The Phylogenetic Relationships of Major Lizard Families Using Mitochondrial Genomes and Selection Pressure Analyses in Anguimorpha
Abstract
1. Introduction
2. Results
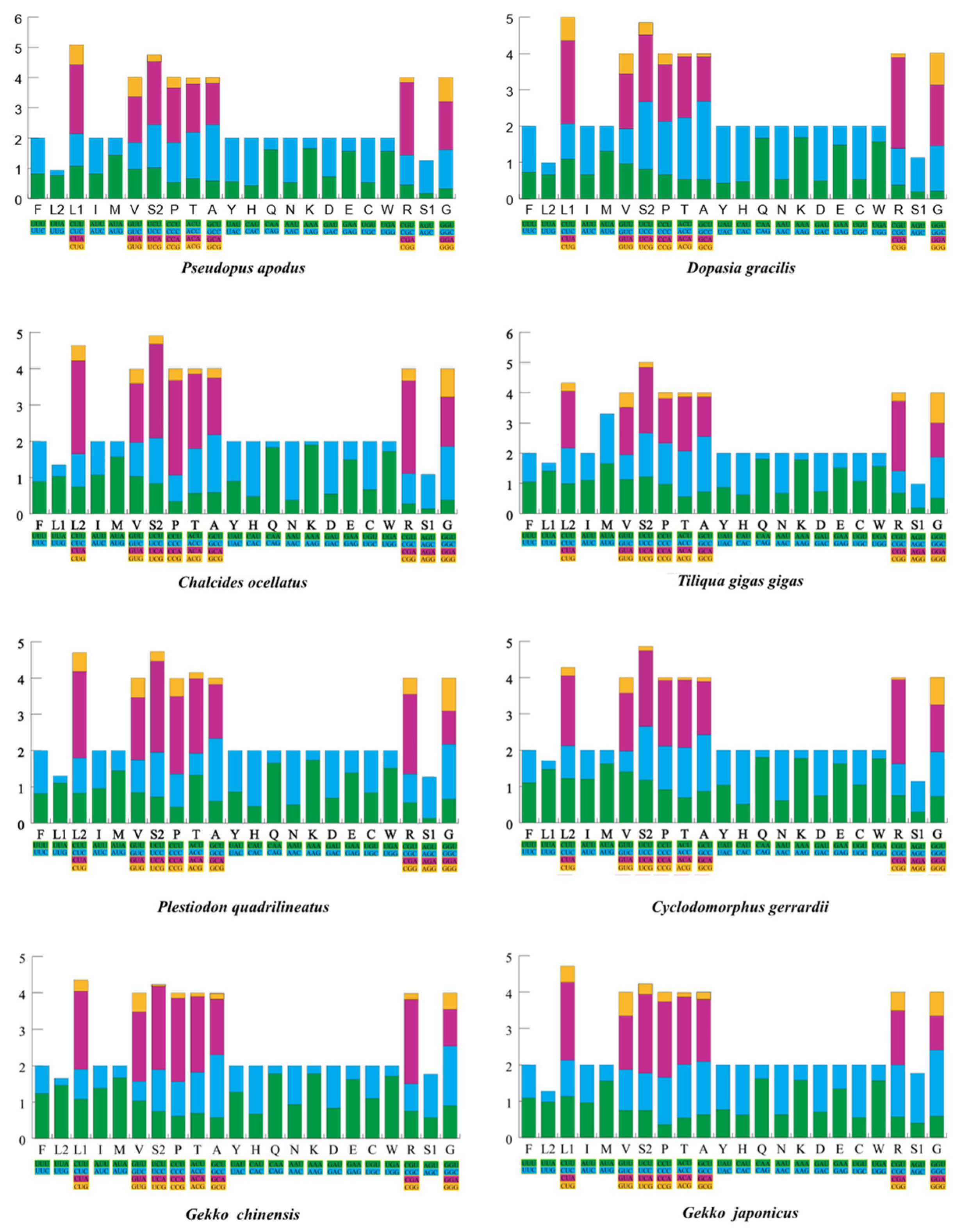
2.1. Basic Features of Mitogenomes
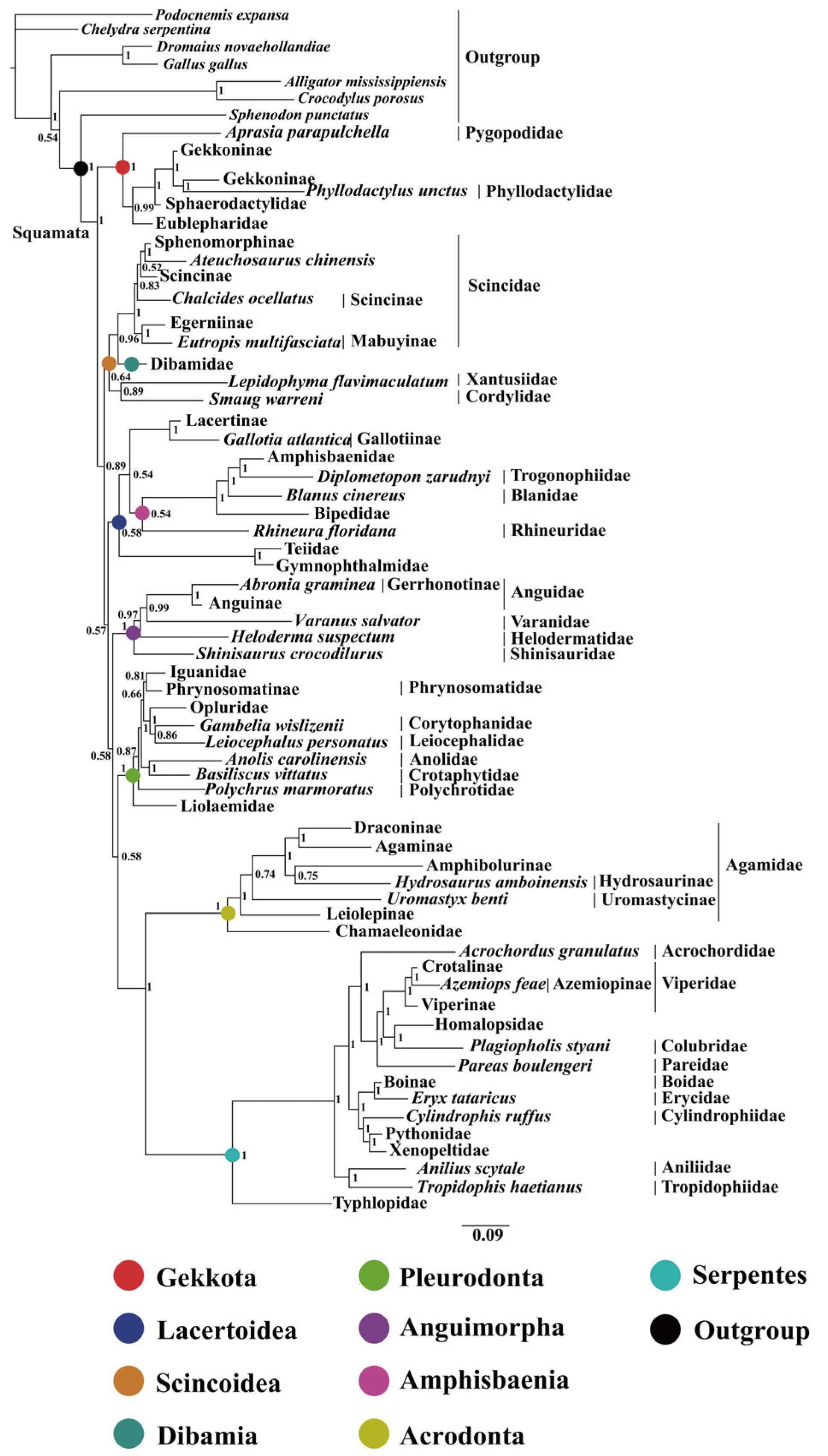
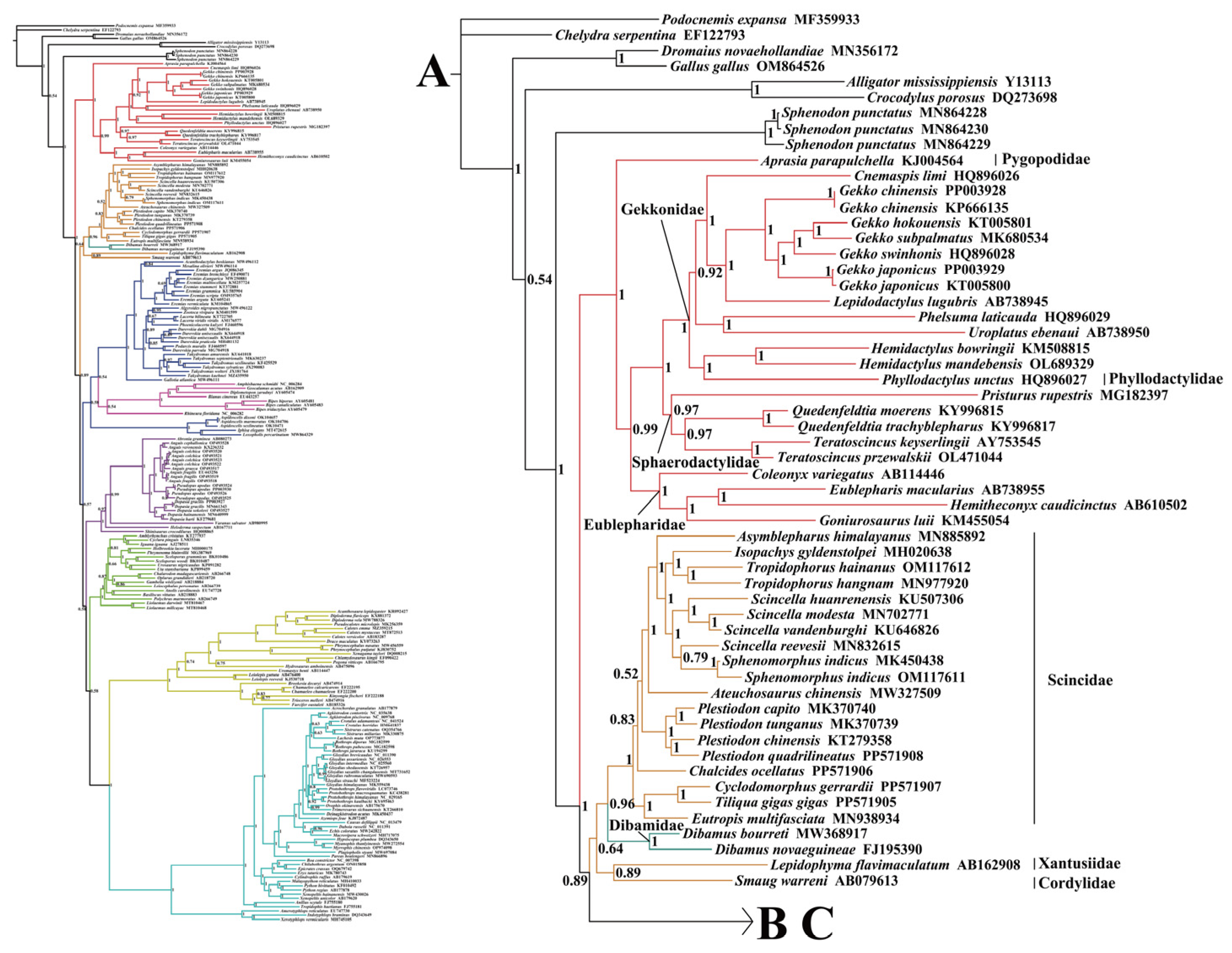
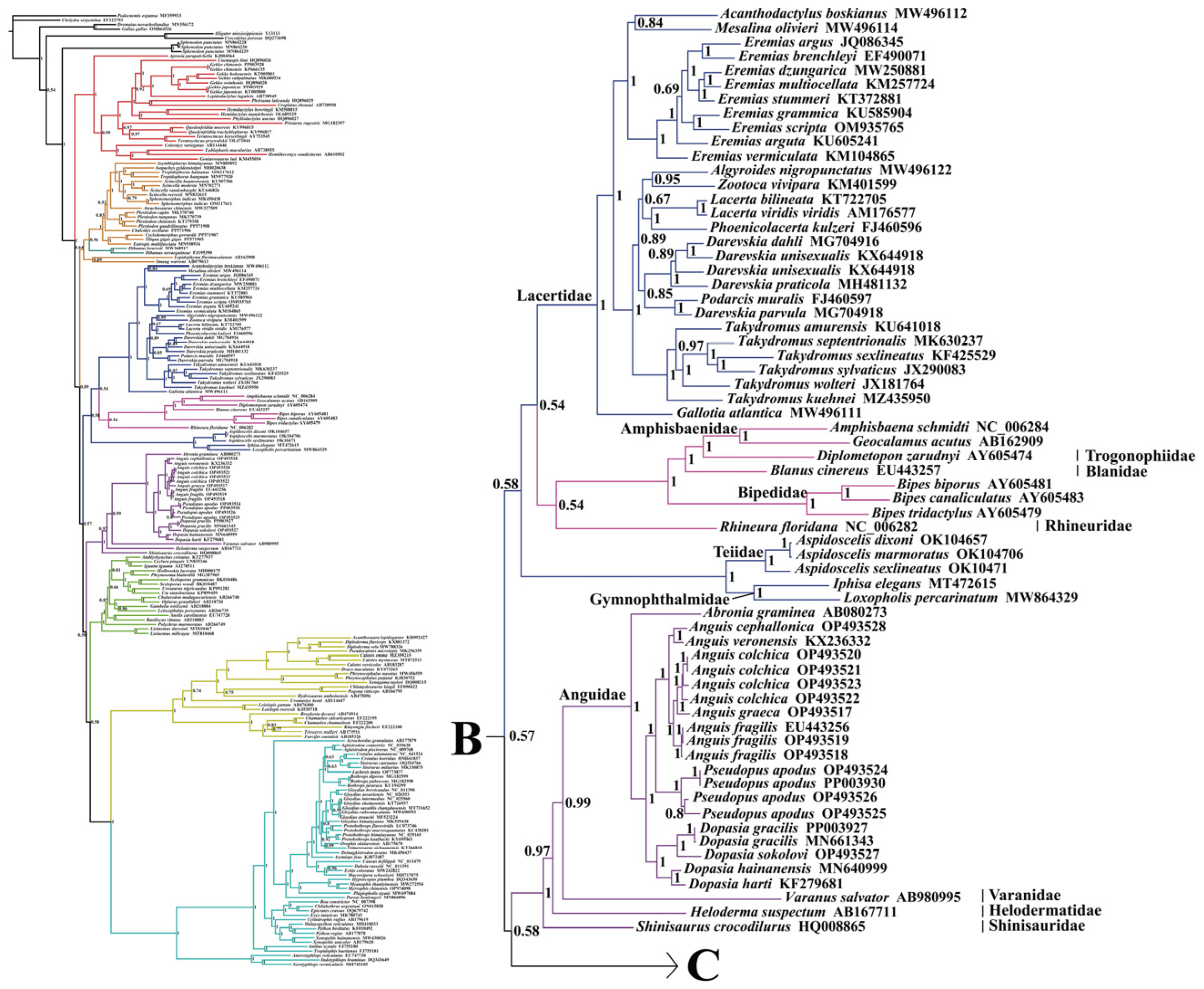
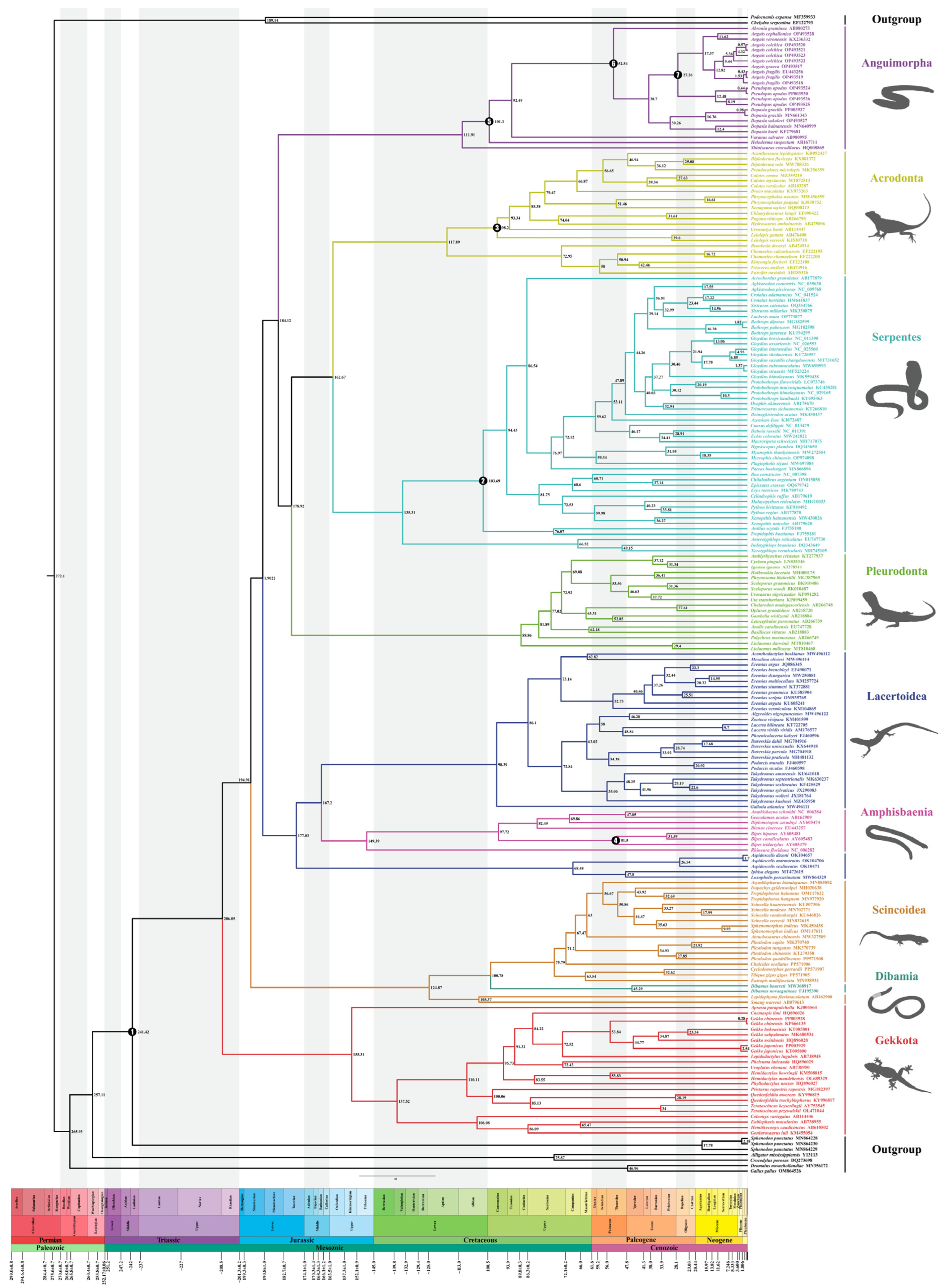
2.2. Phylogenetic Relationships
2.3. Divergence Time Estimation
2.4. Detecting Selective Pressure Within Anguimorpha
3. Discussion
3.1. Comparison of Phylogenetic Relationships in Squamates Based on the Mitogenome
3.2. Analysis of the Phylogenetic Relationships within Anguimorpha
3.3. Analysis of Divergence Time Estimation
3.4. Evolutionary History of Species in Anguinae
3.5. Selective Pressure within Anguimorpha
4. Materials and Methods
4.1. Sample Collection and DNA Extraction
4.2. PCR Amplification and Sequence Capture
4.3. Mitogenome Annotation and Sequence Analyses
4.4. Phylogenetic Analyses
4.5. Divergence Dating Estimation
4.6. Detecting Selective Pressure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uetz, P.; Koo, M.; Aguilar, R.; Brings, E.; Catenazzi, A.; Chang, A.; Chaitanya, R.; Freed, P.; Gross, J.; Hammermann, M. A quarter century of reptile and amphibian databases. Herpetol. Rev. 2021, 52, 246–255. [Google Scholar]
- Pianka, E.R.; Vitt, L.J. Lizards: Windows to the Evolution of Diversity; University of California Press: Berkeley, CA, USA, 2003; Volume 5, p. 346. [Google Scholar]
- Zug, G.R.; Vitt, L.; Caldwell, J.P. Herpetology: An Introductory Biology of Amphibians and Reptiles; Academic Press: San Diego, CA, USA; New York, NY, USA, 2001; p. 630. [Google Scholar]
- Camp, C.L. Classification of the Lizards; Order of the Trustees, American Museum of Natural History: New York, NY, USA, 1923. [Google Scholar]
- Estes, R.; de Queiroz, K.; Gauthier, J. Phylogenetic Relationships within Squamata; Stanford University Press: Redwood City, CA, USA, 1988; pp. 119–270. [Google Scholar]
- Evans, S.E.; Wang, Y. The early cretaceous lizard Dalinghosaurus from China. Acta Palaeontol. Pol. 2005, 50, 725–742. [Google Scholar]
- Conrad, J.L. Phylogeny and systematics of squamata (reptilia) based on morphology. Bull. Am. Mus. Nat. Hist. 2008, 310, 1–182. [Google Scholar] [CrossRef]
- Gauthier, J.A.; Kearney, M.; Maisano, J.A.; Rieppel, O.; Behlke, A.R. Assembling the Squamate Tree of Life: Perspectives from the Phenotype and the Fossil Record. Bull. Peabody Mus. Nat. Hist. 2012, 53, 3–308. [Google Scholar] [CrossRef]
- Pyron, R.A. Novel Approaches for Phylogenetic Inference from Morphological Data and Total-Evidence Dating in Squamate Reptiles (Lizards, Snakes, and Amphisbaenians). Syst. Biol. 2017, 66, 38–56. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.; Hedges, S.B. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. Comptes Rendus Biol. 2009, 332, 129–139. [Google Scholar] [CrossRef]
- Simões, T.R.; Caldwell, M.W.; Talanda, M.; Bernardi, M.; Palci, A.; Vernygora, O.; Bernardini, F.; Mancini, L.; Nydam, R.L. The origin of squamates revealed by a Middle Triassic lizard from the Italian Alps. Nature 2018, 557, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Pereira, S.L. Mitochondrial genome organization and vertebrate phylogenetics. Genet. Mol. Biol. 2000, 23, 745–752. [Google Scholar] [CrossRef]
- Saccone, C.; De Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Brown, W.M.; George Jr, M.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Pichaud, N. Mitochondrial DNA: More than an evolutionary bystander. Funct. Ecol. 2014, 28, 218–231. [Google Scholar] [CrossRef]
- Chong, R.A.; Mueller, R.L. Low metabolic rates in salamanders are correlated with weak selective constraints on mitochondrial genes. Evolution 2013, 67, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sun, J.; Xu, T.; Qiu, J.W.; Qian, P.Y. Phylogenetic relationships and adaptation in deep-sea mussels: Insights from mitochondrial genomes. Int. J. Mol. Sci. 2021, 22, 1900. [Google Scholar] [CrossRef]
- Li, N.; Yang, W.Z.; Fu, J.Z. High-altitude adaptation of genus phrynocephalus based on mitochondrial genome. Sichuan J. Zool. 2015, 34, 810–816. [Google Scholar]
- Hong, Y.H.; Huang, H.M.; Wu, L.; Storey, K.B.; Zhang, J.Y.; Zhang, Y.P.; Yu, D.N. Characterization of two mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with phylogenetic relationships and selection pressure analyses of Hylidae. Animals 2023, 13, 1593. [Google Scholar] [CrossRef]
- Wu, L.; Tong, Y.; Ayivi, S.P.G.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. The complete mitochondrial genomes of three Sphenomorphinae species (Squamata: Scincidae) and the selective pressure analysis on mitochondrial genomes of limbless Isopachys gyldenstolpei. Animals 2022, 12, 2015. [Google Scholar] [CrossRef]
- Georgalis, G.L. Necrosaurus or Palaeovaranus? Appropriate nomenclature and taxonomic content of an enigmatic fossil lizard clade (Squamata). Ann. Paleontol. 2017, 103, 293–303. [Google Scholar] [CrossRef]
- Georgalis, G.L.; Scheyer, T.M. A new species of Palaeopython (Serpentes) and other extinct squamates from the Eocene of Dielsdorf (Zurich, Switzerland). Swiss J. Geosci. 2019, 112, 383–417. [Google Scholar] [CrossRef]
- Kearney, M.; Rieppel, O. An investigation into the occurrence of plicidentine in the teeth of squamate reptiles. Copeia 2006, 2006, 337–350. [Google Scholar] [CrossRef]
- Palci, A.; LeBlanc, A.R.; Panagiotopoulou, O.; Cleuren, S.G.; Mehari Abraha, H.; Hutchinson, M.N.; Evans, A.R.; Caldwell, M.W.; Lee, M.S. Plicidentine and the repeated origins of snake venom fangs. Proc. R. Soc. B 2021, 288, 20211391. [Google Scholar] [CrossRef] [PubMed]
- Georgalis, G.L.; Mennecart, B.; Smith, K.T. First fossil record of Varanus (Reptilia, Squamata) from Switzerland and the earliest occurrences of the genus in Europe. Swiss J. Geosci. 2023, 116, 9. [Google Scholar] [CrossRef]
- Georgalis, G.L.; Čerňanský, A.; Klembara, J. Osteological atlas of new lizards from the Phosphorites du Quercy (France), based on historical, forgotten, fossil material. Geodiversitas 2021, 43, 219–293. [Google Scholar] [CrossRef]
- Čerňanský, A.; Tabuce, R.; Vidalenc, D. Anguimorph lizards from the lower Eocene (MP 10–11) of the Cos locality, Phosphorites du Quercy, France, and the early evolution of Glyptosaurinae in Europe. J. Vertebr. Paleontol. 2022, 42, e2211646. [Google Scholar] [CrossRef]
- Zheng, Y.; Wiens, J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016, 94, 537–547. [Google Scholar] [CrossRef] [PubMed]
- McConkey, E.H. A systematic study of the North American lizards of the genus Ophisaurus. Am. Midl. Nat. 1954, 51, 133–171. [Google Scholar] [CrossRef]
- Pough, F.H.; Andrews, R.; Cadle, J.; Crump, M.; Savitzky, A.; Wells, K. Herpetology; Prentice Hall: Upper Saddle River, NJ, USA, 1998; p. 577. [Google Scholar]
- Greenwood, D.R.; Wing, S.L. Eocene continental climates and latitudinal temperature gradients. Geology 1995, 23, 1044–1048. [Google Scholar] [CrossRef]
- Pross, J.; Contreras, L.; Bijl, P.K.; Greenwood, D.R.; Bohaty, S.M.; Schouten, S.; Bendle, J.A.; Röhl, U.; Tauxe, L.; Raine, J.I. Persistent near-tropical warmth on the Antarctic continent during the early Eocene epoch. Nature 2012, 488, 73–77. [Google Scholar] [CrossRef]
- Čerňanský, A.; Daza, J.D.; Smith, R.; Bauer, A.M.; Smith, T.; Folie, A. A new gecko from the earliest Eocene of Dormaal, Belgium: A thermophilic element of the ‘greenhouse world’. R. Soc. Open Sci. 2022, 9, 220429. [Google Scholar] [CrossRef]
- Georgalis, G.L.; Rabi, M.; Smith, K.T. Taxonomic revision of the snakes of the genera Palaeopython and Paleryx (Serpentes, Constrictores) from the Paleogene of Europe. Swiss J. Palaeontol. 2021, 140, 18. [Google Scholar] [CrossRef]
- Smith, K.T. A new lizard assemblage from the earliest Eocene (zone Wa0) of the Bighorn Basin, Wyoming, USA: Biogeography during the warmest interval of the Cenozoic. J. Syst. Palaeontol. 2009, 7, 299–358. [Google Scholar] [CrossRef]
- Smith, K.T.; Georgalis, G.L. The Diversity and Distribution of Palaeogene Snakes; Cambridge University Press: Cambridge, UK, 2022; Volume 90. [Google Scholar]
- Eberle, J.J.; Greenwood, D.R. Life at the top of the greenhouse Eocene world- A review of the Eocene flora and vertebrate fauna from Canada’s High Arctic. Bull. Geol. Soc. Am. 2012, 124, 3–23. [Google Scholar] [CrossRef]
- Williams, C.J.; LePage, B.A.; Johnson, A.H.; Vann, D.R. Structure, Biomass, and Productivity of a Late Paleocene Arctic Forest. Proc. Acad. Nat. Sci. USA 2009, 158, 107–127. [Google Scholar] [CrossRef]
- Greenwood, D.R.; Basinger, J.F. The paleoecology of high-latitude Eocene swamp forests from Axel Heiberg Island, Canadian High Arctic. Rev. Palaeobot. Palynol. 1994, 81, 83–97. [Google Scholar] [CrossRef]
- Maxbauer, D.P.; Royer, D.L.; LePage, B.A. High Arctic forests during the middle Eocene supported by moderate levels of atmospheric CO2. Geology 2014, 42, 1027–1030. [Google Scholar] [CrossRef]
- Strömberg, C.A.E. Evolution of grasses and grassland ecosystems. Annu. Rev. Earth Planet. Sci. 2011, 39, 517–544. [Google Scholar] [CrossRef]
- Lavin, B.R.; Girman, D.J. Phylogenetic relationships and divergence dating in the glass lizards (Anguinae). Mol. Phylogenet. Evol. 2019, 133, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Klembara, J.; Böhme, M.; Rummel, M. Revision of the anguine lizard Pseudopus laurillardi (Squamata, Anguidae) from the Miocene of Europe, with comments on paleoecology. J. Paleont. 2010, 84, 159–196. [Google Scholar] [CrossRef]
- Klembara, J. A new species of Pseudopus (Squamata, Anguidae) from the early Miocene of Northwest Bohemia (Czech Republic). J. Vertebr. Paléontol. 2012, 32, 854–866. [Google Scholar] [CrossRef]
- Augé, M.L. Evolution des lézards du Paléogène en Europe; Mémoires du Muséum National d’Histoire Naturelle: Paris, France, 2005; Volume 192, pp. 1–369. [Google Scholar]
- Klembara, J.; Hain, M.; Dobiašová, K. Comparative Anatomy of the Lower Jaw and Dentition of Pseudopus apodus and the Interrelationships of Species of Subfamily Anguinae (Anguimorpha, Anguidae). Anat. Rec. 2014, 297, 516–544. [Google Scholar] [CrossRef] [PubMed]
- Georgalis, G.L.; Villa, A.; Delfino, M. Fossil lizards and snakes from Ano Metochi—A diverse squamate fauna from the latest Miocene of northern Greece. Hist. Biol. 2016, 29, 730–742. [Google Scholar] [CrossRef]
- Georgalis, G.L.; Villa, A.; Ivanov, M.; Vasilyan, D.; Delfino, M. Fossil amphibians and reptiles from the Neogene locality of Maramena (Greece), the most diverse European herpetofauna at the Miocene/Pliocene transition boundary. Palaeontol. Electron. 2019, 22, 1–99. [Google Scholar] [CrossRef] [PubMed]
- Klembara, J.; Rummel, M. New material of Ophisaurus, Anguis and Pseudopus (Squamata, Anguidae, Anguinae) from the Miocene of the Czech Republic and Germany and systematic revision and palaeobiogeography of the Cenozoic Anguinae. Geol. Mag. 2018, 155, 20–44. [Google Scholar] [CrossRef]
- Klembara, J.; Čerňanský, A. Revision of the cranial anatomy of Ophisaurus acuminatus Jörg, 1965 (Anguimorpha, Anguidae) from the late Miocene of Germany. Geodiversitas 2020, 42, 539–557. [Google Scholar] [CrossRef]
- Čerňanský, A.; Klembara, J. A skeleton of Ophisaurus (Squamata: Anguidae) from the middle Miocene of Germany, with a revision of the partly articulated postcranial material from Slovakia using micro-computed tomography. J. Vertebr. Paleontol. 2017, 37, e1333515. [Google Scholar] [CrossRef]
- Klembara, J. New finds of anguines (Squamata, Anguidae) from the Early Miocene of Northwest Bohemia (Czech Republic). PalZ 2014, 89, 171–195. [Google Scholar] [CrossRef]
- Holman, J.A. Herpetofauna of the Wood Mountain Formation (Upper Miocene) of Saskatchewan. Can. J. Earth Sci. 1970, 7, 1317–1325. [Google Scholar] [CrossRef]
- Čerňanský, A.; Vasilyan, D.; Georgalis, G.L.; Joniak, P.; Mayda, S.; Klembara, J. First record of fossil anguines (Squamata; Anguidae) from the Oligocene and Miocene of Turkey. Swiss J. Geosci. 2017, 110, 741–751. [Google Scholar] [CrossRef]
- Escalante, I.; Ellis, V.R.; Elias, D.O. Leg loss decreases endurance and increases oxygen consumption during locomotion in harvestmen. J. Comp. Physiol. A 2021, 207, 257–268. [Google Scholar] [CrossRef]
- Gans, C. Tetrapod limblessness: Evolution and functional corollaries. Am. Zool. 1975, 15, 455–467. [Google Scholar] [CrossRef]
- Mehta, R.S.; Wainwright, P.C. Functional morphology of the pharyngeal jaw apparatus in moray eels. Integr. Comp. Biol. 2010, 50, 1091–1105. [Google Scholar] [CrossRef]
- Sharpe, S.S.; Koehler, S.A.; Kuckuk, R.M.; Serrano, M.; Vela, P.A.; Mendelson III, J.; Goldman, D.I. Locomotor benefits of being a slender and slick sand swimmer. J. Exp. Biol. 2015, 218, 1111. [Google Scholar] [CrossRef] [PubMed]
- Morinaga, G.; Bergmann, P.J. Evolution of fossorial locomotion in the transition from tetrapod to snake-like in lizards. Proc. Biol. Sci. 2020, 287, 20200192. [Google Scholar] [CrossRef] [PubMed]
- Georgalis, G.; Smith, K. Constrictores Oppel, 1811–the available name for the taxonomic group uniting boas and pythons. Vertebr. Zool. 2020, 70, 291–304. [Google Scholar]
- Vidal, N.; Delmas, A.S.; Hedges, S.B.; Henderson, R.W.; Powell, R. The Higher-Level Relationships of Alethinophidian Snakes Inferred from Seven Nuclear and Mitochondrial Genes; Henderson, R.W., Powell, R., Eds.; Eagle Mountain Publishing: Eagle Mountain, UT, USA, 2007. [Google Scholar]
- Burbrink, F.T.; Grazziotin, F.G.; Pyron, R.A.; Cundall, D.; Donnellan, S.; Irish, F.; Keogh, J.S.; Kraus, F.; Murphy, R.W.; Noonan, B.; et al. Interrogating Genomic-Scale Data for Squamata (Lizards, Snakes, and Amphisbaenians) Shows no Support for Key Traditional Morphological Relationships. Syst. Biol. 2020, 69, 502–520. [Google Scholar] [CrossRef]
- Joyce, W.G.; Anquetin, J.; Cadena, E.-A.; Claude, J.; Danilov, I.G.; Evers, S.W.; Ferreira, G.S.; Gentry, A.D.; Georgalis, G.L.; Lyson, T.R. A nomenclature for fossil and living turtles using phylogenetically defined clade names. Swiss J. Palaeontol. 2021, 140, 5. [Google Scholar] [CrossRef]
- Townsend, T.; Larson, A.; Louis, E.; Macey, J.R. Molecular phylogenetics of squamata: The position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 2004, 53, 735–757. [Google Scholar] [CrossRef]
- Bohme, M.U.; Fritzsch, G.; Tippmann, A.; Schlegel, M.; Berendonk, T.U. The complete mitochondrial genome of the green lizard Lacerta viridis viridis (Reptilia: Lacertidae) and its phylogenetic position within squamate reptiles. Gene 2007, 394, 69–77. [Google Scholar] [CrossRef]
- Albert, E.M.; San Mauro, D.; García-París, M.; Rüber, L.; Zardoya, R. Effect of taxon sampling on recovering the phylogeny of squamate reptiles based on complete mitochondrial genome and nuclear gene sequence data. Gene 2009, 441, 12–21. [Google Scholar] [CrossRef]
- Kumazawa, Y. Mitochondrial genomes from major lizard families suggest their phylogenetic relationships and ancient radiations. Gene 2007, 388, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Okajima, Y.; Kumazawa, Y. Mitochondrial genomes of acrodont lizards: Timing of gene rearrangements and phylogenetic and biogeographic implications. BMC Evol. Biol. 2010, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Douglas, D.A.; Janke, A.; Arnason, U. A mitogenomic study on the phylogenetic position of snakes. Zool. Scr. 2010, 35, 545–558. [Google Scholar] [CrossRef]
- Simões, T.R.; Vernygora, O.; Caldwell, M.W.; Pierce, S.E. Megaevolutionary dynamics and the timing of evolutionary innovation in reptiles. Nat. Commun. 2020, 11, 3322. [Google Scholar] [CrossRef]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.J.; Manuel, M.; Wörheide, G.; Baurain, D. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011, 9, e1000602. [Google Scholar] [CrossRef]
- Saint, K.M.; Austin, C.C.; Donnellan, S.C.; Hutchinson, M.N. C-mos, a nuclear marker useful for squamate phylogenetic analysis. Mol. Phylogenet. Evol. 1998, 10, 259–263. [Google Scholar] [CrossRef]
- Vidal, N.; Hedges, S.B. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. Comptes Rendus Biol. 2005, 328, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Douglas, M.R.; Schuett, G.W.; Beck, D.D.; Sullivan, B.K. Conservation phylogenetics of helodermatid lizards using multiple molecular markers and a supertree approach. Mol. Phylogenet. Evol. 2010, 55, 153–167. [Google Scholar] [CrossRef]
- Dong, S.; Kumazawa, Y. Complete mitochondrial DNA sequences of six snakes: Phylogenetic relationships and molecular evolution of genomic features. J. Mol. Evol. 2005, 61, 12–22. [Google Scholar] [CrossRef]
- Zhou, K.; Li, H.; Han, D.; Bauer, A.M.; Feng, J. The complete mitochondrial genome of Gekko gecko (Reptilia: Gekkonidae) and support for the monophyly of Sauria including Amphisbaenia. Mol. Phylogenet. Evol. 2006, 40, 887–892. [Google Scholar] [CrossRef]
- Douglas, D.A.; Arnason, U. Examining the utility of categorical models and alleviating artifacts in phylogenetic reconstruction of the Squamata (Reptilia). Mol. Phylogenet. Evol. 2009, 52, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Rui, J.; Wang, Y.; Nie, L. The complete mitochondrial DNA genome of Eremias brenchleyi (Reptilia: Lacertidae) and its phylogeny position within squamata reptiles. Amphibia-Reptilia 2009, 30, 25–35. [Google Scholar] [CrossRef]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Tiu, J. Highly Incomplete Taxa Can Rescue Phylogenetic Analyses from the Negative Impacts of Limited Taxon Sampling. PLoS ONE 2012, 7, e42925. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Burbrink, F.T. Early origin of viviparity and multiple reversions to oviparity in squamate reptiles. Ecol. Lett. 2014, 17, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Tałanda, M.; Fernandez, V.; Panciroli, E.; Evans, S.E.; Benson, R.J. Synchrotron tomography of a stem lizard elucidates early squamate anatomy. Nature 2022, 611, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Okajima, Y.; Kumazawa, Y. Mitogenomic perspectives into iguanid phylogeny and biogeography: Gondwanan vicariance for the origin of Madagascan oplurines. Gene 2009, 441, 28–35. [Google Scholar] [CrossRef]
- Mulcahy, D.G.; Noonan, B.P.; Moss, T.; Townsend, T.M.; Reeder, T.W.; Sites, J.W., Jr.; Wiens, J.J. Estimating divergence dates and evaluating dating methods using phylogenomic and mitochondrial data in squamate reptiles. Mol. Phylogenet. Evol. 2012, 65, 974–991. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wiens, J.J. Do missing data influence the accuracy of divergence-time estimation with BEAST? Mol. Phylogenet. Evol. 2015, 85, 41–49. [Google Scholar] [CrossRef]
- Pyron, R.A. A likelihood method for assessing molecular divergence time estimates and the placement of fossil calibrations. Syst. Biol. 2010, 59, 185–194. [Google Scholar] [CrossRef]
- Wiens, J.J.; Brandley, M.C.; Reeder, T.W. Why does a trait evolve multiple times within a clade? Repeated evolution of snakeline body form in squamate reptiles. Evolution 2006, 60, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Hugall, A.F.; Foster, R.; Lee, M.S.Y. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst. Biol. 2007, 56, 543–563. [Google Scholar] [CrossRef] [PubMed]
- Klembara, J.; Green, B. Anguimorph lizards (Squamata, Anguimorpha) from the middle and late Eocene of the Hampshire Basin of southern England. J. Syst. Palaeontol. 2010, 8, 97–129. [Google Scholar] [CrossRef]
- Gvoždík, V.; Necas, T.; Jablonski, D.; Lemmon, E.M.; Lemmon, A.R.; Jandzik, D.; Moravec, J. Phylogenomics of Anguis and Pseudopus (Squamata, Anguidae) indicates Balkan-Apennine mitochondrial capture associated with the Messinian event. Mol. Phylogenet. Evol. 2023, 180, 107674. [Google Scholar] [CrossRef] [PubMed]
- Augé, M. La faune de Lacertilia (Reptilia, Squamata) de l’Éocène inférieur de Prémontré (Bassin de Paris, France). Geodiversitas 2003, 25, 539–574. [Google Scholar]
- Gvoždík, V.; Benkovský, N.; Crottini, A.; Bellati, A.; Moravec, J.; Romano, A.; Sacchi, R.; Jandzik, D. An ancient lineage of slow worms, genus Anguis (Squamata: Anguidae), survived in the Italian Peninsula. Mol. Phylogenet. Evol. 2013, 69, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Yang, R. A new species of the genus Ophisaurus from Hainan Island. Acta Herpetol. Sin. 1983, 2, 67–69. [Google Scholar]
- Cai, B.; Guo, X.G.; Song, Z.B.; Chen, D.L. The complete mitochondrial genome of the Hainan glass lizard (Dopasia hainanensis) determined by next-generation sequencing. Mitochondrial DNA Part B 2020, 5, 246–247. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.; Pardo, J.D.; Maddin, H.C. Snake-like limb loss in a Carboniferous amniote. Nat. Ecol. Evol. 2022, 6, 614–621. [Google Scholar] [CrossRef]
- Wiens, J.J.; Slingluff, J.L. How lizards turn into snakes: A phylogenetic analysis of body-form evolution in anguid lizards. Evolution 2001, 55, 2302–2318. [Google Scholar]
- Lambertz, M.; Arenz, N.; Grommes, K. Variability in pulmonary reduction and asymmetry in a serpentiform lizard: The sheltopusik, Pseudopus apodus (Pallas, 1775). Vert. Zool. 2018, 68, 21–26. [Google Scholar] [CrossRef]
- Spinner, M.; Bleckmann, H.; Westhoff, G. Morphology and frictional properties of scales of Pseudopus apodus (Anguidae, Reptilia). Zoology 2015, 118, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Dotson, E.M.; Beard, C.B. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata. Insect Mol. Biol. 2001, 10, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Brandt, U. Energy converting NADH: Quinone oxidoreductase (complex I). Annu. Rev. Biochem. 2006, 75, 69–92. [Google Scholar] [CrossRef] [PubMed]
- Wirth, C.; Brandt, U.; Hunte, C.; Zickermann, V. Structure and function of mitochondrial complex I. Biochim. Biophys. Acta BBA 2016, 1857, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Bridges, H.R.; Birrell, J.A.; Hirst, J. The mitochondrial-encoded subunits of respiratory complex I (NADH: Ubiquinone oxidoreductase): Identifying residues important in mechanism and disease. Biochem. Soc. Trans. 2011, 39, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Sazanov, L.A.; Hinchliffe, P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 2006, 311, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, W.; Ren, J.; Peng, C.; Jiang, D.; Li, J. Evolution of phenotype and mitochondrial genome reveals limbless and body-elongated Squamates may change their energy basis for locomotion. Asian Herp. Res. 2021, 12, 213–220L. [Google Scholar]
- Bergmann, P.J.; Mann, S.D.; Morinaga, G.; Freitas, E.S.; Siler, C.D. Convergent evolution of elongate forms in craniates and of locomotion in elongate squamate reptiles. Integr. Comp. Biol. 2020, 60, 190–201. [Google Scholar] [CrossRef]
- Prud’homme, B.; Gompel, N.; Carroll, S.B. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 8605–8612. [Google Scholar] [CrossRef]
- Engel, M.S.; Davis, S.R.; Prokop, J. Insect wings: The evolutionary development of nature’s first flyers. In Arthropod Biology and Evolution; Springer: Berlin/Heidelberg, Germany, 2013; pp. 269–298. [Google Scholar]
- Mehta, R.S.; Ward, A.B.; Alfaro, M.E.; Wainwright, P.C. Elongation of the body in eels. Integr. Comp. Biol. 2010, 50, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, Y.; Endo, H. Mitochondrial genome of the Komodo dragon: Efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res. 2004, 11, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer Premier 5. Biotech. Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Zhang, L.P.; Yu, D.N.; Storey, K.B.; Cheng, H.Y.; Zhang, J.Y. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int. J. Biol. Macromol. 2018, 111, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Bioinf. Methods Protoc. 1999, 132, 71–91. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Guo, X.; Jiang, J. Next-generation sequencing yields a complete mitochondrial genome of the Asian glass lizard (Dopasia gracillis) from the Yungui Plateau in Southwest China. Mitochondrial DNA Part B 2020, 5, 992–993. [Google Scholar] [CrossRef]
- Pan, H.C.; Liu, L.; Li, P.; Li, X.F.; Liu, Z.L. The complete mitochondrial genome of Chinese glass lizard Ophisaurus harti (Squamata: Anguidae). Mitochondrial DNA 2015, 26, 280–281. [Google Scholar] [CrossRef] [PubMed]
- Li, H.M.; Guo, L.; Zeng, D.L.; Guan, Q.X.; Wu, Z.J.; Qin, X.M. Complete mitochondrial genome of Shinisaurus crocodilurus (Squamata: Shinisaurus) and its genetic relationship with related species. Mitochondrial DNA 2012, 23, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Kirchhof, S.; Lyra, M.L.; Rodriguez, A.; Ineich, I.; Mueller, J.; Roedel, M.O.; Trape, J.F.; Vences, M.; Boissinot, S. Mitogenome analyses elucidate the evolutionary relationships of a probable Eocene wet tropics relic in the xerophile lizard genus Acanthodactylus. Sci. Rep. 2021, 11, 4858. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, J.; Zhang, B.; Guo, X. The complete mitochondrial genome of Eremias dzungarica (Reptilia, Squamata, Lacertidae) from the Junggar Basin in Northwest China. Mitochondrial DNA Part B 2021, 6, 2012–2014. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.L.; Yao, Y.T.; Lin, L.H.; Ji, X. The complete mitochondrial genome of Eremias multiocellata (Squamata: Lacertidae). Mitochondrial DNA Part A 2016, 27, 1654–1655. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, D.; Dujsebayeva, T.N.; Liu, J.; Guo, X. Complete mitochondrial genome of Stummer’s Racerunner (Eremias stummeri) from Kazakhstan. Mitochondrial DNA Part A 2016, 27, 4340–4341. [Google Scholar] [CrossRef]
- Tian, L.; Guo, X. Complete mitochondrial genomes of five racerunners (Lacertidae: Eremias) and comparison with other lacertids: Insights into the structure and evolution of the control region. Genes 2022, 13, 726. [Google Scholar] [CrossRef]
- Tong, Q.L.; Yao, Y.T.; Lin, L.H.; Ji, X. The complete mitochondrial genome of Eremias vermiculata (Squamata: Lacertidae). Mitochondrial DNA Part A 2016, 27, 1447–1448. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhu, D.; Zhao, W.G.; Ji, X. The complete mitochondrial genome of the common lizard Zootoca vivipara (Squamata: Lacertidae). Mitochondrial DNA Part A 2016, 27, 1944–1945. [Google Scholar] [CrossRef] [PubMed]
- Murtskhvaladze, M.; Tarkhnishvili, D.; Anderson, C.L.; Kotorashvili, A. Phylogeny of caucasian rock lizards (Darevskia) and other true lizards based on mitogenome analysis: Optimisation of the algorithms and gene selection. PLoS ONE 2020, 15, e0233680. [Google Scholar] [CrossRef] [PubMed]
- Komissarov, A.; Korchagin, V.; Kliver, S.; Dobrynin, P.; Semyenova, S.; Vergun, A.; O’Brien, S.; Ryskov, A. The complete mitochondrial genome of the parthenogenetic Caucasian rock lizard Darevskia unisexualis (Squamata: Lacertidae) contains long tandem repeat formed by 59 bp monomer. Mitochondrial DNA Part B 2016, 1, 875–877. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.W.; Liu, H.; Zhao, W.G.; Liu, P. The complete mitochondrial genome of Takydromus amurensis (Squamata: Lacertidae). Mitochondrial DNA Part B 2016, 1, 214–215. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.N.; Ji, X. The complete mitochondrial genome of Takydromus wolteri (Squamata: Lacertidae). Mitochondrial DNA 2013, 24, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.S.; Chen, J.M.; Huang, S. Mitochondrial genome of the Chung-an ground lizard Takydromus sylvaticus (Reptilia: Lacertidae). Mitochondrial DNA 2014, 25, 319–320. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.S.; Zeng, D.L.; Hou, L.X.; Yang, X.W.; Qin, X.M. Complete mitochondrial genome of Takydromus sexlineatus (Squamata, Lacertidae). Mitochondrial DNA 2015, 26, 465–466. [Google Scholar] [CrossRef] [PubMed]
- Vacher, J.P.; Manzi, S.; Rodrigues, M.T.; Fouquet, A. The complete mitochondrial genome of Iphisa elegans (Reptilia: Squamata: Gymnophthalmidae). Mitochondrial DNA Part B 2020, 5, 3106–3108. [Google Scholar] [CrossRef]
- MacLeod, A.; Irisarri, I.; Vences, M.; Steinfartz, S. The complete mitochondrial genomes of the Galapagos iguanas, Amblyrhynchus cristatus and Conolophus subcristatus. Mitochondrial DNA Part A 2016, 27, 3699–3700. [Google Scholar] [CrossRef]
- Janke, A.; Erpenbeck, D.; Nilsson, M.; Arnason, U. The mitochondrial genomes of the iguana (Iguana iguana) and the caiman (Caiman crocodylus): Implications for amniote phylogeny. Proc. Biol. Sci. 2001, 268, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Castoe, T.A.; Jiang, Z.J.; Gu, W.; Wang, Z.O.; Pollock, D.D. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS ONE 2008, 3, e2201. [Google Scholar] [CrossRef] [PubMed]
- Nogueira Dumans, A.T.; Teixeira, G.W.; Vieira, G.A.; Sarzi, D.S.; Furtado, C.; Bryan Jennings, W.; Prosdocimi, F. Complete mitochondrial genomes for three lizards (Anolis punctatus, Sceloporus woodi, and S. grammicus): A contribution to mitochondrial phylogenomics of Iguanoidea. Mitochondrial DNA Part B 2019, 4, 700–702. [Google Scholar] [CrossRef]
- Bernardo, P.H.; Felipe Aguilera-Miller, E.; Ticul Alvarez-Castaneda, S.; Roberto Mendez-de la Cruz, F.; Murphy, R.W. The complete mitochondrial genome of the black-tailed brush lizard Urosaurus nigricaudus (Reptilia, Squamata, Phrynosomatidae). Mitochondrial DNA Part A 2016, 27, 4023–4025. [Google Scholar] [CrossRef] [PubMed]
- Leache, A.D.; Chavez, A.S.; Jones, L.N.; Grummer, J.A.; Gottscho, A.D.; Linkem, C.W. Phylogenomics of phrynosomatid lizards: Conflicting signals from sequence capture versus restriction site associated DNA sequencing. Genome Biol. Evol. 2015, 7, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.L.; Du, Y.; Yao, Y.T.; Lin, C.X.; Lin, L.H. The complete mitochondrial genome of Acanthosaura lepidogaster (Squamata: Agamidae). Mitochondrial DNA Part A 2017, 28, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Du, Y.; Fang, M.; Li, H.; Lin, L. The mitochondrial genome of Pseudocalotes microlepis (Squamata: Agamidac) and its phylogenetic position in Agamids. Asian Herpetol. Res. 2018, 9, 24–34. [Google Scholar]
- Wang, M.; Jiang, Z.; Wang, J.; Cui, L.; Zhang, M. The complete mitochondrial genome of the blue-crested lizard, Calotes mystaceus (Squamata, Agamidae) in China. Mitochondrial DNA Part B 2020, 5, 3530–3531. [Google Scholar] [CrossRef] [PubMed]
- Amer, S.A.M.; Kumazawa, Y. The mitochondrial genome of the lizard Calotes versicolor and a novel gene inversion in South Asian draconine agamids. Mol. Biol. Evol. 2007, 24, 1330–1339. [Google Scholar] [CrossRef]
- Qiv, Q.B.; Zhu, G.Y.; Yu, X.L.; Du, Y. The complete mitochondrial genome of the Draco maculatus (Squamata: Agamidae). Mitochondrial DNA Part B 2019, 4, 426–427. [Google Scholar]
- Tong, H.; Jin, Y. The complete mitochondrial genome of an agama, Phrynocephalus putjatia (Reptilia, Squamata, Agamidae). Mitochondrial DNA Part A 2016, 27, 1028–1029. [Google Scholar] [CrossRef]
- Macey, J.R.; Schulte, J.A., II; Fong, J.J.; Das, I.; Papenfuss, T.J. The complete mitochondrial genome of an agamid lizard from the Afro-Asian subfamily agaminae and the phylogenetic position of Bufoniceps and Xenagama. Mol. Phylogenet. Evol. 2006, 39, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Ujvari, B.; Dowton, M.; Madsen, T. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol. Lett. 2007, 3, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Amer, S.A.M.; Kumazawa, Y. Mitochondrial genome of Pogona vitticepes (Reptilia; Agamidae): Control region duplication and the origin of Australasian agamids. Gene 2005, 346, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.L.; Du, Y.; Lin, L.H.; Ji, X. The complete mitochondrial genome of Leiolepis reevesii (Sauria, Agamidae). Mitochondrial DNA Part A 2016, 27, 541–542. [Google Scholar] [CrossRef] [PubMed]
- Macey, J.R.; Kuehl, J.V.; Larson, A.; Robinson, M.D.; Ugurtas, I.H.; Ananjeva, N.B.; Rahman, H.; Javed, H.I.; Osmani, R.M.; Doumma, A.; et al. Socotra Island the forgotten fragment of Gondwana: Unmasking chameleon lizard history with complete mitochondrial genomic data. Mol. Phylogenet. Evol. 2008, 49, 1015–1018. [Google Scholar] [PubMed]
- Macey, J.R.; Larson, A.; Ananjeva, N.B.; Papenfuss, T.J. Evolutionary shifts in three major structural features of the mitochondrial genome among iguanian lizards. J. Mol. Evol. 1997, 44, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Koo, K.S.; Kim, I.H.; Park, D. Complete mitochondrial genomes of Scincella vandenburghi and S. huanrenensis (Squamata: Scincidae). Mitochondrial DNA Part B 2016, 1, 237–238. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhong, J.; Ma, L.; Guo, K.; Du, Y. Complete mitochondrial genome of Scincella modesta (Squamata: Scincidae) and phylogenetic analysis. Mitochondrial DNA Part B 2020, 5, 373–374. [Google Scholar] [CrossRef]
- Tang, X.S.; Yang, D.C.; Lin, Y.J.; Dai, L.L. The complete mitochondrial genome of Sphenomorphus indicus (Reptilia: Scincidae). Mitochondrial DNA Part B 2019, 4, 2727–2728. [Google Scholar] [CrossRef]
- Duangjai, S.; Srisodsuk, S.; Chuaynkern, C.; Chuaynkern, Y. Complete mitochondrial genome of Tropidophorus hangnam (Squamata: Scincidae) with phylogenetic analysis. Mitochondrial DNA Part B 2020, 5, 3701–3702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sun, X.; Chen, L.; Xiao, W.; Zhu, X.; Xia, Y.; Chen, J.; Wang, H.; Zhang, B. The complete mitochondrial genome of Eumeces chinensis (Squamata: Scincidae) and implications for Scincidae taxonomy. Mitochondrial DNA Part A 2016, 27, 4691–4692. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lin, Y.; Xiao, Q.; Lin, Y.; Du, Y.; Lin, C.; Ward-Fear, G.; Hu, C.; Qu, Y.; Li, H. Characterization of the complete mitochondrial genome of the many-lined sun skink (Eutropis multifasciata) and comparison with other Scincomorpha species. Genomics 2021, 113, 2526–2536. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Nishida, M. Variations in mitochondrial tRNA gene organization of reptiles as phylogenetic markers. Mol. Biol. Evol. 1995, 12, 759–772. [Google Scholar]
- Hu, J.G.; Peng, L.F.; Tang, X.S.; Huang, S. The complete mitochondrial genome of Takydromus septentrionalis (Reptilia: Lacertidae). Mitochondrial DNA Part B 2019, 4, 2193–2194. [Google Scholar] [CrossRef]
- Lyra, M.L.; Joger, U.; Schulte, U.; Slimani, T.; El Mouden, E.H.; Bouazza, A.; Kuenzel, S.; Lemmon, A.R.; Lemmon, E.M.; Vences, M. The mitochondrial genomes of Atlas Geckos (Quedenfeldtia): Mitogenome assembly from transcriptomes and anchored hybrid enrichment datasets. Mitochondrial DNA Part B 2017, 2, 356–358. [Google Scholar] [CrossRef] [PubMed]
- Tarroso, P.; Simo-Riudalbas, M.; Carranza, S. The complete mitochondrial genome of Pristurus rupestris rupestris. Mitochondrial DNA Part B 2017, 2, 802–803. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Miura, S.; Yamada, C.; Hashiguchi, Y. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genom. 2014, 15, 930. [Google Scholar] [CrossRef]
- Luo, H.; Huang, A.; Li, B.; Ni, Q.; Yao, Y.; Xu, H.; Zeng, B.; Li, Y.; Wei, Z.; Zhang, M. Complete mitochondrial genome of the webbed-toed gecko Gekko subpalmatus (Squamata: Gekkonidae). Mitochondrial DNA Part B 2019, 4, 1725–1726. [Google Scholar] [CrossRef]
- Hao, S.; Ping, J.; Zhang, Y. Complete mitochondrial genome of Gekko chinensis (Squamata, Gekkonidae). Mitochondrial DNA Part A 2016, 27, 4226–4227. [Google Scholar] [CrossRef]
- Smid, J.; Uvizl, M.; Shobrak, M.; Busais, S.; Salim, A.F.A.; AlGethami, R.H.M.; AlGethami, A.R.; Alanazi, A.S.K.; Alsubaie, S.D.; Rovatsos, M.; et al. Diversification of Hemidactylus geckos (Squamata: Gekkonidae) in coastal plains and islands of southwestern Arabia with descriptions and complete mitochondrial genomes of two endemic species to Saudi Arabia. Org. Divers. Evol. 2023, 23, 185–207. [Google Scholar] [CrossRef]
- Yan, J.; Tian, C.; Zhou, J.; Bauer, A.M.; Grismer, L.L.; Zhou, K. Complete mitochondrial genome of the Tioman Island rock gecko, Cnemaspis limi (Sauria, Gekkota, Gekkonidae). Mitochondrial DNA 2014, 25, 181–182. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Tian, C.; Lv, L.; Bauer, A.M.; Zhou, K. Complete mitochondrial genome of the San Lucan gecko, Phyllodactylus unctus (Sauria, Gekkota, Phyllodactylidae), in comparison with Tarentola mauritanica. Mitochondrial DNA 2014, 25, 202–203. [Google Scholar] [CrossRef] [PubMed]
- Macey, J.R.; Fong, J.J.; Kuehl, J.V.; Shafiei, S.; Ananjeva, N.B.; Papenfuss, T.J.; Boore, J.L. The complete mitochondrial genome of a gecko and the phylogenetic position of the Middle Eastern Teratoscincus keyserlingii. Mol. Phylogenet. Evol. 2005, 36, 188–193. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jonniaux, P.; Hashiguchi, Y.; Kumazawa, Y. Mitochondrial genomes of two African geckos of genus Hemitheconyx (Squamata: Eublepharidae). Mitochondrial DNA 2012, 23, 278–279. [Google Scholar] [CrossRef] [PubMed]
- Macey, J.R.; Papenfuss, T.J.; Kuehl, J.V.; Fourcade, H.M.; Boore, J.L. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genomic sequences. Mol. Phylogenet. Evol. 2004, 33, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Castoe, T.A.; de Koning, A.P.J.; Kim, H.-M.; Gu, W.; Noonan, B.P.; Naylor, G.; Jiang, Z.J.; Parkinson, C.L.; Pollock, D.D. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 8986–8991. [Google Scholar] [CrossRef] [PubMed]
- Castoe, T.A.; Gu, W.; de Koning, A.P.J.; Daza, J.M.; Jiang, Z.J.; Parkinson, C.L.; Pollock, D.D. Dynamic Nucleotide Mutation Gradients and Control Region Usage in Squamate Reptile Mitochondrial Genomes. Cytogenet. Genome Res. 2009, 127, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.B.; Cobb, V.A.; Cahoon, A.B. The complete mitochondrial DNA sequence of Crotalus horridus (timber rattlesnake). Mitochondrial DNA 2013, 24, 94–96. [Google Scholar] [CrossRef]
- Huang, R.; Peng, L.; Yang, D.; Yong, Z.; Huang, S. Mitochondrial genome of the Boulenger’s Slug-eating snake Pareas boulengeri (Serpentes: Pareidae). Mitochondrial DNA Part B 2020, 5, 3179–3180. [Google Scholar] [CrossRef]
- Jiang, Z.J.; Castoe, T.A.; Austin, C.C.; Burbrink, F.T.; Herron, M.D.; McGuire, J.A.; Parkinson, C.L.; Pollock, D.D. Comparative mitochondrial genomics of snakes: Extraordinary substitution rate dynamics and functionality of the duplicate control region. BMC Evol. Biol. 2007, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Zhang, Y.; Qian, L.; Sun, P.; Wang, C.; Fang, K.; Pan, T.; Zhang, B.; Rao, D.; Wang, H. The complete mitochondrial genome of Protobothrops kaulbacki (Squamata: Viperidae). Mitochondrial DNA Part B 2017, 2, 201–202. [Google Scholar] [CrossRef] [PubMed]
- Koehler, G.; Khaing, K.P.P.; Than, N.L.; Baranski, D.; Schell, T.; Greve, C.; Janke, A.; Pauls, S.U. A new genus and species of mud snake from Myanmar (Reptilia, Squamata, Homalopsidae). Zootaxa 2021, 4915, 301–325. [Google Scholar] [CrossRef] [PubMed]
- Kornilios, P. The complete mitogenome of the Eurasian blindsnake Xerotyphlops vermicularis (Reptilia, Typhlopidae). Mitochondrial DNA Part B 2019, 4, 1990–1991. [Google Scholar] [CrossRef]
- Liu, Q.; Zhu, F.; Wang, X.; Xiao, R.; Fang, M.; Sun, L.; Li, P.; Guo, P. The complete mitochondrial genome sequence of Gloydius shedaoensis (Squamata: Viperidae). Mitochondrial DNA Part A 2016, 27, 4679–4680. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Chijiwa, T.; Hattori, S.; Terada, K.; Ohno, M.; Fukumaki, Y. The taxonomic position and the unexpected divergence of the Habu viper, Protobothrops among Japanese subtropical islands. Mol. Phylogenet. Evol. 2016, 101, 91–100. [Google Scholar] [CrossRef]
- Sun, X.; Li, Y.; Liu, J.; Sheng, J. The complete mitochondrial genome of the tartar Sand Boa Eryx tataricus. Mitochondrial DNA Part B 2019, 4, 1994–1995. [Google Scholar] [CrossRef]
- Thanou, E.; Kornilios, P. Next-generation sequencing yields the complete mitochondrial genome of the endangered Milos viper Macrovipera schweizeri (Reptilia, Viperidae). Mitochondrial DNA Part B 2018, 3, 1250–1251. [Google Scholar] [CrossRef]
- Wang, D.Q.; Pan, L.L.; Yang, D.C.; Dai, L.L. Complete mitochondrial genome of the sharp-snouted pitviper Deinagkistrodon acutus (Reptilia, Viperidae). Mitochondrial DNA Part B 2019, 4, 2900–2901. [Google Scholar] [CrossRef]
- Wu, S.X. Characterization of the complete mitochondrial genome of Crotalus adamanteus (Eastern diamondback rattlesnake). Mitochondrial DNA Part B 2019, 4, 632–634. [Google Scholar] [CrossRef]
- Wu, Y.; Li, K.; Liu, Q.; Chen, S.; Cai, B. The complete mitochondrial genome of the Asian pitviper Gloydius changdaoensis (Squamata, Viperidae). Mitochondrial DNA Part B 2020, 5, 3294–3295. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Li, H.; Zhou, K. Evolution of the mitochondrial genome in snakes: Gene rearrangements and phylogenetic relationships. BMC Genom. 2008, 9, 569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, X.; Li, Z.; Hu, H.; Zhang, B. Mitochondrial genome of Protobothrops mucrosquamatus (Squamata: Viperidae: Crotalinae). Mitochondrial DNA 2013, 24, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Liu, Q.; Zhong, G.; Xiao, R.; Fang, M.; Guo, P. Complete mitochondrial genome of Sinovipera sichuanensis (Reptilia: Squamata: Viperidae). Mitochondrial DNA Part A 2016, 27, 3666–3667. [Google Scholar] [CrossRef] [PubMed]
- Janke, A.; Arnason, U. The complete mitochondrial genome of Alligator mississippiensis and the separation between recent archosauria (birds and crocodiles). Mol. Biol. Evol. 1997, 14, 1266–1272. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, X.; Ji, X.; Yan, P.; Amato, G. The complete mitochondrial genome of salt-water crocodile (Crocodylus porosus) and phylogeny of crocodilians. J. Genet. Genom. 2007, 34, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Macey, J.R.; Pabinger, S.; Barbieri, C.G.; Buring, E.S.; Gonzalez, V.L.; Mulcahy, D.G.; DeMeo, D.P.; Urban, L.; Hime, P.M.; Prost, S.; et al. Evidence of two deeply divergent co-existing mitochondrial genomes in the Tuatara reveals an extremely complex genomic organization. Commun. Biol. 2021, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Phylogenet. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.E.; Anderson, C.L.; Hipsley, C.A.; Müller, J.; Evans, S.E.; Schoch, R.R. Integration of molecules and new fossils supports a Triassic origin for Lepidosauria (lizards, snakes, and tuatara). BMC Evol. Biol. 2013, 13, 208. [Google Scholar] [CrossRef] [PubMed]
- Gradstein, F.; Ogg, J.G.; Schmitz, M.D.; Ogg, G.M. The Geologic Time Scale 2012; Elsevier: Amsterdam, The Netherlands, 2012; Volume 1, pp. 207–232. [Google Scholar]
- Head, J.J.; Bloch, J.I.; Hastings, A.K.; Bourque, J.R.; Cadena, E.A.; Herrera, F.A.; Polly, P.D.; Jaramillo, C.A. Giant boid snake from the Palaeocene neotropics reveals hotter past equatorial temperatures. Nature 2009, 457, 715–717. [Google Scholar] [CrossRef]
- Wagner, P.; Stanley, E.L.; Daza, J.D.; Bauer, A.M. A new agamid lizard in mid-Cretaceous amber from northern Myanmar. Cretac. Res. 2021, 124, 104813. [Google Scholar] [CrossRef]
- Nydam, R.L. A new taxon of helodermatid-like lizard from the Albian–Cenomanian of Utah. J. Vertebr. Paleontol. 2000, 20, 285–294. [Google Scholar] [CrossRef]
- Brikiatis, L. The De Geer, Thulean and Beringia routes: Key concepts for understanding early Cenozoic biogeography. J. Biogeogr. 2014, 41, 1036–1054. [Google Scholar] [CrossRef]
- Rage, J.; Auge, M. Amphibians and squamate reptiles from the lower Eocene of Silveirinha (Portugal). Ciências Terra (UNL) 2003, 15, 103–116. [Google Scholar]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A. FigTree, version 1.4.0; 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 20 May 2024).
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Hershberg, R.; Petrov, D.A. Selection on codon bias. Annu. Rev. Genet. 2008, 42, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.L.; Chen, C.J.; Arab, D.A.; Du, Z.G.; He, Y.H.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, D115–D119. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]


| Species | Whole Genome | PCGs | ||||||
|---|---|---|---|---|---|---|---|---|
| Length (bp) | A+T% | AT-K | GC-K | Length (bp) | A+T% | AT-K | GC-K | |
| Dopasia gracilis | 15,855 | 54.3 | 0.124 | −0.344 | 11,379 | 53.8 | 0.064 | −0.386 |
| Pseudopus apodus | 16,274 | 55.2 | 0.112 | −0.346 | 11,388 | 54.7 | 0.053 | −0.380 |
| Cyclodomorphus gerrardii | 16,093 | 59.3 | 0.086 | −0.335 | 11,373 | 59.5 | 0.019 | −0.386 |
| Chalcides ocellatus | 16,563 | 57.9 | 0.143 | −0.330 | 11,388 | 57.8 | 0.099 | −0.375 |
| Tiliqua gigas gigas | 16,957 | 57.7 | 0.078 | −0.317 | 11,367 | 57.9 | 0.021 | −0.380 |
| Plestiodon quadrilineatus | 17,391 | 56.5 | 0.120 | −0.322 | 11,382 | 56.1 | 0.076 | −0.357 |
| Gekko chinensis | 17,657 | 60.9 | 0.100 | −0.315 | 11,298 | 60.6 | 0.062 | −0.361 |
| Gekko japonicus | 17,707 | 56.6 | 0.113 | −0.322 | 11,319 | 55.8 | 0.075 | −0.368 |
| Nodes/Clades | Mean Divergence Time (Mya) | 95% HPD Range (Mya) |
|---|---|---|
| Squamata root | 206.05 | 182.08~229.55 |
| Gekkota | 155.31 | 123.30~186.71 |
| (Scincidae + Dibamia) & (Cordylidae + Xantusiidae) | 124.87 | 86.53~194.85 |
| Scincidae | 75.79 | 58.95~93.74 |
| Anguimorpha | 111.91 | 102.35~126.18 |
| Pleurodonta | 88.86 | 70.27~102.38 |
| Acrodonta | 117.89 | 103.23~137.10 |
| Serpentes | 135.31 | 115.23~154.91 |
| Acrodonta & Serpentes | 162.67 | 141.23~183.67 |
| Anguimorpha & Pleurodonta + (Acrodonta + Serpentes) | 184.12 | 160.76~206.52 |
| Amphisbaenia | 149.39 | 121.61~176.25 |
| Lacertoidea (Gymnophthalmidae, Lacertidae, Amphisbaenia) | 167.20 | 151.53~200.53 |
| Dopasia & (Pseudopus + Anguis) | 38.70 | 31.18~46.33 |
| Pseudopus & Anguis | 27.26 | 22.85~30.51 |
| (Dopasia gracilis + Dopasia sokolovi) & (Dopasia hati + Dopasia hainanensis) | 30.26 | 21.94~38.83 |
| Foreground Branch | Model | np | Ln L | Estimates of Parameters | Model Compared | LRT p-Value | Positive Sites | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Anguis | Model A | 49 | −81,825.55313 | Site class | 0 | 1 | 2a | 2b | Model A vs. Model A null | 0.00000 | 1151 E 0.989 *,1216 T 0.972 *, 1259 N 0.959 *,1688 A 0.987 *, 2249 T 0.973 *,2504 F 0.983 *, 3138 S 0.956 *,3187 T 0.991 **, 3464 A 0.979 *,3468 G 0.984 * |
| Dopasia | f | 0.83441 | 0.10720 | 0.05174 | 0.00665 | ||||||
| Pseudopus | ω0 | 0.04478 | 1.00000 | 0.04478 | 1.00000 | ||||||
| Model A null | 48 | −81,845.54159 | ω1 | 0.04478 | 1.00000 | 7.91959 | 7.91959 | ||||
| 1 | Not Allowed | ||||||||||
| Genes | Positive Selection Sites | Amino Acids | BEB Value | Feature Key * | Description | |
|---|---|---|---|---|---|---|
| Foreground | Background | |||||
| ND2 | 90 | E | S\T | 0.989 * | Transmembrane | Helical |
| 155 | T\M | L | 0.972 * | Transmembrane | Helical | |
| 198 | N | T\P | 0.959 * | / | / | |
| ND3 | 21 | A | S | 0.987 * | Transmembrane | Helical |
| ND5 | 12 | T\A | L | 0.973 * | Transmembrane | Helical |
| 267 | F\S | H | 0.983 * | Transmembrane | Helical | |
| ND6 | 72 | S | A\S | 0.956 * | Transmembrane | Helical |
| 119 | T | G\D | 0.991 ** | Transmembrane | Helical | |
| CYTB | 183 | A | L | 0.979 * | Transmembrane | Helical |
| 187 | G | I | 0.984 * | Transmembrane | Helical | |
| Node Figure 6 | Minimum Age of Fossil Constraint (Mya) | Maximum Age of Fossil Constraint (Mya) | Fossil Calibration | Age (Period/Stage) | References |
|---|---|---|---|---|---|
| 1 | 238 | 252 | Rhynchocephalia-Squamata | Middle Triassic | [203,204] |
| 2 | 58.00 | 66.00 | Titanoboa cerrejonensis | Paleocene | [205] |
| 3 | 93.30 | 99.60 | Protodraco monocoli | Late Cretaceous | [206] |
| 4 | 47.80 | 55.80 | Anniealexandria | Early Eocene | [37] |
| 5 | 99.60 | 102.70 | Primaderma | Early Cretaceous | [207] |
| 6 | 48.60 | 57.00 | Gerrhonotus | Lower Eocene | [208,209] |
| 7 | 18.40 | 30.00 | Anguis rarus, Pseudopus ahnikoviensis | Early Neogene | [46,54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, L.; Chen, Y.; He, J.; Guo, Z.; Wu, L.; Storey, K.B.; Zhang, J.; Yu, D. The Phylogenetic Relationships of Major Lizard Families Using Mitochondrial Genomes and Selection Pressure Analyses in Anguimorpha. Int. J. Mol. Sci. 2024, 25, 8464. https://doi.org/10.3390/ijms25158464
Zhan L, Chen Y, He J, Guo Z, Wu L, Storey KB, Zhang J, Yu D. The Phylogenetic Relationships of Major Lizard Families Using Mitochondrial Genomes and Selection Pressure Analyses in Anguimorpha. International Journal of Molecular Sciences. 2024; 25(15):8464. https://doi.org/10.3390/ijms25158464
Chicago/Turabian StyleZhan, Lemei, Yuxin Chen, Jingyi He, Zhiqiang Guo, Lian Wu, Kenneth B. Storey, Jiayong Zhang, and Danna Yu. 2024. "The Phylogenetic Relationships of Major Lizard Families Using Mitochondrial Genomes and Selection Pressure Analyses in Anguimorpha" International Journal of Molecular Sciences 25, no. 15: 8464. https://doi.org/10.3390/ijms25158464
APA StyleZhan, L., Chen, Y., He, J., Guo, Z., Wu, L., Storey, K. B., Zhang, J., & Yu, D. (2024). The Phylogenetic Relationships of Major Lizard Families Using Mitochondrial Genomes and Selection Pressure Analyses in Anguimorpha. International Journal of Molecular Sciences, 25(15), 8464. https://doi.org/10.3390/ijms25158464






