Antimicrobial Susceptibility and Genetic Epidemiology of Extended-Spectrum β-Lactamase-Positive Enterobacterales Clinical Isolates in Central Poland
Abstract
:1. Introduction
2. Results
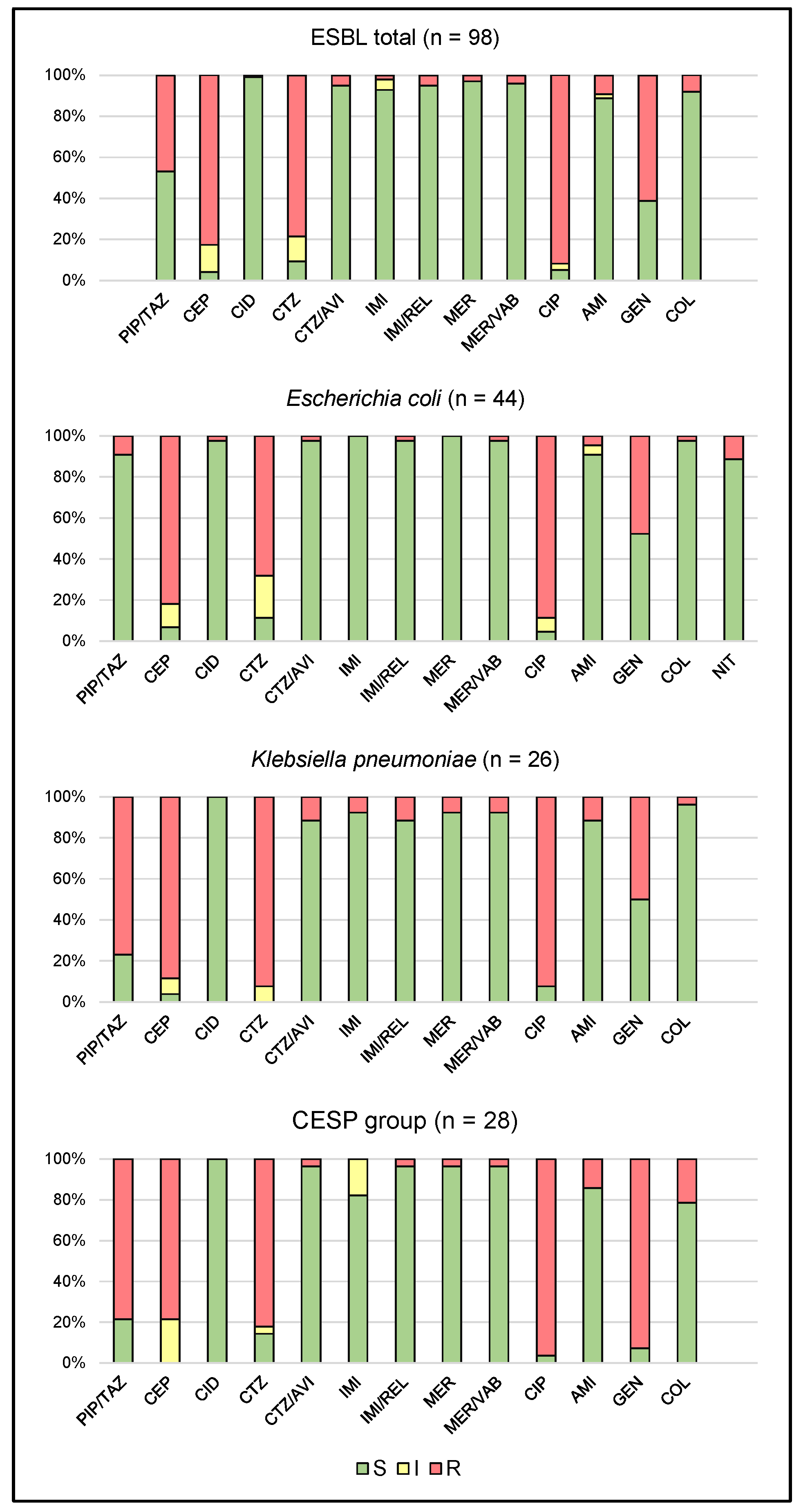
2.1. Antimicrobial Susceptibility
2.2. Genetic Epidemiology
3. Discussion
3.1. Antimicrobial Susceptibility
3.2. Genetic Epidemiology
4. Materials and Methods
4.1. Ethical Issues
4.2. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P.; Coulson, A.F.; Frère, J.M.; Ghuysen, J.M.; Joris, B.; Forsman, M.; Levesque, R.C.; Tiraby, G.; Waley, S.G. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 1991, 276, 269–270. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Hächler, H.; Nüesch-Inderbinen, M.; Stephan, R. Characteristics of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl. Environ. Microbiol. 2013, 79, 3021–3026. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Knothe, H.; Shah, P.; Krcmery, V.; Antal, M.; Mitsuhashi, S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 1983, 11, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Novais, A.; Valverde, A.; Machado, E.; Peixe, L.; Baquero, F.; Coque, T.M. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 144–153. [Google Scholar] [CrossRef] [PubMed]
- Empel, J.; Baraniak, A.; Literacka, E.; Mrówka, A.; Fiett, J.; Sadowy, E.; Hryniewicz, W.; Gniadkowski, M. Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob. Agents Chemother. 2008, 52, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 90–103. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. β-Lactams and β-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Onduru, O.G.; Aboud, S.; Nyirenda, T.S.; Rumisha, S.F.; Mkakosya, R.S. Antimicrobial susceptibility testing profiles of ESBL-producing Enterobacterales isolated from hospital and community adult patients in Blantyre, Malawi. IJID Reg. 2021, 1, 47–52. [Google Scholar] [CrossRef] [PubMed]
- WHO/ECDC, Antimicrobial Resistance Surveillance in Europe 2022–2020 Data. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Joint-WHO-ECDC-AMR-report-2022.pdf (accessed on 10 May 2024).
- Talan, D.A.; Takhar, S.S.; Krishnadasan, A.; Abrahamian, F.M.; Mower, W.R.; Moran, G.J. EMERGEncy ID Net Study Group. Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli infections in patients with pyelonephritis, United States. Emerg. Infect. Dis. 2016, 22, 1594–1603. [Google Scholar] [CrossRef] [PubMed]
- Yahav, D.; Giske, C.G.; Grāmatniece, A.; Abodakpi, H.; Tam, V.H.; Leibovici, L. New β-lactam-β-lactamase inhibitor combinations. Clin. Microbiol. Rev. 2020, 34, e00115-20. [Google Scholar] [CrossRef] [PubMed]
- Kaye, K.S.; Naas, T.; Pogue, J.M.; Rossolini, G.M. Cefiderocol, a siderophore cephalosporin as a treatment option for infections caused by carbapenem-resistant Enterobacterales. Infect. Dis. Ther. 2023, 12, 777–806. [Google Scholar] [CrossRef] [PubMed]
- Boyd, S.E.; Livermore, D.M.; Hooper, D.C.; Hope, W.W. Metallo-β-lactamases: Structure, function, epidemiology, treatment options and the development pipeline. Antimicrob. Agents Chemother. 2020, 64, e00397-20. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef]
- Bassetti, M.; Righi, E. New antibiotics and antimicrobial combination therapy for the treatment of gram-negative bacterial infections. Curr. Opin. Crit. Care 2015, 21, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Caldes, C.; Cisneros-Vásquez, E.; Zambrano, A.; Mosquera-Maza, A.; Calero-Cáceres, W.; Rey, J.; Yamamoto, Y.; Yamamoto, M.; Calvopina, M.; de Waard, J.H. Co-harboring of beta-lactamases and mcr-1 genes in Escherichia coli and Klebsiella pneumoniae from healthy carriers and backyard animals in rural communities in Ecuador. Antibiotics 2023, 12, 856. [Google Scholar] [CrossRef] [PubMed]
- Masoud, S.M.; Abd El-Baky, R.M.; Aly, S.A.; Ibrahem, R.A. Co-existence of certain ESBLs, MBLs and plasmid mediated quinolone resistance genes among MDR E. coli isolated from different clinical specimens in Egypt. Antibiotics 2021, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Kazemian, H.; Heidari, H.; Ghanavati, R.; Ghafourian, S.; Yazdani, F.; Sadeghifard, N.; Pakzad, I. Phenotypic and genotypic characterization of ESBL-, AmpC-, and carbapenemase-producing Klebsiella pneumoniae and Escherichia coli isolates. Med. Princ. Pract. 2019, 28, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Irfan, S.; Azhar, A.; Bashir, A.; Ahmed, S.; Haque, A. High frequency of simultaneous presence of ESBL and carbapenemase producers among nosocomial coliform isolates in Faisalabad, Pakistan. Pak. J. Med. Sci. 2021, 37, 34–39. [Google Scholar] [CrossRef]
- Deku, J.G.; Duedu, K.O.; Ativi, E.; Kpene, G.E.; Feglo, P.K. Occurrence and distribution of extended-spectrum β-lactamase in clinical Escherichia coli isolates at Ho Teaching Hospital in Ghana. Ghana. Med. J. 2021, 55, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Caldes, C.; Guerrero-Freire, S.; Ortuño-Gutiérrez, N.; Sunyoto, T.; Gomes-Dias, C.A.; Ramírez, M.S.; Calero-Caceres, W.; Harries, A.D.; Rey, J.; de Waard, J.H.; et al. Colistin resistance in Escherichia coli and Klebsiella pneumoniae in humans and backyard animals in Ecuador. Rev. Panam. Salud Publica 2023, 47, e48. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Ver. 14.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_14.0_Breakpoint_Tables.pdf (accessed on 10 May 2024).
- Moghaddam, M.N.; Beidokhti, M.H.; Jamehdar, S.A.; Ghahraman, M. Genetic properties of blaCTX-M. and blaPER β-lactamase genes in clinical isolates of Enterobacteriaceae by polymerase chain reaction. Iran. J. Basic. Med. Sci. 2014, 17, 378–383. [Google Scholar] [PubMed]
- Ejaz, H. Dissemination of SHV, TEM and CTX-M. genotypes in Pseudomonas aeruginosa: A pre-eminent reason for therapeutic failure in pediatrics. Ann. Clin. Lab. Sci. 2020, 50, 797–805. [Google Scholar] [PubMed]
- Amirkamali, S.; Naserpour-Farivar, T.; Azarhoosh, K.; Peymani, A. Distribution of the blaOXA, blaVEB-1, and blaGES-1 genes and resistance patterns of ESBL-producing Pseudomonas aeruginosa isolated from hospitals in Tehran and Qazvin, Iran. Rev. Soc. Bras. Med. Trop. 2017, 50, 315–320. [Google Scholar] [CrossRef] [PubMed]

| Gene or Region | Primers (5′ → 3′) | Amplicon Size [bp] | Reference |
|---|---|---|---|
| blaCTX-M | F: ATGTGCACCAGTAARGT R: TGGGTRAARTARGTSACCAGA | 593 | [25] |
| blaSHV | F: ATTTGTCGCTTCTTTACTCGCC R: TTCACCACCATCATTACCGACC | 1027 | [26] |
| blaTEM | F: GTGCGCGGAACCCCTATT R: GGGATTTTGGTCATGAGATTATC | 1083 | [26] |
| blaPER | F: AATTTGGGCTTAGGGCAGAA R: ATGAATGTCATTATAAAAGC | 924 | [26] |
| blaVEB | F: CGACTTCCATTTCCCGATGC R: GGACTCTGCAACAAATACGC | 642 | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brauncajs, M.; Bielec, F.; Macieja, A.; Machnicki, P.; Pastuszak-Lewandoska, D. Antimicrobial Susceptibility and Genetic Epidemiology of Extended-Spectrum β-Lactamase-Positive Enterobacterales Clinical Isolates in Central Poland. Int. J. Mol. Sci. 2024, 25, 8371. https://doi.org/10.3390/ijms25158371
Brauncajs M, Bielec F, Macieja A, Machnicki P, Pastuszak-Lewandoska D. Antimicrobial Susceptibility and Genetic Epidemiology of Extended-Spectrum β-Lactamase-Positive Enterobacterales Clinical Isolates in Central Poland. International Journal of Molecular Sciences. 2024; 25(15):8371. https://doi.org/10.3390/ijms25158371
Chicago/Turabian StyleBrauncajs, Małgorzata, Filip Bielec, Anna Macieja, Piotr Machnicki, and Dorota Pastuszak-Lewandoska. 2024. "Antimicrobial Susceptibility and Genetic Epidemiology of Extended-Spectrum β-Lactamase-Positive Enterobacterales Clinical Isolates in Central Poland" International Journal of Molecular Sciences 25, no. 15: 8371. https://doi.org/10.3390/ijms25158371





