Unearthing Optimal Symbiotic Rhizobia Partners from the Main Production Area of Phaseolus vulgaris in Yunnan
Abstract
:1. Introduction
2. Results
2.1. Physicochemical Characteristics of Soils and the Environment
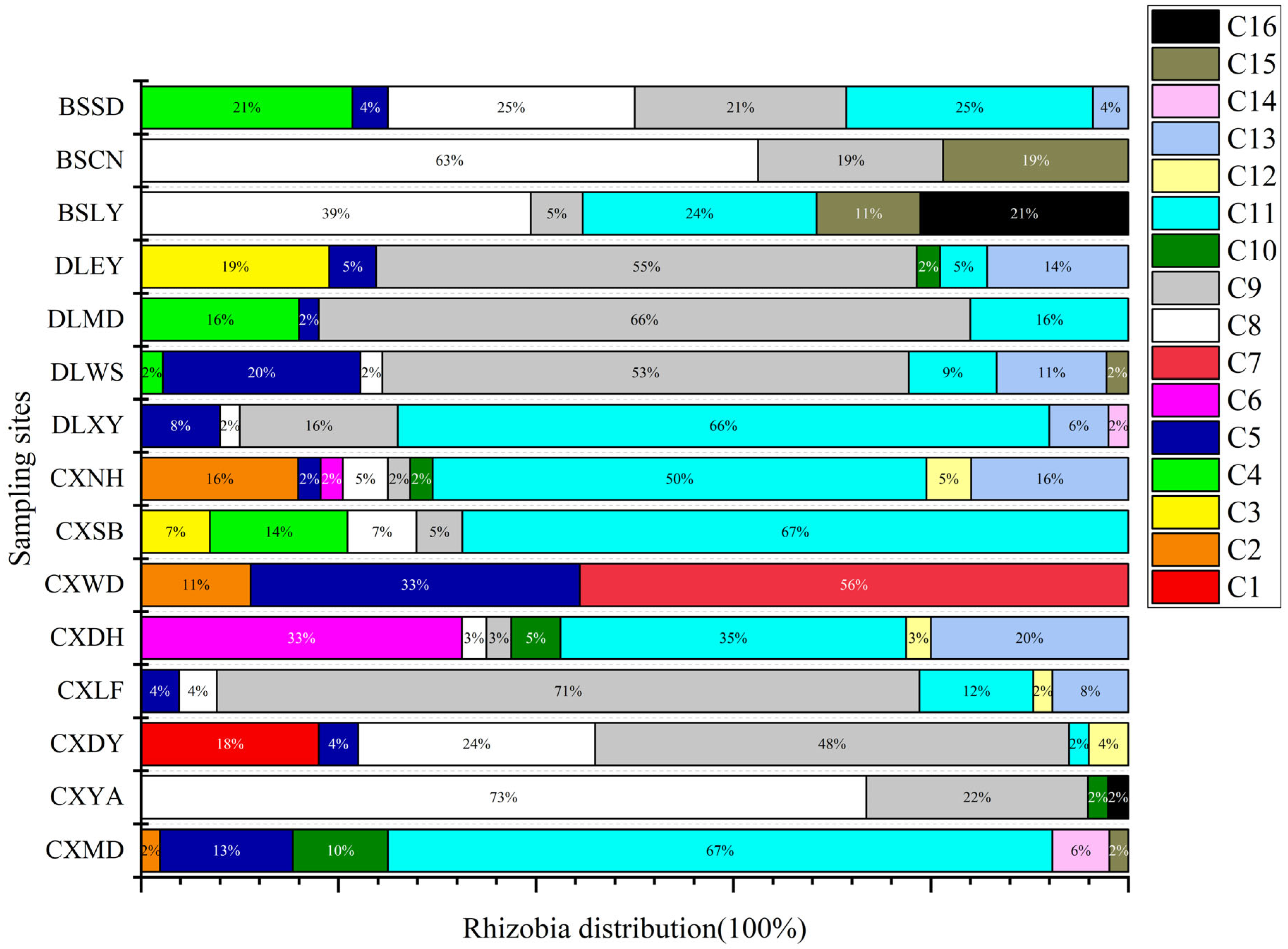
2.2. Rhizobial Collection and IGS PCR-RFLP Analysis
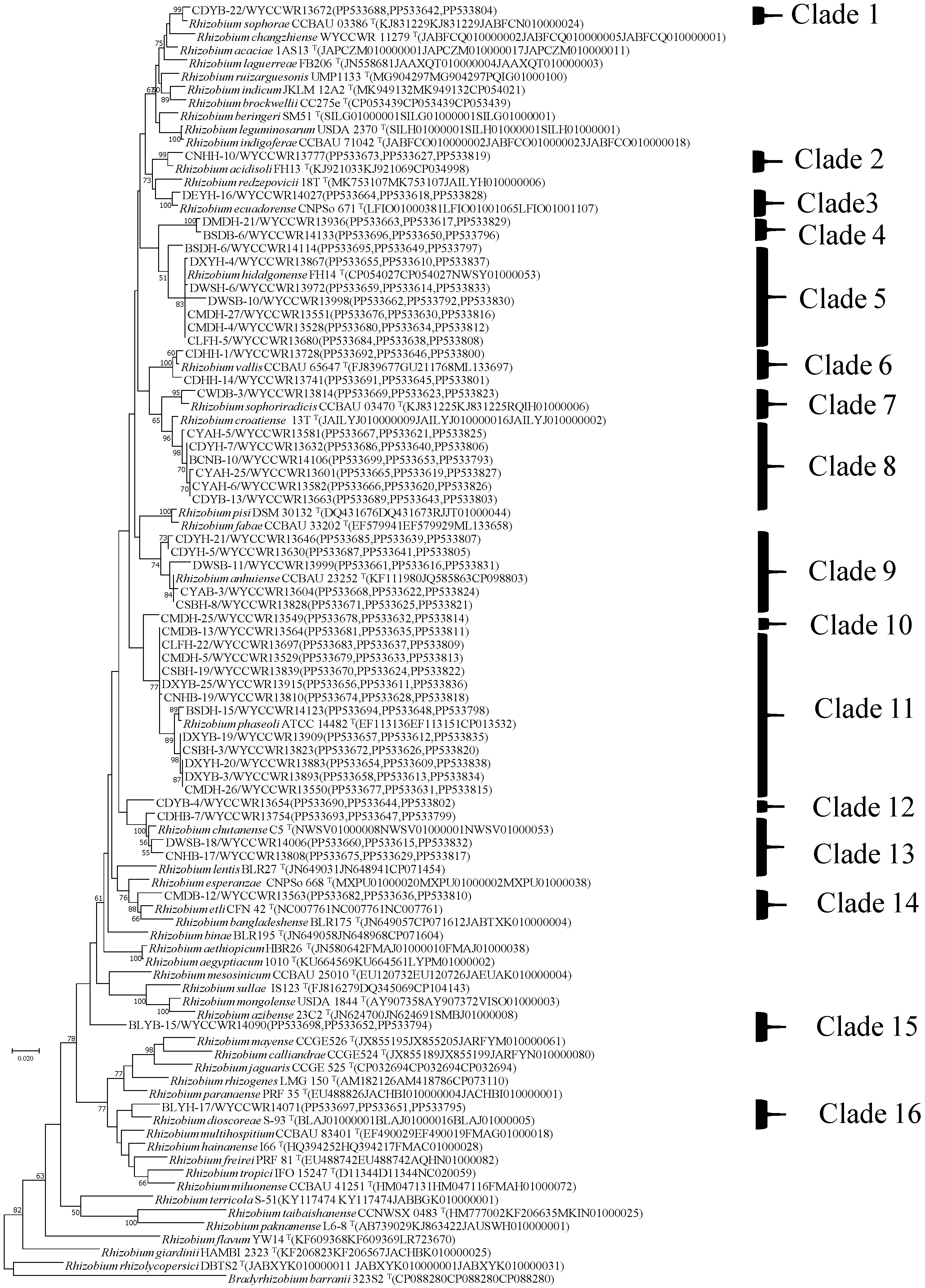
2.3. Identification of Species by Phylogenetic Analysis of Core Genes
2.4. Identification of Symbiovars by Phylogenetic Analysis of Symbiotic Genes
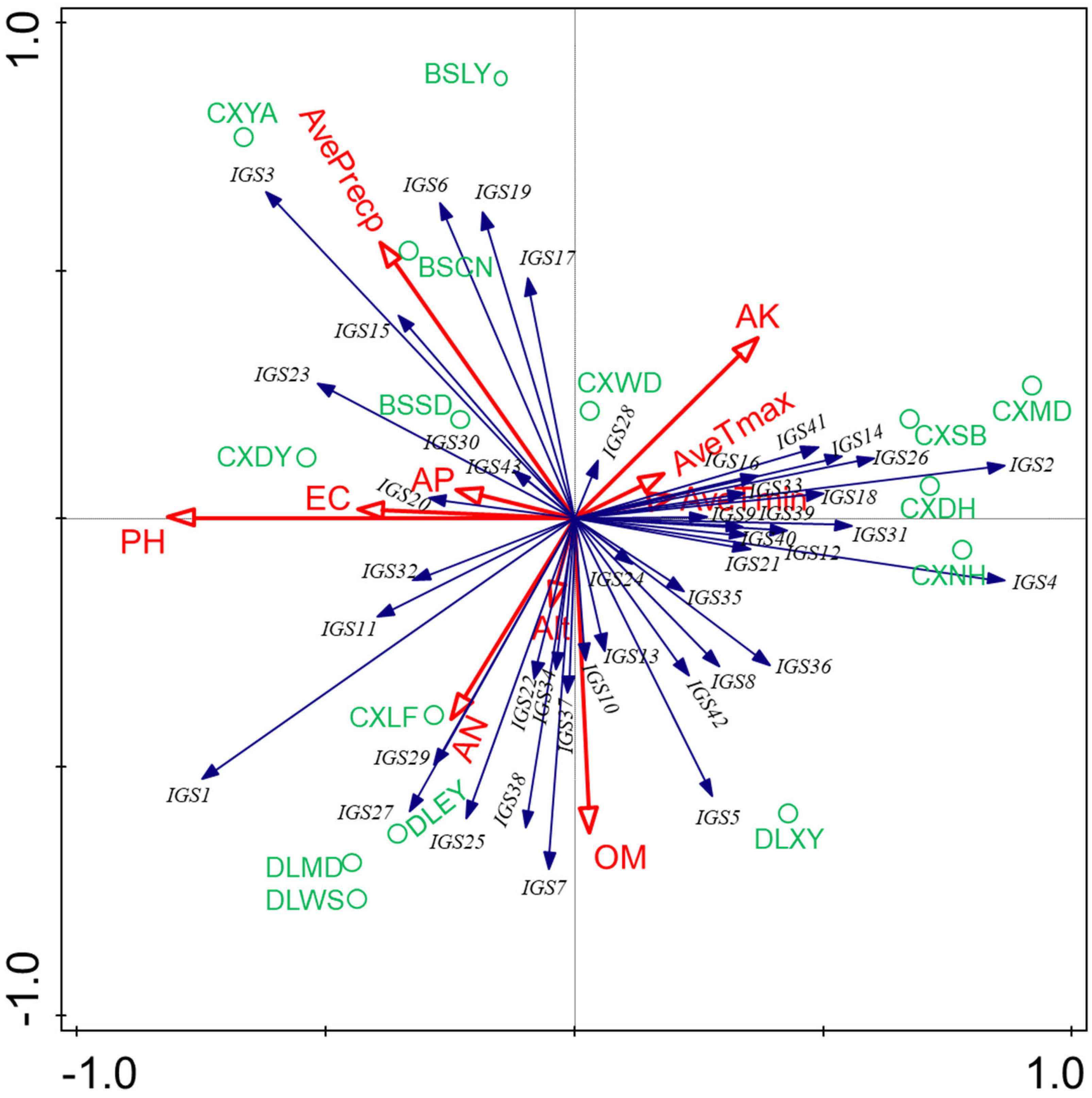
2.5. Correlation Analysis of Rhizobial Distribution with Soil and Environmental Properties
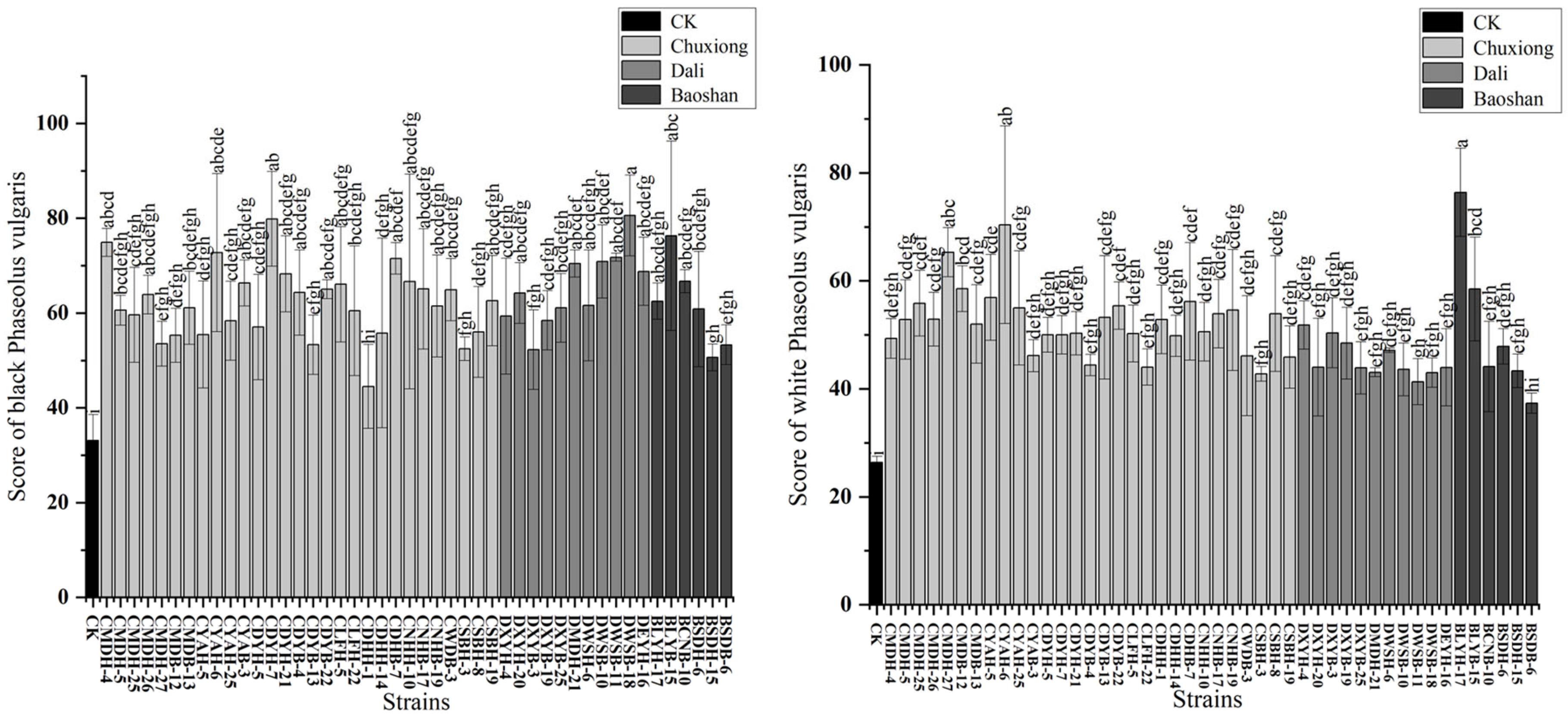
2.6. Symbiotic Efficiency of Rhizobial Representative Strains
2.7. Abiotic Stress Tolerance of Representative Strains
3. Discussion
4. Materials and Methods
4.1. Field Soil Sampling and Soil and Environmental Characteristics
4.2. Rhizobial Isolation and Conservation
4.3. Genomic Characterization of Rhizobial Isolates
4.4. Molecular and Phylogenetic Identification of the Isolates, Alpha-Diversity Estimation
4.5. Correlation Analysis of Soil Properties and Environmental Factors with Rhizobial Communities
4.6. Symbiotic Efficiency Measurements
4.7. Measurements of Abiotic Stress on Rhizobial Strain Growth
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, Z. High yield cultivation and management technology of kidney beans in North China. Agric. Technol. Equip. 2017, 11, 43–45. [Google Scholar]
- Luo, A.; Li, M.; Liu, F.; Fu, Q.; Dong, J. Research progress in genetic breeding of common bean. Vegetables 2023, 36–42. [Google Scholar]
- Grange, L.; Hungria, M.; Graham, P.; Martinez-Romero, E. New insights into the origins and evolution of rhizobia that nodulate common bean (Phaseolus vulgaris) in Brazil. Soil Biol. Biochem. 2007, 39, 867–876. [Google Scholar] [CrossRef]
- Zhang, X.; Blair, M.W.; Wang, S. Genetic diversity of Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeat markers. Theor. Appl. Genet. 2008, 117, 629–640. [Google Scholar] [CrossRef]
- Ma, X.; Li, L. 16S rDNA sequence analysis of Rhizobium strains isolated from Soybeans and kidney beans in Rizhao. Hubei Agric. Sci. 2017, 56, 2938–2941. [Google Scholar] [CrossRef]
- Wu, J. Research progress on common bean genomics study. J. Sichuan Agric. Univ. 2021, 39, 4–10. [Google Scholar] [CrossRef]
- Yao, Y.; Jie, W.; Du, Y.; Zhao, D.; Yan, X. Taxonomy, Identification and application of Rhizobium. Chin. Agric. Sci. Bull. 2020, 36, 100–105. [Google Scholar]
- Khalid, R.; Zhang, Y.; Ali, S.; Sui, X.; Zhang, X.; Amara, U.; Chen, W.; Hayat, R. Rhizobium pakistanensis sp. nov., isolated from groundnut (Arachis hypogaea) nodules grown in rainfed Pothwar, Pakistan. Antonie Van Leeuwenhoek 2014, 107, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Li, Z. The Effects of Intercropping and Dual Inoculation on Maize and Common Bean (Phaseolus vulgaris) Growth. Chin. Agric. Sci. Bull. 2014, 30, 182–188. [Google Scholar]
- Sena, P.T.S.; do Nascimento, T.R.; Lino, J.d.O.S.; Oliveira, G.S.; Ferreira Neto, R.A.; de Freitas, A.D.S.; Fernandes-Júnior, P.I.; Martins, L.M.V. Molecular, physiological, and symbiotic characterization of cowpea rhizobia from soils under different agricultural systems in the semiarid region of Brazil. J. Soil Sci. Plant Nutr. 2020, 20, 1178–1192. [Google Scholar] [CrossRef]
- Kumar, N.; Srivastava, P.; Vishwakarma, K.; Kumar, R.; Kuppala, H.; Maheshwari, S.K.; Vats, S. The Rhizobium–plant symbiosis: State of the art. In Plant Microbe Symbiosis; Varma, A., Tripathi, S., Prasad, R., Eds.; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Yan, Y.; Tian, C.; Yang, J.; Wang, Y.; Lin, M. Establishment of artificial efficiency biological nitrogen fixation system and its agricultural application. Chin. Bull. Life Sci. 2021, 33, 1532–1543. [Google Scholar] [CrossRef]
- Xiang, Z.; Song, Y. Breeding of Rh. phaseoli and its effective application technology. J. Microbiol. 1988, 40–44. [Google Scholar]
- Martínez-Romero, E.; Segovia, L.; Mercante, F.M.; Franco, A.A.; Graham, P.; Pardo, M.A. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int. J. Syst. Bacteriol. 1991, 41, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Roman-Ponce, B.; Zhang, Y.J.; Soledad Vasquez-Murrieta, M.; Sui, X.H.; Chen, W.F.; Alberto Padilla, J.C.; Guo, X.W.; Gao, J.L.; Yan, J.; Wei, G.H.; et al. Rhizobium acidisoli sp nov., isolated from root nodules of Phaseolus vulgaris in acid soils. Int. J. Syst. Evol. Microbiol. 2016, 66, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Rajnovic, I.; Ramírez-Bahena, M.-H.; Kajic, S.; Igual, J.M.; Peix, Á.; Velázquez, E.; Sikora, S. Rhizobium croatiense sp. nov. and Rhizobium redzepovicii sp. nov., two new species isolated from nodules of Phaseolus vulgaris in Croatia. Syst. Appl. Microbiol. 2022, 45, 126317. [Google Scholar] [CrossRef] [PubMed]
- Dall’Agnol, R.F.; Ribeiro, R.A.; Ormeno-Orrillo, E.; Rogel, M.A.; Hungria, M. Rhizobium freirei sp. nov., a symbiont of Phaseolus vulgaris that is very effective at fixing nitrogen. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 11, 4167–4173. [Google Scholar] [CrossRef] [PubMed]
- Aurag, J.; Sasson, A. Tolerance of Rhizobium leguminosarum bv. phaseoli to acidity and drought. World J. Microbiol. Biotechnol. 1992, 8, 532–533. [Google Scholar] [CrossRef]
- Segovia, L.; Young, J.P.W.; Martinez-Romero, E. Reclassification of American Rhizobium leguminosarum Biovar Phaseoli Type I Strains as Rhizobium etli sp. nov. Int. J. Syst. Bacteriol. 1993, 43, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yan, H.; Liu, L.; Chen, W.; Zhang, X.; Verástegui-Valdés, M.M.; Wang, E.; Han, X. Rhizobium hidalgonense sp. nov., a nodule endophytic bacterium of Phaseolus vulgaris in acid soil. Arch. Microbiol. 2016, 199, 97–104. [Google Scholar] [CrossRef]
- Ribeiro, R.A.; Martins, T.B.; Ormeno-Orrillo, E.; Marcon Delamuta, J.R.; Rogel, M.A.; Martinez-Romero, E.; Hungria, M. Rhizobium ecuadorense sp nov., an indigenous N2-fixing symbiont of the Ecuadorian common bean (Phaseolus vulgaris L.) genetic pool. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 9, 3162–3169. [Google Scholar] [CrossRef]
- Amarger, N.; Macheret, V.; Laguerre, G. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris Nodules. Int. J. Syst. Bacteriol. 1997, 47, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, R.A.; Rogel, M.A.; Lopez-Lopez, A.; Ormeno-Orrillo, E.; Barcellos, F.G.; Martinez, J.; Thompson, F.L.; Martinez-Romero, E.; Hungria, M. Reclassification of Rhizobium tropici type A strains as Rhizobium leucaenae sp nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Romero, E. Diversity of Rhizobium-Phaseolus vulgaris symbiosis: Overview and perspectives. Plant Soil 2003, 252, 11–23. [Google Scholar] [CrossRef]
- Mnasri, B.; Liu, T.Y.; Saidi, S.; Chen, W.F.; Chen, W.X.; Zhang, X.X.; Mhamdi, R. Rhizobium azibense sp nov., a nitrogen fixing bacterium isolated from root-nodules of Phaseolus vulgaris. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 5, 1501–1506. [Google Scholar] [CrossRef] [PubMed]
- Aserse, A.A.; Rasanen, L.A.; Assefa, F.; Hailemariam, A.; Lindstrom, K. Phylogeny and genetic diversity of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia. Syst. Appl. Microbiol. 2012, 35, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Lopez, A.; Rogel-Hernandez, M.A.; Barois, I.; Ortiz Ceballos, A.I.; Martinez, J.; Ormeno-Orrillo, E.; Martinez-Romero, E. Rhizobium grahamii sp nov., from nodules of Dalea leporina, Leucaena leucocephala and Clitoria ternatea, and Rhizobium mesoamericanum sp nov., from nodules of Phaseolus vulgaris, siratro, cowpea and Mimosa pudica. Int. J. Syst. Evol. Microbiol. 2012, 62, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Luis Zurdo-Pineiro, J.; Garcia-Fraile, P.; Rivas, R.; Peix, A.; Leon-Barrios, M.; Willems, A.; Francisco Mateos, P.; Martinez-Molina, E.; Velazquez, E.; van Berkum, P. Rhizobia from Lanzarote, the Canary Islands, that nodulate Phaseolus vulgaris have characteristics in common with Sinorhizobium meliloti Isolates from Mainland Spain. Appl. Environ. Microbiol. 2009, 75, 2354–2359. [Google Scholar] [CrossRef] [PubMed]
- Mnasri, B.; Saïdi, S.; Chihaoui, S.-A.; Mhamdi, R. Sinorhizobium americanum symbiovar mediterranense is a predominant symbiont that nodulates and fixes nitrogen with common bean (Phaseolus vulgaris L.) in a Northern Tunisian field. Syst. Appl. Microbiol. 2012, 35, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Sadowsky, M.J.; Cregan, P.B.; Keyser, H.H. Nodulation and nitrogen fixation efficacy of Rhizobium fredii with Phaseolus vulgaris genotypes. Appl. Environ. Microbiol. 1988, 54, 1907–1910. [Google Scholar] [CrossRef]
- Wang, F.; Wang, E.T.; Wu, L.J.; Sui, X.H.; Li, Y.; Chen, W.X. Rhizobium vallis sp. nov., isolated from nodules of three leguminous species. Int. J. Syst. Evol. Microbiol. 2011, 61, 2582–2588. [Google Scholar] [CrossRef]
- Huo, Y.; Tong, W.; Wang, J.; Wang, F.; Bai, W.; Wang, E.; Shi, P.; Chen, W.; Wei, G. Rhizobium chutanense sp. nov., isolated from root nodules of Phaseolus vulgaris in China. Int. J. Syst. Evol. Microbiol. 2019, 69, 2049–2056. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Diversity and Population Structures of Rhizobia Nodulated with Phaseolus vulgaris in Two Ecoregions of China. Ph.D. Thesis, Northwest A&F University, Yangling, China, 2015. [Google Scholar]
- Wang, L.; Cao, Y.; Wang, E.T.; Qiao, Y.J.; Jiao, S.; Liu, Z.S.; Zhao, L.; Wei, G.H. Biodiversity and biogeography of rhizobia associated with common bean (Phaseolus vulgaris L.) in Shaanxi Province. Syst. Appl. Microbiol. 2016, 39, 211–219. [Google Scholar] [CrossRef]
- Tong, W. Comparative Genomics and Phylogeny Ofrhizobia Isolated from Phaseolus vulgaris. Ph.D. Thesis, Northwest A&F University, Yangling, China, 2019. [Google Scholar]
- Anyango, B.; Wilson, K.J.; Beynon, J.L.; Giller, K.E. Diversity of rhizobia nodulating Phaseolus vulgaris L. in two Kenyan Soils with contrasting pHs. Appl. Environ. Microbiol. 1995, 61, 4016–4021. [Google Scholar]
- Efstathiadou, E.; Savvas, D.; Tampakaki, A.P. Genetic diversity and phylogeny of indigenous rhizobia nodulating faba bean (Vicia faba L.) in Greece. Syst. Appl. Microbiol. 2020, 43, 126149. [Google Scholar] [CrossRef] [PubMed]
- Xia, C. Identification of Common Phaseolus vulgaris Germplasm Resources Based on Phenotype and SSR Molecular Markers. Master’s Thesis, Heilongjiang Bayi Agricultural Reclamation University, Daqing, China, 2020. [Google Scholar]
- Satyanandam, T.; Babu, K.; Yellamanda, B.; Kumar, K.V.; Rosaiah, G.; Vijayalakshmi, M. Diversity of indigenous symbiotic nitrogen fixing bacteria from blackgram Vigna mungo (L.) Hepper cultivated in rice fallows. Legume Res. 2022, 45, 994–999. [Google Scholar] [CrossRef]
- Wang, X.; Yang, J.; Xu, J.; Az, G. Isolation and back-grafting test of rhizobia from kidney bean. Xinjiang Agric. Sci. 2001, 76–77. [Google Scholar] [CrossRef]
- Savy, D.; Cozzolino, V. Novel fertilising products from lignin and its derivatives to enhance plant development and increase the sustainability of crop production. J. Clean. Prod. 2022, 366, 132832. [Google Scholar] [CrossRef]
- Xiang, Z.; Song, Y. Breeding and application of rhizobia vulgaris. Liaoning Agric. Sci. 1985, 35–39. [Google Scholar]
- Wei, G.; Ma, Z. Application of rhizobia-legume symbiosis for remediation of heavy-metal contaminated soils. Acta Microbiol. Sin. 2010, 50, 1421–1430. [Google Scholar] [CrossRef]
- Yu, B.; Liu, Y. Recent progress in study on salt tolerance in soybean. Soybean Sci. 2000, 19, 154–159. [Google Scholar]
- Han, M. Drought and saline tolerance of Vicia faba Rhizobium. J. Qinghai Univ. 2019, 37, 35–41. [Google Scholar] [CrossRef]
- Chi, Y.C.; Wang, J.H.; Fan, T.Q.; Yu, S.L.; Jiang, C.J.; Zhou, G.; Li, H.Z. Preliminary study on NaCl-tolerance and drought-tolerance of bradyrhizobial strains (Arachis) isolated from Shandong. J. Peanut Sci. 2008, 21–25. [Google Scholar]
- Wu, P.; He, Q.y.; Li, Z.p.; Shi, J.; Zhu, C.w.; Sheng, W. Screening of Rhizobium from soybean in North Anhui for strong resistance to stresses. J. Anhui Sci. Technol. Univ. 2012, 26, 15–22. [Google Scholar]
- Cheng, B.; Wang, J.; Shi, H.b.; Zhang, H.m.; Zhang, R.q.; Zhen, C.; Zhai, B. Effects of glyphosate and Rhizobium on the quality traits and nitrogen fixation of Medicago sativa. Chin. J. Grassl. 2021, 43, 47–53. [Google Scholar] [CrossRef]
- Chen, Z.; Jin, Y.; Yao, X.; Wei, X.; Li, X.; Li, C.; White, J.F.; Nan, Z. Gene analysis reveals that leaf litter from Epichloe endophyte-infected perennial ryegrass alters diversity and abundance of soil microbes involved in nitrification and denitrification. Soil Biol. Biochem. 2021, 154, 108123. [Google Scholar] [CrossRef]
- Xu, J. Screening and Molecular Marker of High Efficient Symbiotic Rhizobia on Kidney Bean. Master’s Thesis, China Agricultural University, Beijing, China, 2004. [Google Scholar]
- Jiao, Y.S.; Yan, H.; Ji, Z.J.; Liu, Y.H.; Sui, X.H.; Wang, E.T.; Guo, B.L.; Chen, W.X.; Chen, W.F. Rhizobium sophorae sp nov and Rhizobium sophoriradicis sp nov., nitrogen-fixing rhizobial symbionts of the medicinal legume Sophora flavescens. Int. J. Syst. Evol. Microbiol. 2015, 65, 497–503. [Google Scholar] [CrossRef]
- Chen, Y.X.; Zou, L.; Penttinen, P.; Chen, Q.; Li, Q.Q.; Wang, C.Q.; Xu, K.W. Faba Bean (Vicia faba L.) nodulating rhizobia in Panxi, China, are diverse at species, plant growth promoting ability, and symbiosis related gene levels. Front. Microbiol. 2018, 9, 1338. [Google Scholar] [CrossRef]
- Zhang, J.; Shang, Y.; Peng, S.; Chen, W.; Wang, E.; de Lajudie, P.; Li, B.; Guo, C.; Liu, C. Rhizobium sophorae, Rhizobium laguerreae, and two novel Rhizobium genospecies associated with Vicia sativa L. in Northwest China. Plant Soil 2019, 442, 113–126. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Wang, N.; Yang, T.; Brunel, B.; Andrews, M.; Zong, X.; Wang, E. Rhizobium sophorae is the dominant rhizobial symbiont of Vicia faba L. In North China. Syst. Appl. Microbiol. 2022, 45, 126291. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, N.; Li, S.; Brunel, B.; Wang, J.; Feng, Y.; Yang, T.; Zong, X. Genotypic composition and performance of pea-nodulating rhizobia from soils outside the native plant-host range. Front. Microbiol. 2023, 14, 1201140. [Google Scholar] [CrossRef]
- El Idrissi, M.M.; Lamin, H.; Bouhnik, O.; Lamrabet, M.; Alami, S.; Jabrone, Y.; Bennis, M.; Bedmar, E.J.; Abdelmoumen, H. Characterization of Pisum sativum and Vicia faba microsymbionts in Morocco and definition of symbiovar viciae in Rhizobium acidisoli. Syst. Appl. Microbiol. 2020, 43, 126084. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Zheng, W.T.; Everall, I.; Young, J.P.W.; Zhang, X.X.; Tian, C.F.; Sui, X.H.; Wang, E.T.; Chen, W.X. Rhizobium anhuiense sp nov., isolated from effective nodules of Vicia faba and Pisum sativum. Int. J. Syst. Evol. Microbiol. 2015, 65, 2960–2967. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, E.T.; Liu, Y.; Li, X.; Yu, B.; Ren, C.; Liu, W.; Li, Y.; Xie, Z. Rhizobium anhuiense as the predominant microsymbionts of Lathyrus maritimus along the Shandong Peninsula seashore line. Syst. Appl. Microbiol. 2016, 39, 384–390. [Google Scholar] [CrossRef]
- Zou, L.; Chen, Y.X.; Penttinen, P.; Lan, Q.; Wang, K.; Liu, M.; Peng, D.; Zhang, X.; Chen, Q.; Zhao, K.; et al. Genetic diversity and symbiotic efficiency of nodulating rhizobia isolated from root nodules of faba bean in one field. PLoS ONE 2016, 11, e0167804. [Google Scholar] [CrossRef]
- Rouhrazi, K.; Khodakaramian, G.; Velazquez, E. Phylogenetic diversity of rhizobial species and symbiovars nodulating Phaseolus vulgaris in Iran. FEMS Microbiol. Lett. 2016, 363, fnw024. [Google Scholar] [CrossRef]
- Zinga, M.K.; Jaiswal, S.K.; Dakora, F.D. Presence of diverse rhizobial communities responsible for nodulation of common bean (Phaseolus vulgaris) in South African and Mozambican soils. FEMS Microbiol. Ecol. 2017, 93, fiw236. [Google Scholar] [CrossRef] [PubMed]
- Ormeno-Orrillo, E.; Aguilar-Cuba, Y.; Zuniga-Davila, D. Draft Genome Sequence of Rhizobium sophoriradicis H4, a nitrogen-fixing bacterium associated with the leguminous plant Phaseolus vulgaris on the coast of Peru. Genome Announc. 2018, 6, e00241-18. [Google Scholar] [CrossRef]
- Ribeiro, R.A.; Ormeno-Orrillo, E.; Dall’Agnol, R.F.; Graham, P.H.; Martinez-Romero, E.; Hungria, M. Novel Rhizobium lineages isolated from root nodules of the common bean (Phaseolus vulgaris L.) in Andean and Mesoamerican areas. Res. Microbiol. 2013, 164, 740–748. [Google Scholar] [CrossRef]
- Costa, M.R.; Chibeba, A.M.; Mercante, F.M.; Hungria, M. Polyphasic characterization of rhizobia microsymbionts of common bean Phaseolus vulgaris (L.) isolated in Mato Grosso do Sul, a hotspot of Brazilian biodiversity. Symbiosis 2018, 76, 163–176. [Google Scholar] [CrossRef]
- Gunununu, R.P.; Mohammed, M.; Jaiswal, S.K.; Dakora, F.D. Phylogeny and symbiotic effectiveness of indigenous rhizobial microsymbionts of common bean (Phaseolus vulgaris L.) in Malkerns, Eswatini. Sci. Rep. 2023, 13, 17029. [Google Scholar] [CrossRef]
- Wang, E.T.; Rogel, M.A.; Garcia-de los Santos, A.; Martinez-Romero, J.; Cevallos, M.A.; Martinez-Romero, E. Rhizobium etli bv. mimosae, a novel biovar isolated from Mimosa affinis. Int. J. Syst. Bacteriol. 1999, 49 Pt 4, 1479–1491. [Google Scholar] [CrossRef] [PubMed]
- Diouf, A.; de Lajudie, P.; Neyra, M.; Kersters, K.; Gillis, M.; Martinez-Romero, E.; Gueye, M. Polyphasic characterization of rhizobia that nodulate Phaseolus vulgaris in West Africa (Senegal and Gambia). Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 1, 159–170. [Google Scholar] [CrossRef]
- Aguilar, O.M.; Lopez, M.V.; Riccillo, P.M. The diversity of rhizobia nodulating beans in Northwest Argentina as a source of more efficient inoculant strains. J. Biotechnol. 2001, 91, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Tamimi, S.A.; Young, J.P.W. Rhizobium etli is the dominant common bean nodulating rhizobia in cultivated soils from different locations in Jordan. Appl. Soil Ecol. 2004, 26, 193–200. [Google Scholar] [CrossRef]
- Moura, F.T.; Helene, L.C.F.; Ribeiro, R.A.; Nogueira, M.A.; Hungria, M. The outstanding diversity of rhizobia microsymbionts of common bean (Phaseolus vulgaris L.) in Mato Grosso do Sul, central-western Brazil, revealing new Rhizobium species. Arch. Microbiol. 2023, 205, 325. [Google Scholar] [CrossRef] [PubMed]
- Mouhsine, B.; Prell, J.; Filali-Maltouf, A.; Priefer, U.B.; Aurag, J. Diversity, phylogeny and distribution of bean rhizobia in salt-affected soils of North-West Morocco. Symbiosis 2007, 43, 83–96. [Google Scholar]
- Beyene, D.; Kassa, S.; Ampy, F.; Asseffa, A.; Gebremedhin, T.; van Berkum, P. Ethiopian soils harbor natural populations of rhizobia that form symbioses with common bean (Phaseolus vulgaris L.). Arch. Microbiol. 2004, 181, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Shamseldin, A.; Werner, D. Presence of Rhizobium etli bv. phaseoli and Rhizobium gallicum bv. gallicum in Egyptian soils. World J. Microbiol. Biotechnol. 2007, 23, 285–289. [Google Scholar] [CrossRef]
- Youseif, S.H.; Abd El-Megeed, F.H.; Ageez, A.; Cocking, E.C.; Saleh, S.A. Phylogenetic multilocus sequence analysis of native rhizobia nodulating faba bean (Vicia faba L.) in Egypt. Syst. Appl. Microbiol. 2014, 37, 560–569. [Google Scholar] [CrossRef]
- Grange, L.; Hungria, M. Genetic diversity of indigenous common bean (Phaseolus vulgaris) rhizobia in two Brazilian ecosystems. Soil Biol. Biochem. 2004, 36, 1389–1398. [Google Scholar] [CrossRef]
- He, Y.; Guo, L.; Zhang, H.; Huang, G. Symbiotic effectiveness of pea-rhizobia associations and the implications for farming systems in the western Loess Plateau, China. Afr. J. Biotechnol. 2011, 10, 3540–3548. [Google Scholar]
- Zhang, J.; Shang, Y.; Wang, E.; Chen, W.; de Lajudie, P.; Li, B.; Guo, C.; Yang, X.; Zheng, J.; Liu, C. Mesorhizobium jarvisii sv. astragali as predominant microsymbiont for Astragalus sinicus L. in acidic soils, Xinyang, China. Plant Soil 2018, 433, 201–212. [Google Scholar] [CrossRef]
- Han, L.L.; Wang, E.T.; Han, T.X.; Liu, J.; Sui, X.H.; Chen, W.F.; Chen, W.X. Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil 2009, 324, 291–305. [Google Scholar] [CrossRef]
- Appunu, C.; Sasirekha, N.; Prabavathy, V.R.; Nair, S. A significant proportion of indigenous rhizobia from India associated with soybean (Glycine max L.) distinctly belong to Bradyrhizobium and Ensifer genera. Biol. Fertil. Soils 2009, 46, 57–63. [Google Scholar] [CrossRef]
- Shang, Y. The Diversity Analysis of Legume Green Manure Symbiotic Rhizobia and Mining of Dominant Species Resources in China. Master’s Thesis, Zhengzhou University of Light Industry, Zhengzhou, China, 2020. [Google Scholar]
- Terefework, Z.; Kaijalainen, S.; Lindström, K. AFLP fingerprinting as a tool to study the genetic diversity of Rhizobium galegae isolated from Galega orientalis and Galega officinalis. J. Biotechnol. 2001, 91, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, G.; Mavingui, P.; Allard, M.R.; Charnay, M.P.; Louvrier, P.; Mazurier, S.I.; Rigottier-Gois, L.; Amarger, N. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: Application to Rhizobium leguminosarum and its different biovars. Appl. Environ. Microbiol. 1996, 62, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, G.; Allard, M.R.; Revoy, F.; Amarger, N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA Genes. Appl. Environ. Microbiol. 1994, 60, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Vinuesa, P.; Silva, C.; Werner, D.; Martínez-Romero, E. Population genetics and phylogenetic inference in bacterial molecular systematics:: The roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenetics Evol. 2005, 34, 29–54. [Google Scholar] [CrossRef]
- Sijilmassi, B.; Filalimaltouf, A.; Boulahyaoui, H.; Kricha, A.; Boubekri, K.; Udupa, S.; Kumar, S.; Amri, A. Assessment of Genetic Diversity and Symbiotic Efficiency of Selected Rhizobia Strains Nodulating Lentil (Lens culinaris Medik.). Plants 2021, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, G.; Nour, S.M.; Macheret, V.; Sanjuan, J.; Drouin, P.; Amarger, N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 2001, 147, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Sarita, S.; Sharma, P.K.; Priefer, U.B.; Prell, J. Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol. Ecol. 2005, 54, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Higgins, D.G.; Jeanmougin, F.; Gibson, T.J.; Plewniak, F.; Thompson, J.D. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools; Oxford University Press: Oxford, UK, 1997. [Google Scholar]
- Jost, L. Entropy and diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Braak, T.C.J.F.; Smilauer, P. Canoco Reference Manual and User’s Guide: Software for Ordination, Version 5.0; Microcomputer Power: Ithaca, NY, USA, 2012; p. 2012. [Google Scholar]
- Zhang, F.; Cui, Z.; Chen, X.; Ju, X.; Shen, J.; Chen, Q.; Liu, X.; Zhang, W.; Mi, G.; Fan, M.; et al. Chapter one—Integrated Nutrient Management for Food Security and Environmental Quality in China. In Advances in Agronomy; Sparks, D.L., Ed.; Elsevier: Amsterdam, The Netherlands, 2012; Volume 116, pp. 1–40. [Google Scholar]

| IGS RFLP Type | Isolate Number | Representative Isolate | MLSA Similarity (%) with b: | Species Identification (Clade in Figure 1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WYCCWR No./Origin Site a | Rso | Rac | Rec | Rhi | Rva | Rsr | Rcr | Ran | Rph | Rch | Ret | |||
| 20 | 9 | WYCCWR13672/CDYB-22 | 99.1 | 95.8 | 96.3 | 94.9 | 95.1 | 95.4 | 95.1 | 95.1 | 94.9 | 94.2 | 93.4 | Rhizobium sophorae (C1) |
| 18 | 9 | WYCCWR13777/CNHH-10 | 95.2 | 98.6 | 96.3 | 95.5 | 95.3 | 95 | 94.9 | 95 | 95 | 95.7 | 94.3 | Rhizobium acidisoli (C2) |
| 13 | 11 | WYCCWR14027/DEYH-16 | 95.9 | 96.8 | 99 | 95.7 | 95.1 | 94.6 | 95 | 94.9 | 94.7 | 95.1 | 93.7 | Rhizobium ecuadorense (C3) |
| 10 | 16 | WYCCWR13936/DMDH-21 | 94.3 | 94.6 | 93.9 | 95.3 | 94 | 93.6 | 93.9 | 93.6 | 94.4 | 95.6 | 94.8 | Rhizobium genosp. I (C4) |
| 30 | 5 | WYCCWR14133/BSDB-6 | 94.2 | 94.7 | 94 | 95.2 | 94 | 93.5 | 93.9 | 94 | 94.5 | 95.6 | 94.6 | |
| 43 | 1 | WYCCWR14114/BSDH-6 | 94.7 | 95.3 | 95.2 | 97.7 | 95 | 95.3 | 95.7 | 94.1 | 96.5 | 95.2 | 93.4 | Rhizobium hidalgonense (C5) |
| 25 | 6 | WYCCWR13998/DWSB-10 | 94 | 95.2 | 95.1 | 98.5 | 94.5 | 94.1 | 93.9 | 95.7 | 94 | 94.7 | 93.3 | |
| 42 | 1 | WYCCWR13867/DXYH-4 | 94.5 | 95.6 | 95.7 | 99.8 | 95.1 | 94.4 | 94.3 | 94.3 | 94.2 | 94.9 | 93.8 | |
| 37 | 3 | WYCCWR13972/DWSH-6 | 94.6 | 95.7 | 95.8 | 99.8 | 95.2 | 94.6 | 94.5 | 94.5 | 94.4 | 95.1 | 93.6 | |
| 41 | 1 | WYCCWR13551/CMDH-27 | 94.6 | 95.7 | 95.8 | 100 | 95.2 | 94.6 | 94.5 | 94.5 | 94.4 | 95.1 | 93.6 | |
| 8 | 16 | WYCCWR13528/CMDH-4 | 94.6 | 95.7 | 95.8 | 100 | 95.2 | 94.6 | 94.5 | 94.5 | 94.4 | 95.1 | 93.6 | |
| 32 | 4 | WYCCWR13680/CLFH-5 | 94.6 | 95.7 | 95.8 | 100 | 95.2 | 94.6 | 94.5 | 94.5 | 94.4 | 95.1 | 93.6 | |
| 39 | 2 | WYCCWR13741/CDHH-14 | 95.1 | 95.1 | 94.6 | 95 | 98.9 | 95.1 | 96.3 | 94.4 | 95.1 | 95.5 | 93.4 | Rhizobium vallis (C6) |
| 12 | 12 | WYCCWR13728/CDHH-1 | 95.1 | 95.2 | 94.9 | 95 | 99.3 | 95.1 | 96.3 | 94.3 | 95.1 | 95.5 | 93.4 | |
| 28 | 5 | WYCCWR13814/CWDB-3 | 95.6 | 95.4 | 95 | 95.1 | 95.3 | 98.5 | 96.3 | 94.9 | 95.2 | 95.4 | 93.5 | Rhizobium sophoriradicis (C7) |
| 23 | 8 | WYCCWR13632/CDYH-7 | 95.1 | 95.4 | 94.6 | 93.9 | 96.2 | 96.8 | 98.6 | 94 | 95.8 | 94.8 | 93.9 | Rhizobium croatiense (C8) |
| 3 | 44 | WYCCWR14106/BCNB-10 | 95.1 | 95.4 | 94.6 | 93.9 | 96.2 | 96.8 | 98.6 | 94 | 95.8 | 94.8 | 93.9 | |
| WYCCWR13582/CYAH-6 | 95.1 | 95.3 | 94.6 | 93.9 | 96.2 | 96.6 | 98.4 | 94 | 95.8 | 94.8 | 93.9 | |||
| 15 | 10 | WYCCWR13581/CYAH-5 | 95.1 | 95.1 | 94.6 | 93.8 | 96.2 | 96.9 | 98.6 | 94 | 95.9 | 94.6 | 93.9 | |
| 6 | 19 | WYCCWR13601/CYAH-25 | 95.2 | 95.4 | 94.7 | 94 | 96.3 | 96.7 | 98.5 | 94 | 95.9 | 94.7 | 94 | |
| 21 | 9 | WYCCWR13663/CDYB-13 | 95.1 | 95.3 | 94.6 | 93.9 | 96.2 | 96.6 | 98.4 | 94 | 95.8 | 94.8 | 93.9 | |
| 1 | 153 | WYCCWR13630/CDYH-5 | 95.1 | 95.8 | 95.1 | 95.1 | 94.9 | 95.3 | 95.1 | 98.6 | 95.1 | 95 | 94.3 | Rhizobium anhuiense (C9) |
| WYCCWR13646/CDYH-21 | 95.1 | 95.7 | 95.1 | 94.7 | 94.6 | 95.1 | 94.7 | 98.9 | 94.7 | 94.6 | 94.1 | |||
| 29 | 5 | WYCCWR13999/DWSB-11 | 94.2 | 95.1 | 94.9 | 96.3 | 94.8 | 94.5 | 94 | 98.2 | 94.5 | 94.7 | 93.4 | Rhizobium anhuiense (C9) |
| 11 | 13 | WYCCWR13604/CYAB-3 | 94.9 | 95.3 | 94.9 | 94.4 | 94.5 | 94.2 | 94.1 | 99.7 | 94.5 | 94.4 | 93.7 | |
| 34 | 4 | WYCCWR13828/CSBH-8 | 95 | 95.3 | 95 | 94.5 | 94.5 | 94.6 | 94.3 | 100 | 94.6 | 94.6 | 93.9 | |
| 14 | 10 | WYCCWR13549/CMDH-25 | 94.6 | 95.6 | 95.3 | 97 | 95.2 | 94.6 | 94.6 | 95.1 | 96.3 | 95.8 | 94.6 | Rhizobium genosp. II (C10) |
| 2 | 63 | WYCCWR13564/CMDB-13 | 94.6 | 95.1 | 94.7 | 94.7 | 95 | 95.3 | 95.8 | 94.7 | 98.4 | 96 | 94.4 | Rhizobium phaseoli (C11) |
| 40 | 2 | WYCCWR13810/CNHB-19 | 94.5 | 95.1 | 94.6 | 94.6 | 95 | 95.1 | 95.8 | 94.7 | 98.2 | 96 | 94.2 | |
| 4 | 38 | WYCCWR13550/CMDH-26 | 94.5 | 94.7 | 94.2 | 93.9 | 95 | 95.5 | 96.3 | 94.6 | 98.9 | 95.3 | 94.2 | |
| WYCCWR13893/DXYB-3 | 94.5 | 94.7 | 94.2 | 93.9 | 95 | 95.5 | 96.3 | 94.6 | 98.9 | 95.3 | 94.2 | |||
| 38 | 3 | WYCCWR13883/DXYH-20 | 94.5 | 94.7 | 94.2 | 93.9 | 95 | 95.5 | 96.3 | 94.6 | 98.9 | 95.3 | 94.2 | |
| 22 | 9 | WYCCWR14123/BSDH-15 | 94.7 | 95.1 | 94.5 | 94.1 | 95.2 | 95.7 | 96.6 | 94.6 | 99.5 | 95.4 | 94.4 | |
| 36 | 3 | WYCCWR13915/DXYB-25 | 94.6 | 95.1 | 94.7 | 94.7 | 95 | 95.3 | 95.8 | 94.7 | 98.4 | 96 | 94.4 | |
| 7 | 17 | WYCCWR13909/DXYB-19 | 94.6 | 94.7 | 94.4 | 94 | 95 | 95.7 | 96.3 | 94.6 | 99.1 | 95.3 | 94.4 | |
| 16 | 10 | WYCCWR13839/CSBH-19 | 94.6 | 95.1 | 94.7 | 94.7 | 95 | 95.3 | 95.8 | 94.7 | 98.4 | 96 | 94.4 | |
| 33 | 4 | WYCCWR13823/CSBH-3 | 94.6 | 94.7 | 94.4 | 94 | 95 | 95.7 | 96.3 | 94.6 | 99.1 | 95.3 | 94.4 | |
| 26 | 5 | WYCCWR13529/CMDH-5 | 94.6 | 95.1 | 94.7 | 94.7 | 95 | 95.3 | 95.8 | 94.7 | 98.4 | 96 | 94.4 | |
| 9 | 16 | WYCCWR13697/CLFH-22 | 94.6 | 95.1 | 94.7 | 94.7 | 95 | 95.3 | 95.8 | 94.7 | 98.4 | 96 | 94.4 | |
| 24 | 6 | WYCCWR13654/CDYB-4 | 93.9 | 94 | 94 | 94.3 | 95.1 | 94.2 | 94.5 | 94.1 | 95 | 96.3 | 95.1 | Rhizobium genosp. III (C12) |
| 27 | 5 | WYCCWR14006/DWSB-18 | 94 | 95.4 | 94.6 | 95.1 | 95.8 | 94.3 | 94.3 | 94 | 94.7 | 98.6 | 94.7 | Rhizobium chutanense (C13) |
| 35 | 3 | WYCCWR13808/CNHB-17 | 94 | 95.5 | 94.6 | 95.1 | 95.3 | 94.2 | 94.2 | 93.9 | 95 | 98.6 | 94.5 | |
| 5 | 26 | WYCCWR13754/CDHB-7 | 94.5 | 95.9 | 95.1 | 95.1 | 95.2 | 94.6 | 94.6 | 94.6 | 95.1 | 98.6 | 94.8 | |
| 31 | 4 | WYCCWR13563/CMDB-12 | 93 | 93.9 | 93 | 92.8 | 93.4 | 93.1 | 93.8 | 92.8 | 94.3 | 94.7 | 97.1 | Rhizobium etli (C14) |
| 17 | 9 | WYCCWR14090/BLYB-15 | 93.3 | 93.3 | 92.9 | 92.4 | 93.4 | 94.5 | 93.9 | 92.3 | 94.5 | 94.6 | 94.2 | Rhizobium genosp. IV (C15) |
| 19 | 9 | WYCCWR14071/BLYH-17 | 91.8 | 91.5 | 91.7 | 91.1 | 91.7 | 92.3 | 91.9 | 91.1 | 92.9 | 92.9 | 92.6 | Rhizobium genosp. V (C16) |
| IGS type number: 43 | Total isolate number: 608 | Total representatives: 46 | Total species: 16 | |||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, J.; Feng, Y.; Brunel, B.; Zong, X. Unearthing Optimal Symbiotic Rhizobia Partners from the Main Production Area of Phaseolus vulgaris in Yunnan. Int. J. Mol. Sci. 2024, 25, 8511. https://doi.org/10.3390/ijms25158511
Zhang J, Wang J, Feng Y, Brunel B, Zong X. Unearthing Optimal Symbiotic Rhizobia Partners from the Main Production Area of Phaseolus vulgaris in Yunnan. International Journal of Molecular Sciences. 2024; 25(15):8511. https://doi.org/10.3390/ijms25158511
Chicago/Turabian StyleZhang, Junjie, Jingqi Wang, Yufeng Feng, Brigitte Brunel, and Xuxiao Zong. 2024. "Unearthing Optimal Symbiotic Rhizobia Partners from the Main Production Area of Phaseolus vulgaris in Yunnan" International Journal of Molecular Sciences 25, no. 15: 8511. https://doi.org/10.3390/ijms25158511






