Native and Alien Antarctic Grasses as a Habitat for Fungi
Abstract
1. Introduction
2. Results
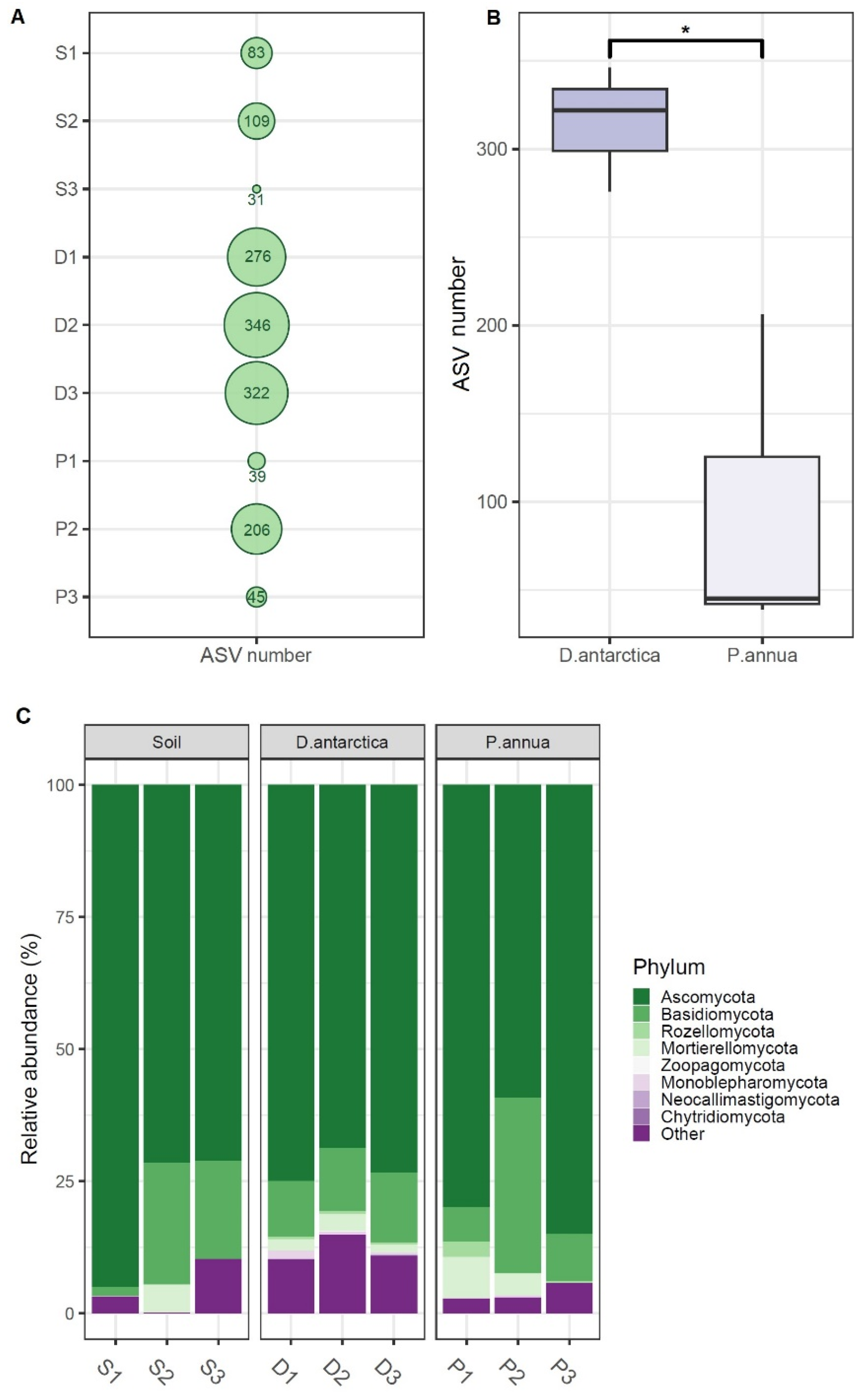
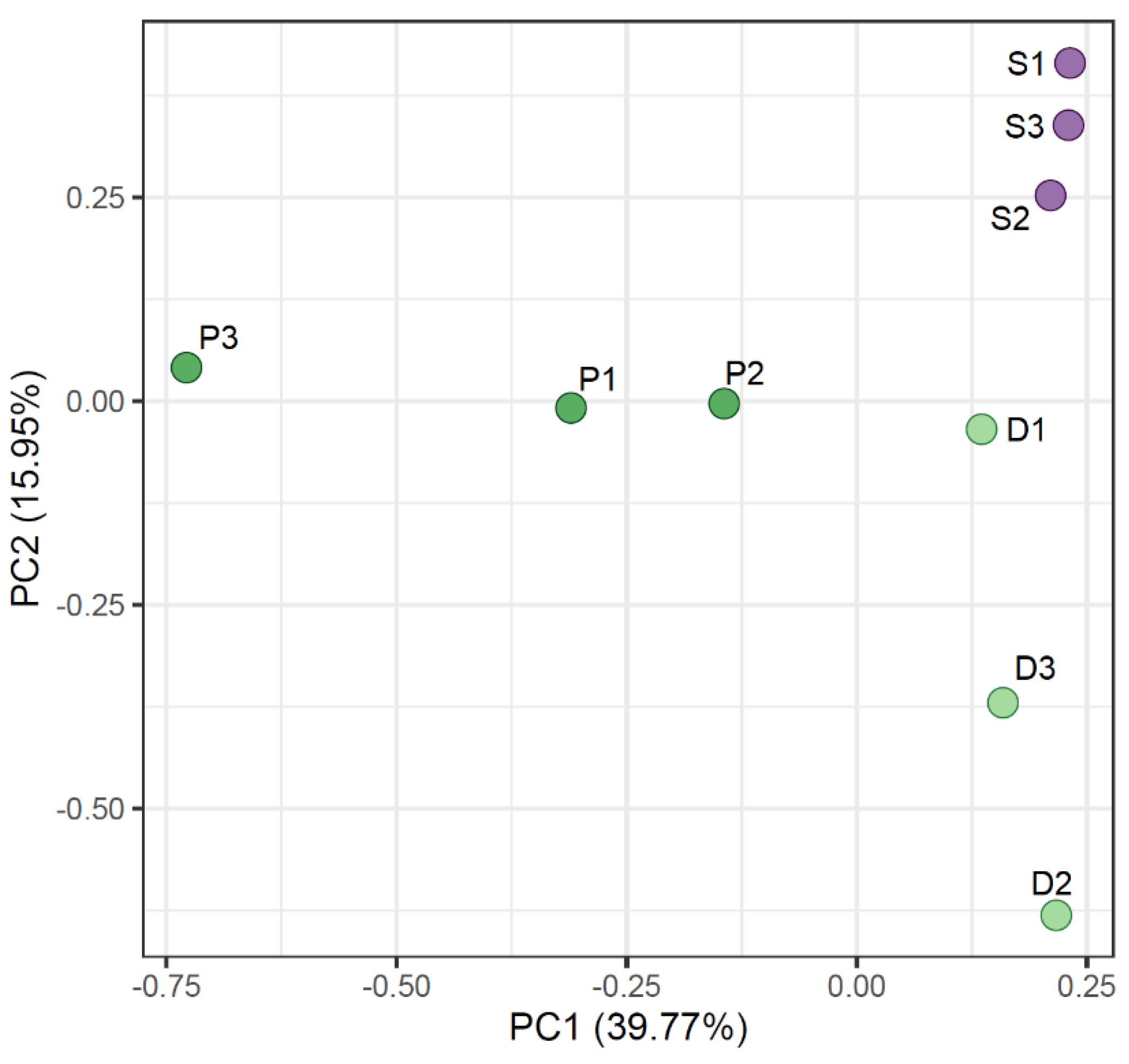
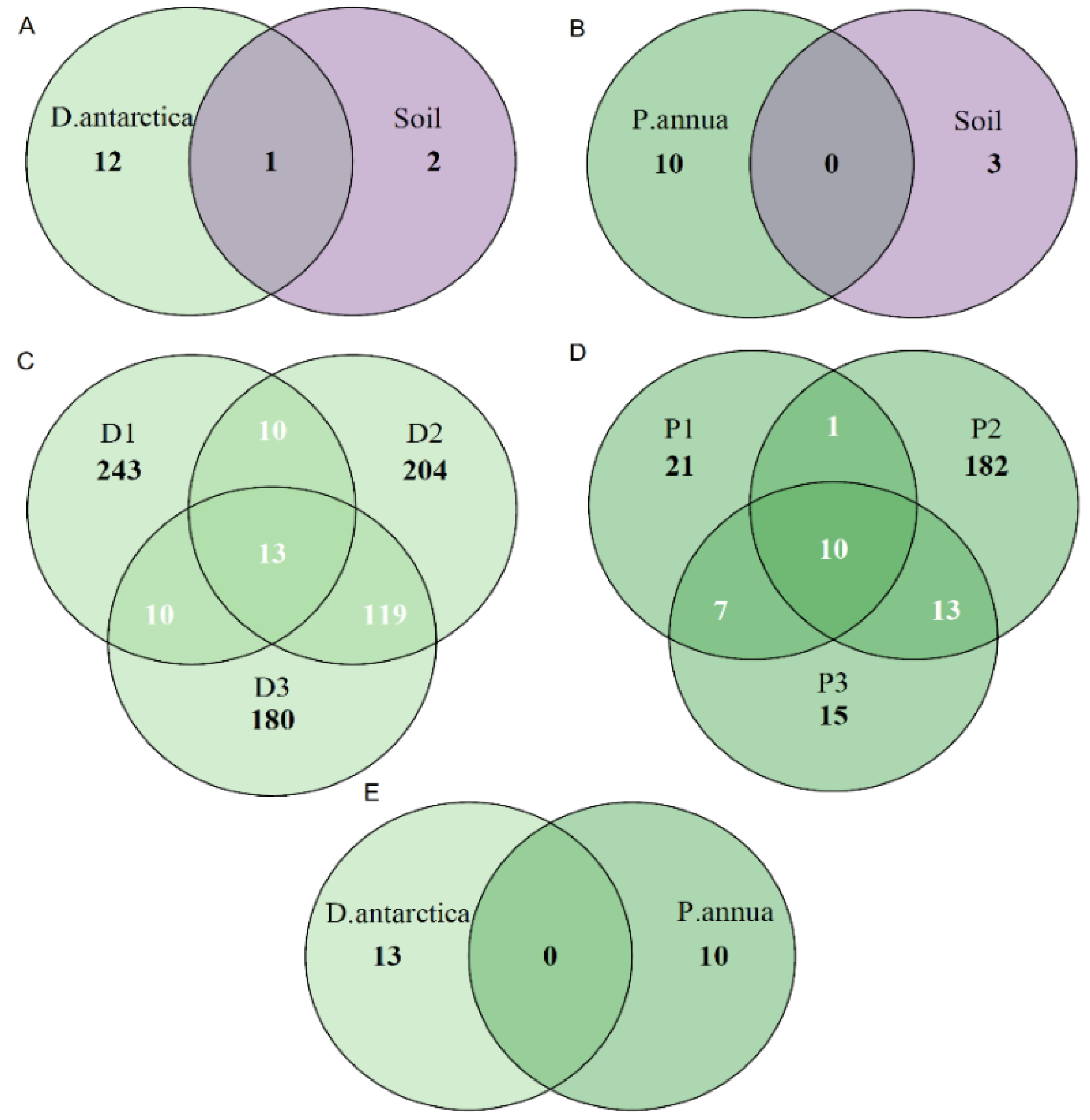
2.1. Fungal Diversity in Antarctic Grasses and the Rhizosphere Soil
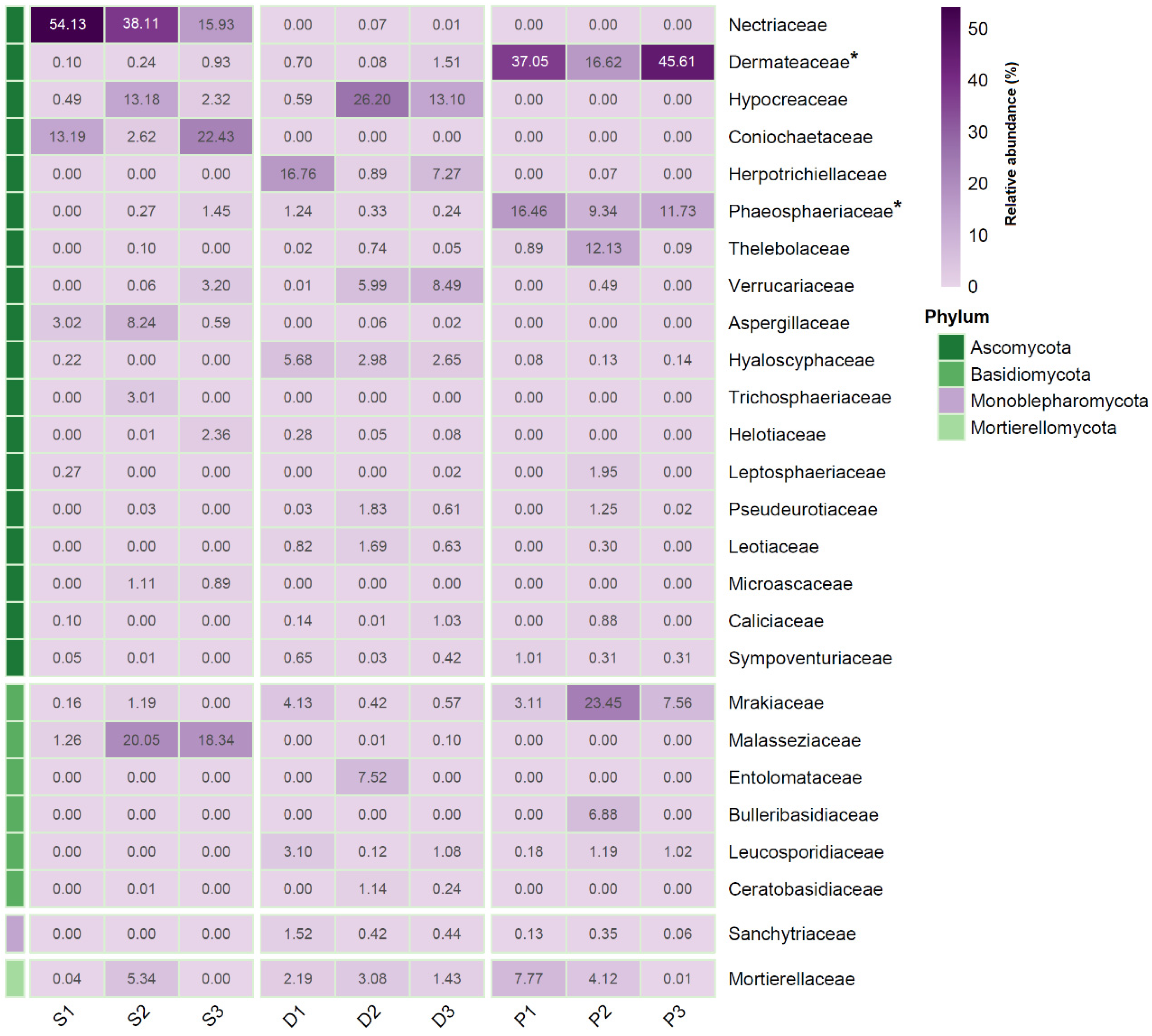
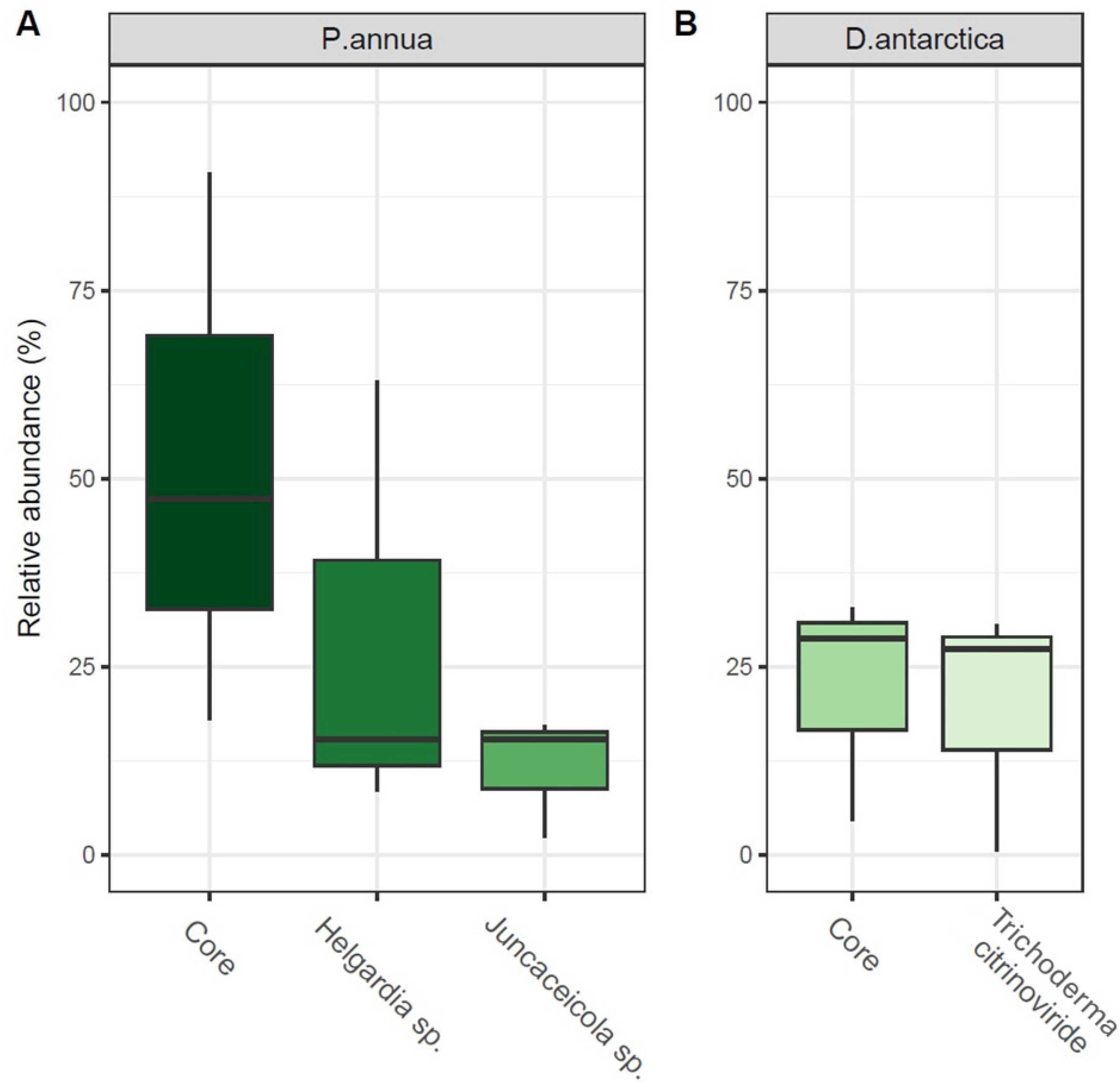
2.2. Fungi Identified to the Species Level from Grass Roots and Their Characteristics
2.3. Cultivable Fungi Inhabiting Grasses in Antarctica and Their Hydrolytic Activity
3. Discussion
4. Material and Methods
4.1. Sites and Sampling
4.2. DNA Extraction, ITS1 Amplicon Library Preparation, and Sequencing
4.3. Cultivation of Fungi from Roots and Soil and DNA Extraction
4.4. Screening for Phosphate Solubilization Ability
4.5. Evaluation of Proteolytic Activity of Fungi
4.6. Screening for Chitinase, Xylanase, Amylase, Cellulase, and Pectinase Activity
4.7. Data Curation and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol. Plant 2017, 10, 2247–2258. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, H.; Zou, C.; Li, Y.; Chen, Y.; Wang, Z.; Jiang, A.; Zhao, P.; Wang, M.; Ahammed, G.J. Combined inoculation with multiple arbuscular mycorrhizal fungi improves growth, nutrient uptake and photosynthesis in cucumber seedlings. Front. Microbiol. 2017, 8, 2516. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, D.P.; Belesky, D.P. Adaptations of endophyte-infected cool-season grasses to environmental stresses: Mechanisms of drought and mineral stress tolerance. Crop Sci. 2000, 40, 923–940. [Google Scholar] [CrossRef]
- Thangavel, P.; Anjum, N.A.; Muthukumar, T.; Sridevi, G.; Vasudhaven, P.; Maruthupandian, A. Arbuscular mycorrhizae: Natural modulators of plant–nutrient relation and growth in stressful environments. Arch. Microbiol. 2022, 204, 264. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Fontana, D.C.; de Paula, S.; Torres, A.G.; de Souza, V.H.M.; Pascholati, S.F.; Schmidt, D.; Neto, D.D. Endophytic fungi: Biological control and induced resistance to phytopathogens and abiotic stresses. Pathogens 2021, 10, 570. [Google Scholar] [CrossRef]
- Avio, L.; Pellegrino, E.; Bonari, E.; Giovannetti, M. Functional diversity of arbuscular mycorrhizal fungal isolates in relation to extraradical mycelial networks. New Phytol. 2006, 172, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Helber, N.; Wippel, K.; Sauer, N.; Schaarschmidt, S.; Hause, B.; Requena, N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp. is crucial for the symbiotic relationship with plants. Plant Cell 2011, 23, 3812–3823. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D.; Wang, E. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef]
- Lee, E.-H.; Eo, J.-K.; Ka, K.-H.; Eom, A.-H. Diversity of arbuscular mycorhizal fungi and their role in ecosystems. Mycobiology 2013, 41, 121–125. [Google Scholar] [CrossRef]
- Znój, A.; Grzesiak, J.; Gawor, J.; Gromadka, R.; Chwedorzewska, K.J. Bacterial Communities Associated with Poa annua Roots in Central European (Poland) and Antarctic Settings (King George Island). Microorganisms 2021, 9, 811. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.H. Cold adaptation in Arctic and Antarctic fungi. New Physiol. 2001, 151, 341–353. [Google Scholar] [CrossRef]
- Olech, M. Human impact on terrestrial ecosystems in west Antarctica. Proc. NIPR Symp. Polar Biol. 1996, 9, 299–306. [Google Scholar]
- Chwedorzewska, K. Poa annua L. in Antarctic: Searching for the source of introduction. Polar Biol. 2008, 33, 263–268. [Google Scholar] [CrossRef]
- Chwedorzewska, K.; Bednarek, P.T. Genetic and epigenetic variation in a cosmopolitan grass Poa annua from Antarctic and Polish populations. Polish Polar Res. 2012, 33, 63–80. [Google Scholar] [CrossRef]
- Crous, P.W.; Braun, U.; McDonald, B.A.; Lennox, C.L.; Edwards, J.; Mann, R.C.; Zaveri, A.; Linde, C.C.; Dyer, P.S.; Groenewald, J.Z. Redefining genera of cereal pathogens: Oculimacula, Rhynchosporium and Spermospora. Fungal Syst. Evol. 2020, 7, 67–98. [Google Scholar] [CrossRef] [PubMed]
- Palicova, J.; Matusinsky, P.; Dumalasova, V.; Hanzalova, A.; Svacinova, I.; Chrpova, J. The use of Real-Time PCR for the pathogen quantification in breeding winter wheat varieties resistant to eyespot. Plants 2022, 11, 1495. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Grung, C.R.; Holdenrieder, O.; Sieber, T.N. Cryptic speciation and community structure of Herpotrichia juniperi, the causal agent of brown felt blight of conifers. Mycol. Res. 2009, 113, 887–896. [Google Scholar] [CrossRef]
- Barahona, S.; Yuivar, Y.; Socias, G.; Alcaino, J.; Cifuentes, V.; Baeza, M. Identification and characterization of yeasts isolated from sedimentary rocks of Union Glacier at the Antarctica. Extremophiles 2016, 20, 479–491. [Google Scholar] [CrossRef]
- Zhang, F.; Huo, Y.; Cobb, A.B.; Luo, G.; Zhou, J.; Yang, G.; Wilson, G.W.T.; Zhang, Y. Trichoderma biofertilizer links to altered soil chemistry, altered microbial communities, and improved grassland biomass. Front. Microbiol. 2018, 9, 848. [Google Scholar] [CrossRef]
- Ozimek, E.; Jaroszuk-Ściseł, J.; Bohacz, J.; Korniłłowicz-Kowalska, T.; Tyśkiewicz, R.; Słomka, A.; Nowak, A.; Hanaka, A. Synthesis of indoleacetic acid, gibberellic acid and ACC-deaminase by Mortierella strains promote winter wheat seedlings growth under different conditions. Int. J. Mol. Sci. 2018, 19, 3218. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M.; Neubert, K.; Mendgen, K.W.; Wirsel, S.G.R. Niche differentiation of two sympatric species of Microdochium colonizing the roots of common reed. BMC Microbiol. 2011, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.F.I.; Flores, F.J.; Koch, A.R.; Garzon, C.D.; Walker, N.R. Multiplex end-point PCR for the detection of three species of Ophiosphaerella causing spring dead spot of Bermudagrass. Plant Dis. 2019, 103, 2010–2014. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.H.D.; Kristanti, R.A. Bioremediation of perfluorochemicals: Current state and the way forward. Bioprocess Biosyst. Eng. 2022, 45, 1093–1109. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Li, X.-Q.; Wang, Z.-M.; Zheng, L.; Yu, Y.-Y.; Dai, J.-J.; Shi, D.-Y. Bioactivity-guided screening of antimicrobial secondary metabolites from Antarctic cultivable fungus Acrostalagmus luteoalbus CH-6 combined with molecular networking. Mar. Drugs 2022, 20, 334. [Google Scholar] [CrossRef] [PubMed]
- Badet, T.; Peyraud, R.; Raffaele, S. Common protein sequence signatures associate with Sclerotinia borealis lifestyle and secretion in fungal pathogens of the Sclerotiniaceae. Front. Plant Sci. 2015, 6, 776. [Google Scholar] [CrossRef] [PubMed]
- Dhume, G.M.; Maharana, A.K.; Tsuji, M.; Srivastave, A.K.; Singh, S.M. Cold-tolerant endoglucanase producing ability of Mrakia robertii A2-3 isolated from cryoconites, Hamtha glacier, Himalaya. J. Basic. Microbiol. 2019, 59, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Laich, F.; Chavex, R.; Vaca, I. Leucosporidium escuderoi f.a., sp. nov., a basidiomycetous yeast associated with an Antarctic marine sponge. Antonie Van Leeuwenhoek 2014, 105, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Vero, S.; Garmendia, G.; Gonzalez, M.B.; Bentancur, O.; Wisniewski, M. Evaluation of yeasts obtained from Antarctic soil samples as biocontrol agents for the management of postharvest diseases of apple (Malus × domestica). FEMS Yeast Res. 2013, 13, 189–199. [Google Scholar] [CrossRef]
- Perini, L.; Andrejasic, K.; Gostincar, G.; Gunde-Cimerman, N.; Zalar, P. Greenland and Svalbard glaciers host unknown basidiomycetes: The yeast Camptobasidium arcticum sp. nov. and the dimorphic Psychromyces glacialis gen. and sp. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 004655. [Google Scholar] [CrossRef]
- Porto, B.A.; da Silva, T.H.; Machado, M.R.; Soares de Oliveira, F.S.; Rosa, C.A.; Rosa, L.H. Diversity and distribution of cultivable fungi present in acid sulphate soils in chronosequence under para-periglacial conditions in King George Island, Antarctic. Extremophiles 2020, 24, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, D.; Luan, Y.; Gu, Q.; Zhu, T. Cytotoxic metabolites from the Antarctic psychrophilic fungus Oidiodendron truncatum. J. Nat. Prod. 2012, 75, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Rusman, Y.; Wilson, J.M.; Held, B.W.; Balnchette, R.A.; Anderson, B.N.; Lupfer, C.R.; Salomon, C.E. Antifungal norditerpene oidiolactones from the fungus Oidiodendron truncatum, a potential biocontrol agent for white-nose syndrome in bats. J. Nat. Prod. 2020, 83, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Held, B.W.; Salomon, C.E.; Blanchette, R.A. Diverse subterranean fungi of an underground iron ore mine. PLoS ONE 2020, 15, e0234208. [Google Scholar] [CrossRef] [PubMed]
- Curry, T.M.; Pena, M.J.; Urbanowicz, B.R. An update on xylan structure, biosynthesis, and potential commercial applications. Cell Surf. 2023, 9, 100101. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C. Cellulose synthesis in high plants. Annu. Rev. Cell Dev. Biol. 2006, 22, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opi. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Gago, J.; Nadal, M.; Clemente-Moreno, M.J.; Figueroa, C.M.; Medeiros, D.B.; Cubo-Ribas, N.; Cavieres, L.A.; Gulias, J.; Fernie, A.R.; Flexas, J.; et al. Nutrient availability regulates Deschampsia antarctica photosynthetic and stress tolerance performance in Antarctica. J. Exp. Bot. 2023, 74, 2620–2637. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, B.A.; Jumpponen, A.; Trappe, J.M. Utilization of major detrital substrates by dark-septate root endophytes. Mycologia 2000, 92, 230–232. [Google Scholar] [CrossRef]
- Roberts, P.; Newsham, K.K.; Bardgett, R.D.; Farrar, J.F.; Jones, D.L. Vegetation cover regulates the quantity, quality and temporal dynamics of dissolved organic carbon and nitrogen in Antarctic soils. Polar Biol. 2009, 32, 999–1008. [Google Scholar] [CrossRef]
- Wang, T.; Tian, Z.; Tunlid, A.; Persson, P. Nitrogen acquisition from mineral-associated proteins by an ectomycorrhizal fungus. New Phytol. 2020, 228, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Santiago, I.F.; Rosa, C.A.; Rosa, L.H. Endophytic symbiont yeasts associated with the Antarctic angiosperms Deschampsia antarctica and Colobanthus quitensis. Polar Biol. 2017, 40, 177–183. [Google Scholar] [CrossRef]
- Mosyakin, S.L.; Bezusko, L.G.; Mosyakin, A.S. Origins of native vascular plants of Antarctica: Comments from a historical phytogeography viewpoint. Cytol. Genet. 2007, 41, 308–316. [Google Scholar] [CrossRef][Green Version]
- Koyama, A.; Maherali, H.; Antunes, P.M. Plant geographic origin and phylogeny as potential drivers of community structure in root-inhabiting fungi. J. Ecol. 2019, 107, 1720–1736. [Google Scholar] [CrossRef]
- Chauhan, P.; Sharma, N.; Tapwal, A.; Kumar, A.; Verma, G.S.; Meena, M.; Swapnil, P. Soil microbiome: Diversity, benefits and interactions with plants. Sustainability 2023, 15, 14643. [Google Scholar] [CrossRef]
- Upson, R.; Newsham, K.K.; Read, D.J. Root-fungal associations of Colobanthus quitensis and Deschampsia antarctica in the maritime and subantarctic. Arctic Antarct. Alp. Res. 2008, 40, 592–599. [Google Scholar] [CrossRef]
- Guo, Q.; Shi, L.; Wang, X.; Li, D.; Yin, Z.; Zhang, J.; Ding, G.; Chen, L. Structures and biological activities of secondary metabolites from the Trichoderma genus (covering 2018–2022). J. Agric. Food Chem. 2023, 71, 13612–13632. [Google Scholar] [CrossRef]
- Lombardi, N.; Vitale, S.; Turrà, D.; Reverberi, M.; Fanelli, C.; Vinale, F.; Lorito, M. Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol. Plant-Microbe Int. 2018, 31, 982–994. [Google Scholar] [CrossRef]
- Meteyer, C.U.; Dutheil, J.Y.; Keel, M.K.; Boyles, J.G.; Stukenbrock, E.H. Plant pathogens provide clues to the potential origin of bat white-nose syndrome Pseudogymnoascus destructans. Virulence 2022, 13, 1020–1031. [Google Scholar] [CrossRef]
- Molina-Montenegro, M.A.; Ballesteros, G.I.; Acuña-Rodríguez, I.S.; Pertierra, L.R.; Greve, M.; Richardson, D.M.; Newsham, K.K. The “Trojan horse” strategy: Seed fungal endophyte symbiosis helps to explain the invasion success of the grass, Poa annua, in Maritime Antarctica. Divers. Distrib. 2023, 29, 1432–1444. [Google Scholar] [CrossRef]
- Znój, A.; Grzesiak, J.; Gawor, J.; Gromadka, R.; Chwedorzewska, K.J. Highly specialized bacterial communities within three distinct rhizocompartments of Antarctic hairgrass (Deschampsia antarctica Desv.). Polar Biol. 2022, 45, 833–844. [Google Scholar] [CrossRef]
- Garcia, R.; Müller, R. The family Polyangiaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin, Germany, 2014. [Google Scholar]
- Chwedorzewska, K.J.; Giełwanowska, I.; Olech, M.; Molina-Montenegro, M.A.; Wódkiewicz, M.; Galera, H. Poa annua L. in the maritime Antarctic: An overview. Polar Rec. 2015, 51, 637–643. [Google Scholar] [CrossRef]
- Olech, M.; Chwedorzewska, K.J. Short note: The first appearance and establishment of an alien vascular plant in natural habitats on the forefield of a retreating glacier in Antarctica. Antarct. Sci. 2011, 23, 153–154. [Google Scholar] [CrossRef]
- Galera, H.; Wódkiewicz, M.; Czyż, E.; Łapiński, S.; Kowalska, M.E.; Pasik, M.; Chwedorzewska, K.J. First step to eradication of Poa annua L. from point Thomas oasis (King George Island, south Shetlands, Antarctica). Polar Biol. 2017, 40, 939–945. [Google Scholar] [CrossRef]
- Zdanowski, M.K.; Żmuda-Baranowska, M.J.; Borsuk, P.; Świątecki, A.; Górniak, D.; Wolicka, D.; Grzesiak, J. Culturable bacteria community development in postglacial soils of Ecology Glacier, King George Island, Antarctica. Polar Biol. 2013, 36, 511–527. [Google Scholar] [CrossRef]
- Araya, M.A.; Valenzuela, T.; Inostroza, N.G.; Maruyama, F.; Jorquera, M.A.; Acuña, J.J. Isolation and characterization of cold-tolerant hyper-ACC-degrading bacteria from the rhizosphere, endosphere, and phyllosphere of antarctic vascular plants. Microorganisms 2020, 8, 1788. [Google Scholar] [CrossRef]
- Tian, Y.; Gao, L. Bacterial diversity in the rhizosphere of cucumbers grown in soils covering a wide range of cucumber cropping histories and environmental conditions. Microb. Ecol. 2014, 68, 794–806. [Google Scholar] [CrossRef] [PubMed]
- De Andrade, G.A.K.; Cañón, E.R.P.; Alves, R.P.; Schmitz, D.; Schünemann, A.L.; De Albuquerque, M.P.; Putzke, J.; Pereira, A.B.; De Carvalho Victoria, F. First Record of Juncaceicola as Endophytic Fungi Associated with Deschampsia antarctica Desv. Diversity 2018, 10, 107. [Google Scholar] [CrossRef]
- Upson, R.; Newsham, K.K.; Bridge, P.D.; Pearce, D.A.; Read, D.J. Taxonomic affinities of dark septate root endophytes of Colobanthus quitensis and Deschampsia antarctica, the two native Antarctic vascular plant species. Fungal Ecol. 2009, 2, 184–196. [Google Scholar] [CrossRef]
- Koljalg, U.; Nilson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Martin, K.J.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.A.; Balint, M.; Greshake, B.; Bandow, C.; Rombke, J.; Schmitt, I. Illumina metabarcoding of a soil fungal community. Soil. Biol. Biochem. 2013, 65, 128–132. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 17, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Balint, M.; Schidt, P.A.; Sharma, R.; Thines, M.; Schmidt, I. An Illumina metabarcoding pipeline for fungi. Ecol. Evol. 2014, 4, 2642–2653. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- King, A.D., Jr.; Hocking, A.D.; Pitt, J.I. Dichloran-rose Bengal medium for enumeration and isolation of molds from foods. Appl. Environm. Microbiol. 1979, 37, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Castellanos, M.L.; Rodriquez-Segura, Z.; Villabobos, F.J.; Hernandez, L.; Lina, L.; Nunez-Valdez, M.E. Pathogenicity of isolates of Serratia marcescens towards larvae of the scarab Phyllophaga balnchardi (Coleoptera). Pathogens 2015, 4, 210–228. [Google Scholar] [CrossRef] [PubMed]
- Verma, K.; Garg, N. Detection of chitinase on chitin agar plates. Int. J. Sci. Res. 2019, 8, 1186–1189. [Google Scholar]
- Nair, M.G.; Sindhu, R.; Shashidhar, S. Fungal xylanase production under solid state and submerged fermentation conditions. Afr. J. Microbiol. Res. 2008, 2, 82–86. [Google Scholar]
- Aarti, C.; Khusro, A.; Agastian, P.; Darwich, N.M.; Al Farraj, D.A. Molecular diversity and hydrolytic enzymes production abilities of soil bacteria. Saudi J. Biol. Sci. 2020, 27, 3235–3248. [Google Scholar] [CrossRef]
- Sahay, S.; Hamid, B.; Singh, P.; Ranjan, K.; Chauhan, D.; Rana, R.S.; Chaurse, V.K. Evaluation of pectinolytic activities for oenological uses from psychrotrophic yeasts. Lett. Appl. Microbiol. 2013, 57, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 July 2024).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Niu, G.; Chen, T.; Shen, Q.; Yuan, J.; Liu, Y.-X. The best practice for microbiome analysis using R. Protein Cell 2023, 14, 713–725. [Google Scholar] [CrossRef] [PubMed]


| Fungal Species | Phylum | Characteristics | Reference |
|---|---|---|---|
| D. antarctica roots | |||
| Helgardia anguioides-like (Oculimacula) | Ascomycota | causes eyespot disease of wheat | [16,17] |
| Herpotrichia juniperi | Ascomycota | causal agent of brown felt blight of conifers | [18] |
| Sporobolomyces roseus | Basidiomycota | synthesizes carotenoids and mycosporines to absorb UV radiation; secretes antimicrobial protein | [19] |
| Ophiosphaerella sp. | Ascomycota | grass pathogen | [20] |
| P. annua roots | |||
| Mortierella antarctica-like | Mortierellomycota | promotes winter wheat seedling growth | [21] |
| Microdochium phragmitis | Ascomycota | endophyte colonizing roots of common reed | [22] |
| Ophiosphaerella sp. | Ascomycota | causes spring dead spot of bermudagrass | [23] |
| Soil—Ecology Glacier foreland | |||
| Cladosporium herbarum | Ascomycota | source of antimicrobial compounds; effective in bioremediation of perfluorochemicals | [24,25] |
| Botrytis cinerea | Ascomycota | cosmopolitan broad host-range plant pathogen | [26] |
| Mrakia robertii | Basidiomycota | producer of cold-tolerant enzymes | [27] |
| Leucosporidium sp. Camptobasidium gelus | Basidiomycota | Leucosporidium sp.—yeast from Antarctic marine sponge, can biocontrol B. cinerea infection Camptobasidium gelus-yeast found in Svalbard and Greenland glacial | [28,29,30] |
| Leucosporidiales sp. | Basidiomycota | ||
| Soil—Arctowski Station 2 | |||
| Mortierella fimbricystis | Mortierellomycota | tolerates low pH, low temperature, high concentration of metals, and low organic matter | [31] |
| Oidiodendron truncatum | Ascomycota | producer of antifungal metabolites | [32,33] |
| Pseudoeurotium sp. | Ascomycota | candidate for bioremediation of polluted soil | [24] |
| Pseudoeurotium sp. | Ascomycota | candidate for bioremediation of polluted soil | [24] |
| Linnemannia hialina (Mortierella hyalina) | Mortierellomycota | identified in iron ore mine as heavy metal-tolerating | [34] |
| Cladosporium tenuissimum | Ascomycota | candidate for bioremediation of polluted soil | [24] |
| Site | Geographical Coordinates | Distance to the Sea | Sample ID | Landform and Habitat |
|---|---|---|---|---|
| Arctowski Station 1 | 62°09′35″ S 58°28′26″ W | 120 m | S1, D1, P1 | Scientific station area; soil mechanically altered by human activities; Skeletic Eutric Fluvisol (Turbic). The site is strongly influenced by marine aerosols; it is moist and located further from sources of nutrients, with large human influence. |
| Arctowski Station 2 | 62°09′33″ S 58°28′25″ W | 100 m | S2, D2, P2 | Scientific station area; soil mechanically altered by human activities; Skeletic Eutric Fluvisol (Turbic). The site is strongly influenced by marine aerosols; it is moist and close to animal breeding colonies, with large human influence, especially related to using heavy vehicles. |
| Ecology Glacier foreland | 62°10′05″ S 58°27′46″ W | 20 m | S3, D3, P3 | Fluted moraine; Eutric Skeletic Protic Regosol (Turbic). Dry site, sheltered from sources of nutrients, with little influence of marine aerosols. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piłsyk, S.; Perlińska-Lenart, U.; Janik, A.; Skalmowska, P.; Znój, A.; Gawor, J.; Grzesiak, J.; Kruszewska, J.S. Native and Alien Antarctic Grasses as a Habitat for Fungi. Int. J. Mol. Sci. 2024, 25, 8475. https://doi.org/10.3390/ijms25158475
Piłsyk S, Perlińska-Lenart U, Janik A, Skalmowska P, Znój A, Gawor J, Grzesiak J, Kruszewska JS. Native and Alien Antarctic Grasses as a Habitat for Fungi. International Journal of Molecular Sciences. 2024; 25(15):8475. https://doi.org/10.3390/ijms25158475
Chicago/Turabian StylePiłsyk, Sebastian, Urszula Perlińska-Lenart, Anna Janik, Patrycja Skalmowska, Anna Znój, Jan Gawor, Jakub Grzesiak, and Joanna S. Kruszewska. 2024. "Native and Alien Antarctic Grasses as a Habitat for Fungi" International Journal of Molecular Sciences 25, no. 15: 8475. https://doi.org/10.3390/ijms25158475
APA StylePiłsyk, S., Perlińska-Lenart, U., Janik, A., Skalmowska, P., Znój, A., Gawor, J., Grzesiak, J., & Kruszewska, J. S. (2024). Native and Alien Antarctic Grasses as a Habitat for Fungi. International Journal of Molecular Sciences, 25(15), 8475. https://doi.org/10.3390/ijms25158475






