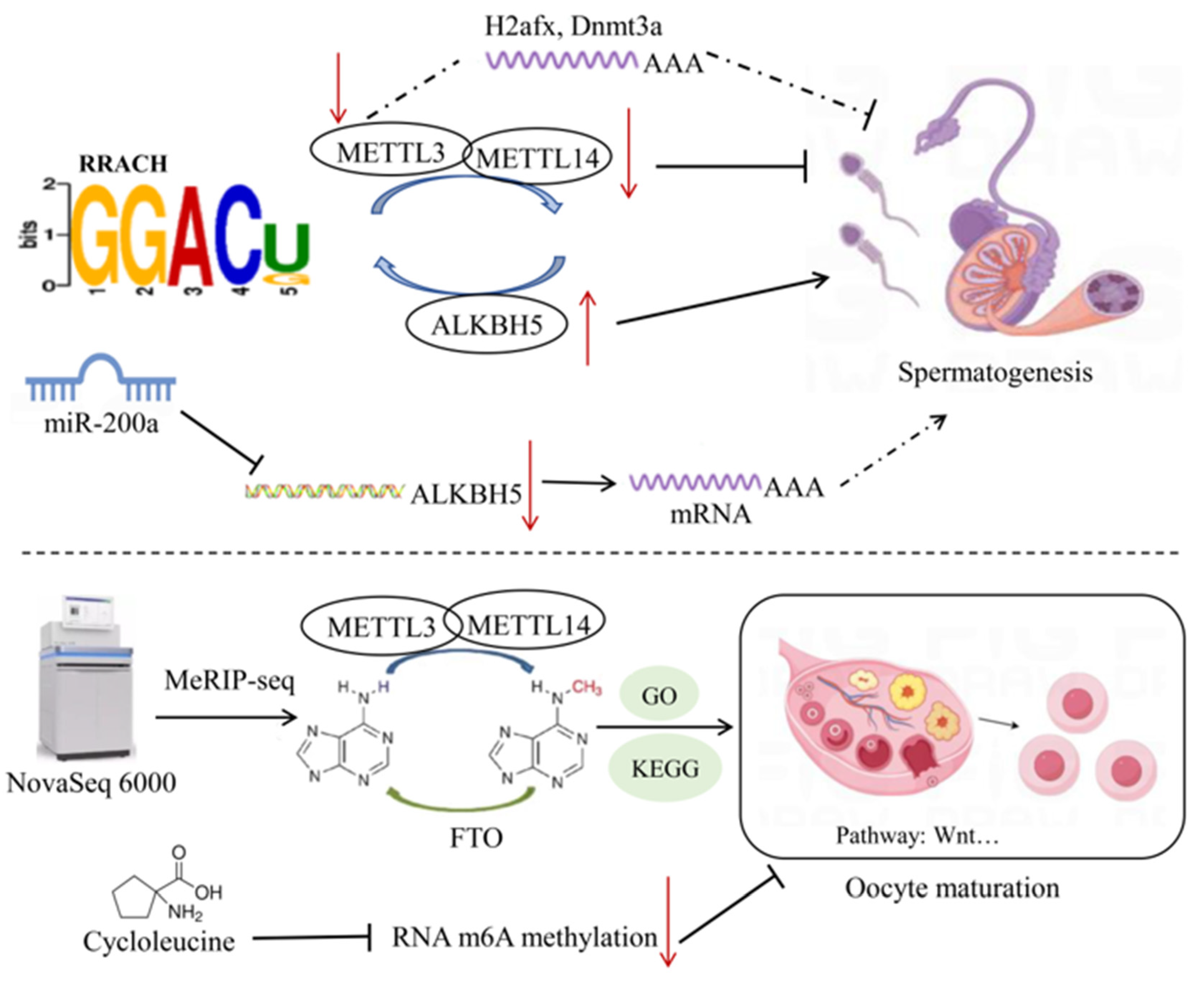
4. The Functional Role of m6A Modifications in Reproductive Traits
4.1. Regulation of Spermatogenesis by m6A Modifications
4.2. Regulation of Animal Oocyte Maturation via m6A Modifications
5. Summary and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2010, 39 (Suppl. S1), D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Ru, W.; Zhang, X.; Yue, B.; Qi, A.; Shen, X.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Insight into m6A methylation from occurrence to functions. Open Biol. 2020, 10, 200091. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G. N 6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Liu, H.; Zhang, S.; da Silva, S.; Zhang, L.; Meng, J.; Cui, X.; Yuan, H.; Sorel, O.; Zhang, S. Viral and cellular N6-methyladenosine and N6, 2=-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 2018, 3, 108–120. [Google Scholar] [CrossRef]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef]
- Yoon, K.-J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.-S.; Zhu, Y.; Zheng, L. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 2017, 171, 877–889.e17. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Chen, J. The biogenesis and precise control of RNA m6A methylation. Trends Genet. 2020, 36, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhang, X.; Weng, Y.-L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Tong, M.-H. m6A mRNA modification regulates mammalian spermatogenesis. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Bansal, H.; Yihua, Q.; Iyer, S.P.; Ganapathy, S.; Proia, D.; Penalva, L.; Uren, P.; Suresh, U.; Carew, J.; Karnad, A. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 2014, 28, 1171–1174. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M. The N 6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 2018, 22, 191–205.e9. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Qing, Y.; Horne, D.; Huang, H.; Chen, J. The roles and implications of RNA m6A modification in cancer. Nat. Rev. Clin. Oncol. 2023, 20, 507–526. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.-H.; Guo, T.; Wang, M.; Liu, J.-H.; Zheng, L.-M.; He, Y. RNA m6A modification facilitates DNA methylation during maize kernel development. Plant Physiol. 2024, 194, 2165–2182. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, S.; Yu, L.; Xiao, Y.; Zhang, S.; Wang, X.; Xu, Y.; Yu, H.; Li, Y.; Yang, J. RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol. 2021, 39, 1581–1588. [Google Scholar] [CrossRef]
- He, P.C.; He, C. m6A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Zheng, X.; Guo, Y.; Cao, J.; Zhang, Y.; Wen, S.; Gao, W.; Wu, Y. Regulatory role and mechanism of m6A RNA modification in human metabolic diseases. Mol. Ther.-Oncolytics 2021, 22, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Y.; Zhang, J.; Zhu, J.-S. The role of m 6 A RNA methylation in human cancer. Mol. Cancer 2019, 18, 1–9. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vanacova, S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017, 45, 11356–11370. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Y.; Han, J.; Yang, Y.; Chen, Y.; Tang, Z.; Gao, F. Longitudinal epitranscriptome profiling reveals the crucial role of N6-methyladenosine methylation in porcine prenatal skeletal muscle development. J. Genet. Genom. 2020, 47, 466–476. [Google Scholar] [CrossRef]
- Deng, K.; Fan, Y.; Liang, Y.; Cai, Y.; Zhang, G.; Deng, M.; Wang, Z.; Lu, J.; Shi, J.; Wang, F. FTO-mediated demethylation of GADD45B promotes myogenesis through the activation of p38 MAPK pathway. Mol. Ther.-Nucleic Acids 2021, 26, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Cao, J.; Sun, Y.; Zhou, H.; Zhu, Q.; Dai, D.; Zhan, S.; Guo, J.; Zhong, T.; Wang, L. METTL3 Promotes the Differentiation of Goat Skeletal Muscle Satellite Cells by Regulating MEF2C mRNA Stability in a m6A-Dependent Manner. Int. J. Mol. Sci. 2023, 24, 14115. [Google Scholar] [CrossRef]
- Ru, W. Function and Mechanism of METTL3 Regulating Bovine Myoblast Development through m6a Modification; Northwest A&F University: Xi’an, China, 2022. (In Chinese) [Google Scholar]
- Yang, X.; Mei, C.; Ma, X.; Du, J.; Wang, J.; Zan, L. m6A Methylases Regulate Myoblast Proliferation, Apoptosis and Differentiation. Animals 2022, 12, 773. [Google Scholar] [CrossRef]
- Ding, H.; Lin, Y.; Zhang, T.; Zhang, S.; Wu, Y.; Duan, Y.; Gong, Y.; Xie, K.; Wang, J.; Dai, G.; et al. Study on the Expression of m6A Methylation in Chicken Muscle Growth and Development. China Anim. Husb. Vet. Med. 2021, 48, 1525–1534. (In Chinese) [Google Scholar]
- Shu, J.; Shan, Y.; Ji, G.; Zhang, M.; Tu, Y.; Liu, Y.; Ju, X.; Sheng, Z.; Tang, Y.; Li, H.; et al. Relationship Between Expression Levels of Guangxi Partridge Chicken m6A Methyltransferase Genes, Myofiber Types and Myogenic Differentiation. Sci. Agric. Sin. 2022, 55, 589–601. (In Chinese) [Google Scholar]
- Diao, L.-T.; Xie, S.-J.; Lei, H.; Qiu, X.-S.; Huang, M.-C.; Tao, S.; Hou, Y.-R.; Hu, Y.-X.; Sun, Y.-J.; Zhang, Q. METTL3 regulates skeletal muscle specific miRNAs at both transcriptional and post-transcriptional levels. Biochem. Biophys. Res. Commun. 2021, 552, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tan, B.; Xiao, L.; Zhao, X.; Zeng, J.; Hong, L.; Yang, J.; Cai, G.; Zheng, E.; Wu, Z. Comprehensive analysis of long noncoding RNA modified by m6A methylation in oxidative and glycolytic skeletal muscles. Int. J. Mol. Sci. 2022, 23, 4600. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Dai, R.; Meng, G.; Dingkao, R.; Wang, X.; Ren, W.; Ma, X.; Wu, X.; Chu, M.; La, Y. Transcriptome-Wide Study of mRNAs and lncRNAs Modified by m6A RNA Methylation in the Longissimus Dorsi Muscle Development of Cattle-Yak. Cells 2022, 11, 3654. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, M.; Yuan, R.; Zhou, Z.; Zhang, J.; Kong, S.; Lin, D.; Lian, L.; Li, J.; Zhang, X. MYH1G-AS is a chromatin-associated lncRNA that regulates skeletal muscle development in chicken. Cell. Mol. Biol. Lett. 2024, 29, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, L.; Chen, J.; Wang, Y. mRNA m6A methylation downregulates adipogenesis in porcine adipocytes. Biochem. Biophys. Res. Commun. 2015, 459, 201–207. [Google Scholar] [CrossRef]
- Yao, Y.; Bi, Z.; Wu, R.; Zhao, Y.; Liu, Y.; Liu, Q.; Wang, Y.; Wang, X. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2–dependent manner. FASEB J. 2019, 33, 7529–7544. [Google Scholar] [CrossRef]
- Wu, R.; Yao, Y.; Jiang, Q.; Cai, M.; Liu, Q.; Wang, Y.; Wang, X. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int. J. Obes. 2018, 42, 1378–1388. [Google Scholar] [CrossRef]
- Wu, R.; Guo, G.; Bi, Z.; Liu, Y.; Zhao, Y.; Chen, N.; Wang, F.; Wang, Y.; Wang, X. m6A methylation modulates adipogenesis through JAK2-STAT3-C/EBPβ signaling. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 796–806. [Google Scholar] [CrossRef]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2020, 16, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Luo, Y.; Jia, G.; Liu, G.; Zhao, H.; Huang, Z. FTO promotes adipogenesis through inhibition of the Wnt/β-catenin signaling pathway in porcine intramuscular preadipocytes. Anim. Biotechnol. 2017, 28, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ren, X.; Li, L.; Lu, S.; Chen, T.; Tan, L.; Liu, M.; Luo, Q.; Liang, S.; Nie, Q. Integrative analyses of mRNA expression profile reveal the involvement of IGF2BP1 in chicken adipogenesis. Int. J. Mol. Sci. 2019, 20, 2923. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Huang, W.; Wang, Z.; Nie, Q. m6A demethylase FTO regulate CTNNB1 to promote adipogenesis of chicken preadipocyte. J. Anim. Sci. Biotechnol. 2022, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, J.; Ma, X.; Zan, L. Expression Analyses of m6A Methylase Genes in Bovine Adipogenesis. Biotechnol. Bull. 2022, 38, 70–79. (In Chinese) [Google Scholar]
- Qi, K.; Dou, Y.; Zhang, Z.; Wei, Y.; Song, C.; Qiao, R.; Li, X.; Yang, F.; Wang, K.; Li, X. Expression Profile and Regulatory Properties of m6A-Modified circRNAs in the Longissimus Dorsi of Queshan Black and Large White Pigs. Animals 2023, 13, 2190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Nie, J.; Chen, Y.; Li, X.; Chen, H. Connecting the Dots: N6-Methyladenosine (m6A) Modification in Spermatogenesis. Adv. Biol. 2023, 7, 2300068. [Google Scholar] [CrossRef] [PubMed]
- Landfors, M.; Nakken, S.; Fusser, M.; Dahl, J.-A.; Klungland, A.; Fedorcsak, P. Sequencing of FTO and ALKBH5 in men undergoing infertility work-up identifies an infertility-associated variant and two missense mutations. Fertil. Steril. 2016, 105, 1170–1179.e5. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Liu, Z.; Cai, Y.; Deng, M.; Li, D.; Leng, Q.; Shi, L.; Tang, Y.; Wang, F.; Wan, Y. Expression pattern of alkB homolog 5 in goat testis and its role in spermatogonial stem cells. Cell Tissue Res. 2022, 387, 131–142. [Google Scholar] [CrossRef]
- Lin, Z.; Hsu, P.J.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.-J.; Zhang, X.; Zhou, Y.; Zhang, T. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef] [PubMed]
- Zhong, D.; Zhang, L.; Huang, K.; Chen, M.; Chen, Y.; Liu, Q.; Shi, D.; Li, H. circRNA-miRNA-mRNA network analysis to explore the pathogenesis of abnormal spermatogenesis due to aberrant m6A methylation. Cell Tissue Res. 2023, 392, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, X.; Zhang, P.; Li, F.; Zhang, L.; Li, X.; Huang, T.; Zheng, Y.; Yu, T.; Zhang, T. Transcriptome-wide dynamics of m 6 A mRNA methylation during porcine spermatogenesis. Genom. Proteom. Bioinform. 2023, 21, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Yang, Y.; Feng, G.-H.; Sun, B.-F.; Chen, J.-Q.; Li, Y.-F.; Chen, Y.-S.; Zhang, X.-X.; Wang, C.-X.; Jiang, L.-Y. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. The Relationship between the m6A Demethylase ALKBH5 and Yattle Male Sterility and the Targeted Regulation Analysis of MiR-200a; Southwest University: Chongqing, China, 2021. (In Chinese) [Google Scholar]
- Mu, H.; Zhang, T.; Yang, Y.; Zhang, D.; Gao, J.; Li, J.; Yue, L.; Gao, D.; Shi, B.; Han, Y. METTL3-mediated mRNA N 6-methyladenosine is required for oocyte and follicle development in mice. Cell Death Dis. 2021, 12, 989. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yan, Y.; Wang, X.; Liu, Q.; Zhang, Y.; Cao, Z. Advances in mA Methylation Regulating Mammalian Gamete Maturation and Embryo Development. J. Agric. Biotechnol. 2023, 31, 1534–1546. (In Chinese) [Google Scholar]
- Zuccotti, M.; Merico, V.; Cecconi, S.; Redi, C.A.; Garagna, S. What does it take to make a developmentally competent mammalian egg? Hum. Reprod. Update 2011, 17, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Kidder, G.M.; Vanderhyden, B.C. Bidirectional communication between oocytes and follicle cells: Ensuring oocyte developmental competence. Can. J. Physiol. Pharmacol. 2010, 88, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhang, D.; Wang, Y.; Tong, X.; Avalos, L.F.C.; Khan, I.M.; Gao, D.; Xu, T.; Zhang, L.; Knott, J.G. Identification and functional annotation of m6A methylation modification in granulosa cells during antral follicle development in pigs. Anim. Reprod. Sci. 2020, 219, 106510. [Google Scholar] [CrossRef] [PubMed]
- Gershon, E.; Dekel, N. Newly identified regulators of ovarian folliculogenesis and ovulation. Int. J. Mol. Sci. 2020, 21, 4565. [Google Scholar] [CrossRef]
- Guo, S.; Wang, X.; Cao, M.; Wu, X.; Xiong, L.; Bao, P.; Chu, M.; Liang, C.; Yan, P.; Pei, J. The transcriptome-wide N6-methyladenosine (m6A) map profiling reveals the regulatory role of m6A in the yak ovary. BMC Genom. 2022, 23, 358. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L. The avian ovary and follicle development: Some comparative and practical insights. Turk. J. Vet. Anim. Sci. 2014, 38, 660–669. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, C.; Zhu, G. Profiling of RNA N6-methyladenosine methylation during follicle selection in chicken ovary. Poult. Sci. 2019, 98, 6117–6124. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S.; Zhai, Y.; Han, Y.; Huang, R.; An, X.; Dai, X.; Li, Z. Cycloleucine negatively regulates porcine oocyte maturation and embryo development by modulating N6-methyladenosine and histone modifications. Theriogenology 2022, 179, 128–140. [Google Scholar] [CrossRef]
- Li, Z.; Ruan, Z.; Feng, Y.; Wang, Y.; Zhang, J.; Lu, C.; Shi, D.; Lu, F. METTL3-mediated m6A methylation regulates granulosa cells autophagy during follicular atresia in pig ovaries. Theriogenology 2023, 201, 83–94. [Google Scholar] [CrossRef]
- Chen, B.; Yuan, C.; Guo, T.; Liu, J.; Yang, B.; Lu, Z. Molecular Mechanism of m6A Methylation Modification Genes METTL3 and FTO in Regulating Heat Stress in Sheep. Int. J. Mol. Sci. 2023, 24, 11926. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Xu, A.; Zhao, B.; Xia, Y.; He, Y.; Xue, H.; Li, S. Dietary selenomethionine reduced oxidative stress by resisting METTL3-mediated m6A methylation level of Nrf2 to ameliorate LPS-induced liver necroptosis in laying hens. J. Nutr. Biochem. 2024, 125, 109563. [Google Scholar] [CrossRef]
- Zhang, T.; Yu, B.; Cai, Z.; Jiang, Q.; Fu, X.; Zhao, W.; Wang, H.; Gu, Y.; Zhang, J. Regulatory role of N6-methyladenosine in intramuscular fat deposition in chicken. Poult. Sci. 2023, 102, 102972. [Google Scholar] [CrossRef]
- Chen, B.; Liu, S.; Zhang, W.; Xiong, T.; Zhou, M.; Hu, X.; Mao, H.; Liu, S. Profiling analysis of N6-methyladenosine mRNA methylation reveals differential m6A patterns during the embryonic skeletal muscle development of ducks. Animals 2022, 12, 2593. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Wei, Y.; Zhang, Z.; Li, C.; Song, C.; Liu, Y.; Qi, K.; Li, X.; Li, X.; Qiao, R. Transcriptome-wide analysis of RNA m6A methylation regulation of muscle development in Queshan Black pigs. BMC Genom. 2023, 24, 239. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, S.; Wang, G.; Zhang, S.; Bao, W.; Wu, S. Expression analysis of m 6 A-related genes in various tissues of Meishan pigs at different developmental stages. Rev. Bras. De Zootec. 2023, 52, e20210149. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Yan, Y.; Zhu, C.; Wang, L.; Fu, S.; Zuo, F.; Zhang, G.-W. N6-methyladenosine RNA demethylase ALKBH5 is testis-specifically downregulated in hybrid male sterile dzo and is a target gene of bta-miR-200a. Theriogenology 2022, 187, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-K.; Yu, X.-X.; Liu, Y.-H.; Li, X.; Liu, X.-M.; Wang, P.-C.; Liu, S.; Miao, J.-K.; Du, Z.-Q.; Yang, C.-X. Reduced nucleic acid methylation impairs meiotic maturation and developmental potency of pig oocytes. Theriogenology 2018, 121, 160–167. [Google Scholar] [CrossRef]
- Madsen, M.B.; Birck, M.M.; Fredholm, M.; Cirera, S. Expression studies of the obesity candidate gene FTO in pig. Anim. Biotechnol. 2009, 21, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Jevsinek Skok, D.; Kunej, T.; Kovac, M.; Malovrh, S.; Potocnik, K.; Petric, N.; Zgur, S.; Dovc, P.; Horvat, S. FTO gene variants are associated with growth and carcass traits in cattle. Anim. Genet. 2016, 47, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, D.; Zhang, X.; Li, F.; Xu, D.; Zhao, L.; Li, X.; Zhang, Y.; Wang, J.; Yang, X. Expression features of the ovine FTO gene and association between FTO polymorphism and tail fat deposition related-traits in Hu sheep. Gene 2022, 826, 146451. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Nie, Q.; Lamont, S.; Zhang, X. Variation in sequence and expression of the avian FTO, and association with glucose metabolism, body weight, fatness and body composition in chickens. Int. J. Obes. 2012, 36, 1054–1061. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Li, W.; Luo, H. Bioinformatic analysis and polymorphism of FTO gene in duck. Genom. Appl. Biol. 2016, 35, 2375–2379. [Google Scholar]
- Jin, J.; Xu, C.; Wu, S.; Wu, Z.; Wu, S.; Sun, M.; Bao, W. m6A Demethylase ALKBH5 Restrains PEDV Infection by Regulating GAS6 Expression in Porcine Alveolar Macrophages. Int. J. Mol. Sci. 2022, 23, 6191. [Google Scholar] [CrossRef]
- Zhang, Q.; Cheng, B.; Jiang, H.; Zhang, H.; Li, H. N 6-methyladenosine demethylase ALKBH5: A novel regulator of proliferation and differentiation of chicken preadipocytes: ALKBH5 regulated preadipocyte proliferation and differentiation. Acta Biochim. Biophys. Sin. 2022, 54, 55. [Google Scholar] [CrossRef]
- Gong, H.; Gong, T.; Liu, Y.; Wang, Y.; Wang, X. Profiling of N6-methyladenosine methylation in porcine longissimus dorsi muscle and unravelling the hub gene ADIPOQ promotes adipogenesis in an m6A-YTHDF1–dependent manner. J. Anim. Sci. Biotechnol. 2023, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ning, Y.; Abbas Raza, S.H.; Mei, C.; Zan, L. MEF2C expression is regulated by the post-transcriptional activation of the METTL3-m6A-YTHDF1 axis in myoblast differentiation. Front. Vet. Sci. 2022, 9, 900924. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ma, Y.; Li, Q.; Liu, E.; Jin, M.; Zhang, L.; Wei, C. The role of N6-methyladenosine RNA methylation in the heat stress response of sheep (Ovis aries). Cell Stress Chaperones 2019, 24, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ding, J.; Chen, M.; Chen, K.; Zou, Y.; Xu, X.; Zhang, D.; Yu, X.; Ding, Z. Transcriptome-wide N6-methyladenosine modification profiling of mRNAs during infection of Newcastle disease virus in chicken macrophages. Virus Res. 2023, 323, 198993. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhou, X.; Li, C.; Shen, C.; He, G.; Chen, T.; Cao, M.; Chen, X.; Zhang, B.; Chen, L. YTHDF2 as a Mediator in BDNF-Induced Proliferation of Porcine Follicular Granulosa Cells. Int. J. Mol. Sci. 2024, 25, 2343. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, H.; Lan, X.; Cao, X.; Pan, C. The InDel variants of sheep IGF2BP1 gene are associated with growth traits. Anim. Biotechnol. 2023, 34, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.-h.; Chen, Y.; Bai, D.-P.; Mai, L.-j.; Fan, Q.-M.; Shi, Y.-Z.; Chen, C.; Li, A. Differential expression of MSTN, IGF2BP1, and FABP2 across different embryonic ages and sexes in white Muscovy ducks. Gene 2022, 829, 146479. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, Y.; Liu, Y.; Zhang, Y.; Chen, S.; Li, G.; Wang, X.; Wang, H.; Song, J.; Gong, S. Novel IGF2BP1 splice variants, expression and their association with growth traits in goose. Br. Poult. Sci. 2022, 63, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wan, Y.; Zhang, Z.; Jiang, Y.; Gu, Z.; Ma, X.; Nie, S.; Yang, J.; Lang, J.; Cheng, W. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics 2021, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hu, H.; Tian, Y.; Li, J.; Scheben, A.; Zhang, C.; Li, Y.; Wu, J.; Yang, L.; Fan, X. The chicken pan-genome reveals gene content variation and a promoter region deletion in IGF2BP1 affecting body size. Mol. Biol. Evol. 2021, 38, 5066–5081. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.; Ma, H.; Zeng, R.; Liu, R.; Wang, P.; Jin, X.; Zhao, Y. Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol. Brain 2020, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-X.; Chen, X.; Xia, L.-P.; Zhang, J.-X.; Pan, Z.-Z.; Ma, X.-D.; Han, K.; Chen, J.-W.; Judde, J.-G.; Deas, O. N 6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Xie, Y.; Yu, T.; Liu, N.; Wang, Z.; Woolsey, R.J.; Tang, Y.; Zhang, X.; Qin, W.; Zhang, Y. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020, 30, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Romano, K.A.; Martinez-del Campo, A.; Kasahara, K.; Chittim, C.L.; Vivas, E.I.; Amador-Noguez, D.; Balskus, E.P.; Rey, F.E. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 2017, 22, 279–290.e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Chen, W.; Shi, H.; Eren, A.M.; Morozov, A.; He, C.; Luo, G.-Z.; Pan, T. Transcriptome-wide reprogramming of N6-methyladenosine modification by the mouse microbiome. Cell Res. 2019, 29, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, L.; Li, M.; Zhang, Y.; Sun, M.; Wang, L.; Lin, J.; Cui, Y.; Chen, Q.; Jin, C. Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis. Nat. Commun. 2022, 13, 1248. [Google Scholar] [CrossRef] [PubMed]
- Sorci, M.; Ianniello, Z.; Cruciani, S.; Larivera, S.; Ginistrelli, L.C.; Capuano, E.; Marchioni, M.; Fazi, F.; Fatica, A. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018, 9, 796. [Google Scholar] [CrossRef]
- Zhou, Z.; Lv, J.; Yu, H.; Han, J.; Yang, X.; Feng, D.; Wu, Q.; Yuan, B.; Lu, Q.; Yang, H. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol. Cancer 2020, 19, 104. [Google Scholar] [CrossRef]


| Type | m6A Regulator | Cellular Localization | Function | Species | Reference |
|---|---|---|---|---|---|
| m6A writers | METTL3 | Nucleus | Catalyzes m6A modification | pig, sheep, cattle, chicken, | [31,67,68,69] |
| METTL14 | Nucleus | Assists METTL3 to recognize the subtract | pig, sheep, cattle, chicken, duck | [31,68,70,71,72] | |
| METTL16 | Nucleus | Catalyzes m6A modification | pig | [73] | |
| WTAP | Nucleus | Promotes METTL3-METTL14 heterodimer to the nuclear speckle | pig, cattle, chicken, duck | [70,71,74,75] | |
| m6A erasers | FTO | Nucleus and Cytoplasm | Removes m6A modification | pig, cattle, sheep, chicken, duck | [76,77,78,79,80] |
| ALKBH5 | Nucleus | Removes m6A modification | pig, cattle, chicken | [74,81,82] | |
| m6A readers | YTHDC1 | Nucleus | Promotes RNA splicing and translocation | ||
| YTHDF1 | Cytoplasm | Promotes mRNA translation | pig, cattle, sheep, chicken | [83,84,85,86] | |
| YTHDF2 | Cytoplasm | Reduces mRNA stability | pig, cattle, sheep, chicken | [84,85,86,87] | |
| YTHDF3 | Cytoplasm | Mediates the translation or degradation | |||
| YTHDC2 | Cytoplasm | Enhances the translation of target RNA | |||
| IGF2BP1/2/3 | Nucleus and Cytoplasm | Enhances mRNA stability | pig, sheep, chicken, duck, goose | [27,44,88,89,90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, T.; Xu, M.; Du, X.; Wang, Y.; Loor, J.J.; Lei, L.; Gao, W.; Du, X.; Song, Y.; Liu, G.; et al. Research Progress on the Role of M6A in Regulating Economic Traits in Livestock. Int. J. Mol. Sci. 2024, 25, 8365. https://doi.org/10.3390/ijms25158365
Ren T, Xu M, Du X, Wang Y, Loor JJ, Lei L, Gao W, Du X, Song Y, Liu G, et al. Research Progress on the Role of M6A in Regulating Economic Traits in Livestock. International Journal of Molecular Sciences. 2024; 25(15):8365. https://doi.org/10.3390/ijms25158365
Chicago/Turabian StyleRen, Tuanhui, Meng Xu, Xinyu Du, Yanxi Wang, Juan J. Loor, Lin Lei, Wenwen Gao, Xiliang Du, Yuxiang Song, Guowen Liu, and et al. 2024. "Research Progress on the Role of M6A in Regulating Economic Traits in Livestock" International Journal of Molecular Sciences 25, no. 15: 8365. https://doi.org/10.3390/ijms25158365
APA StyleRen, T., Xu, M., Du, X., Wang, Y., Loor, J. J., Lei, L., Gao, W., Du, X., Song, Y., Liu, G., & Li, X. (2024). Research Progress on the Role of M6A in Regulating Economic Traits in Livestock. International Journal of Molecular Sciences, 25(15), 8365. https://doi.org/10.3390/ijms25158365










