Chitinase Gene FoChi20 in Fusarium oxysporum Reduces Its Pathogenicity and Improves Disease Resistance in Cotton
Abstract
1. Introduction
2. Results
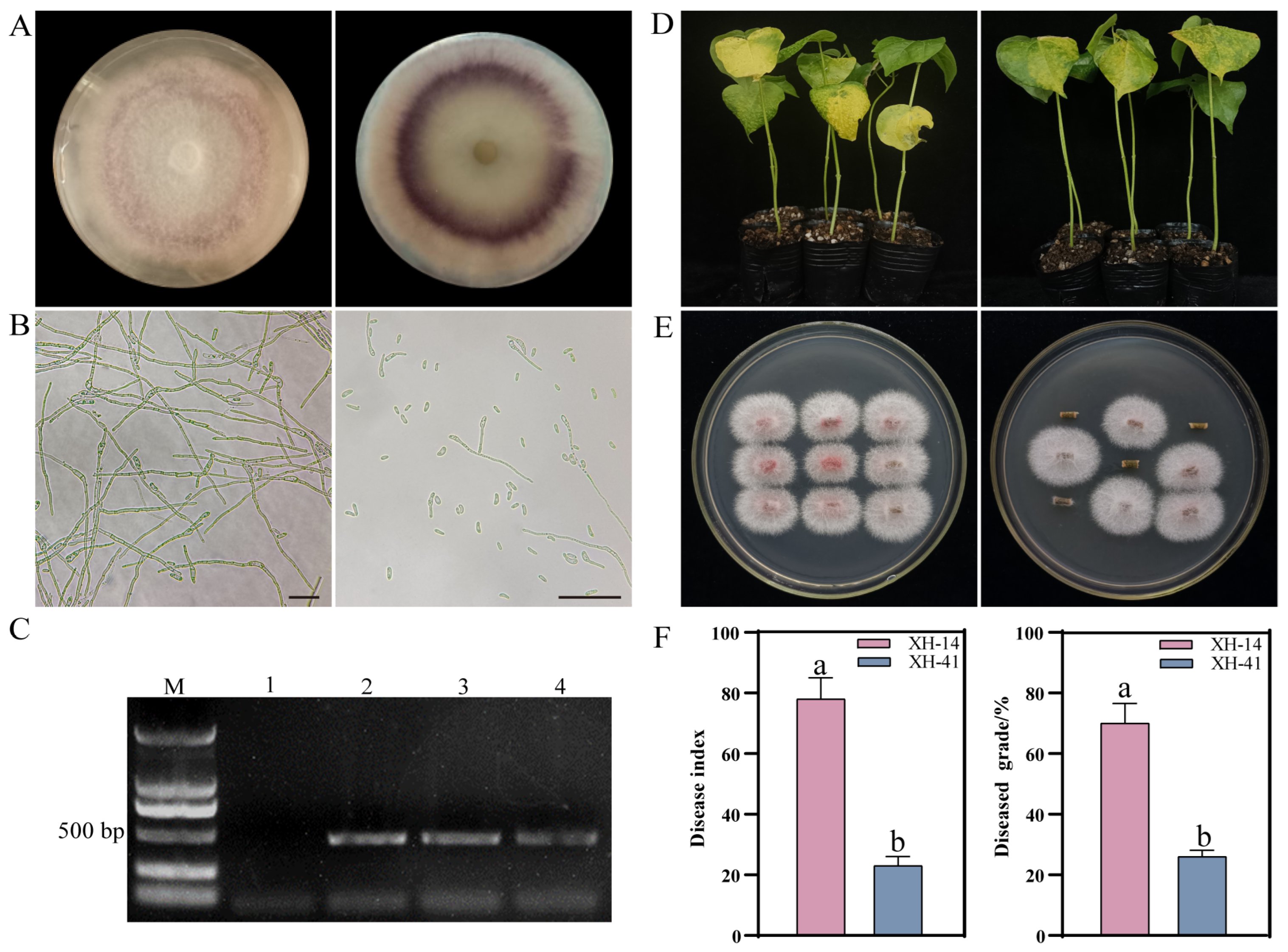
2.1. Identification of Disease Resistance in Cotton and Restoration Culture
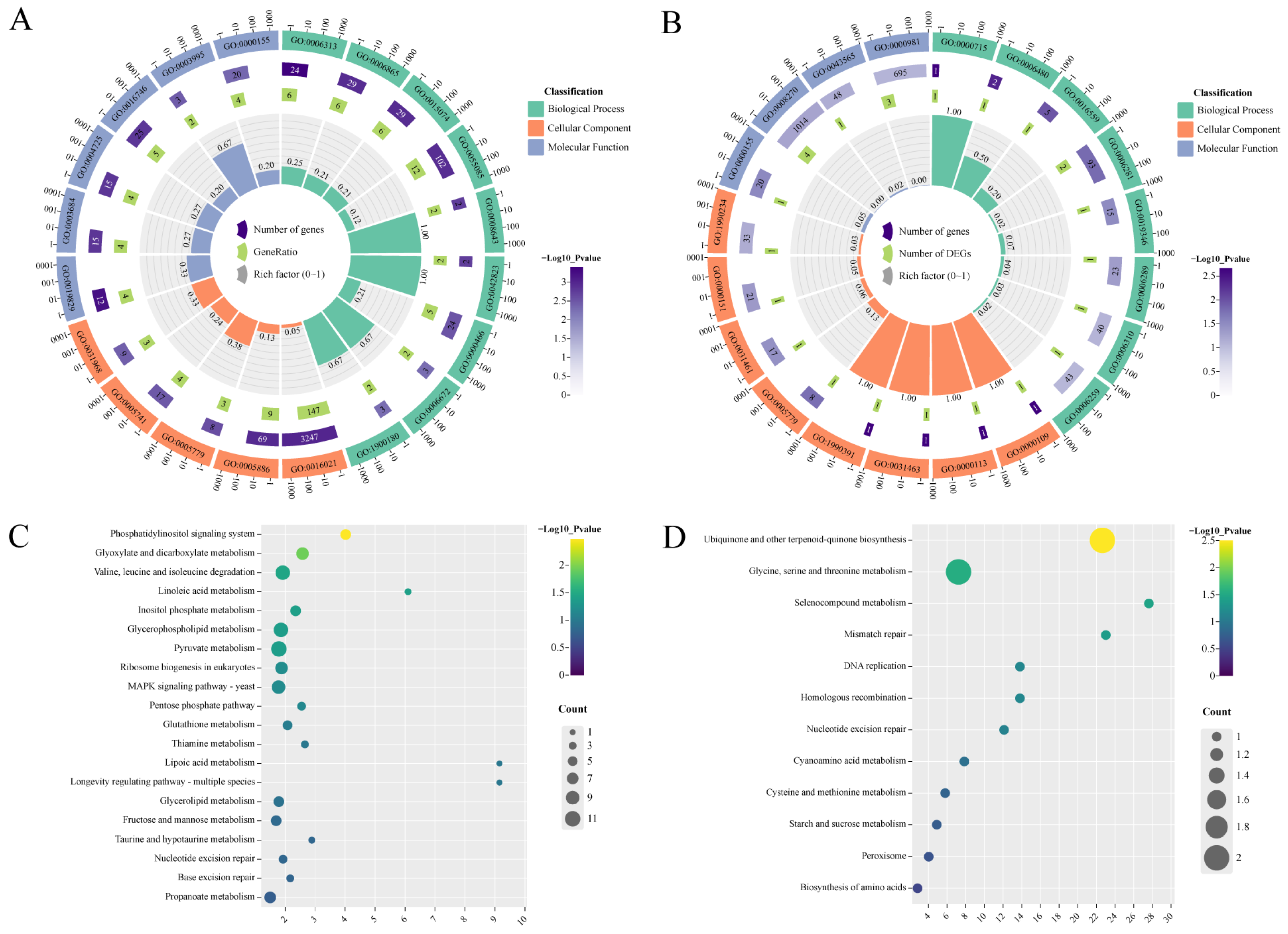
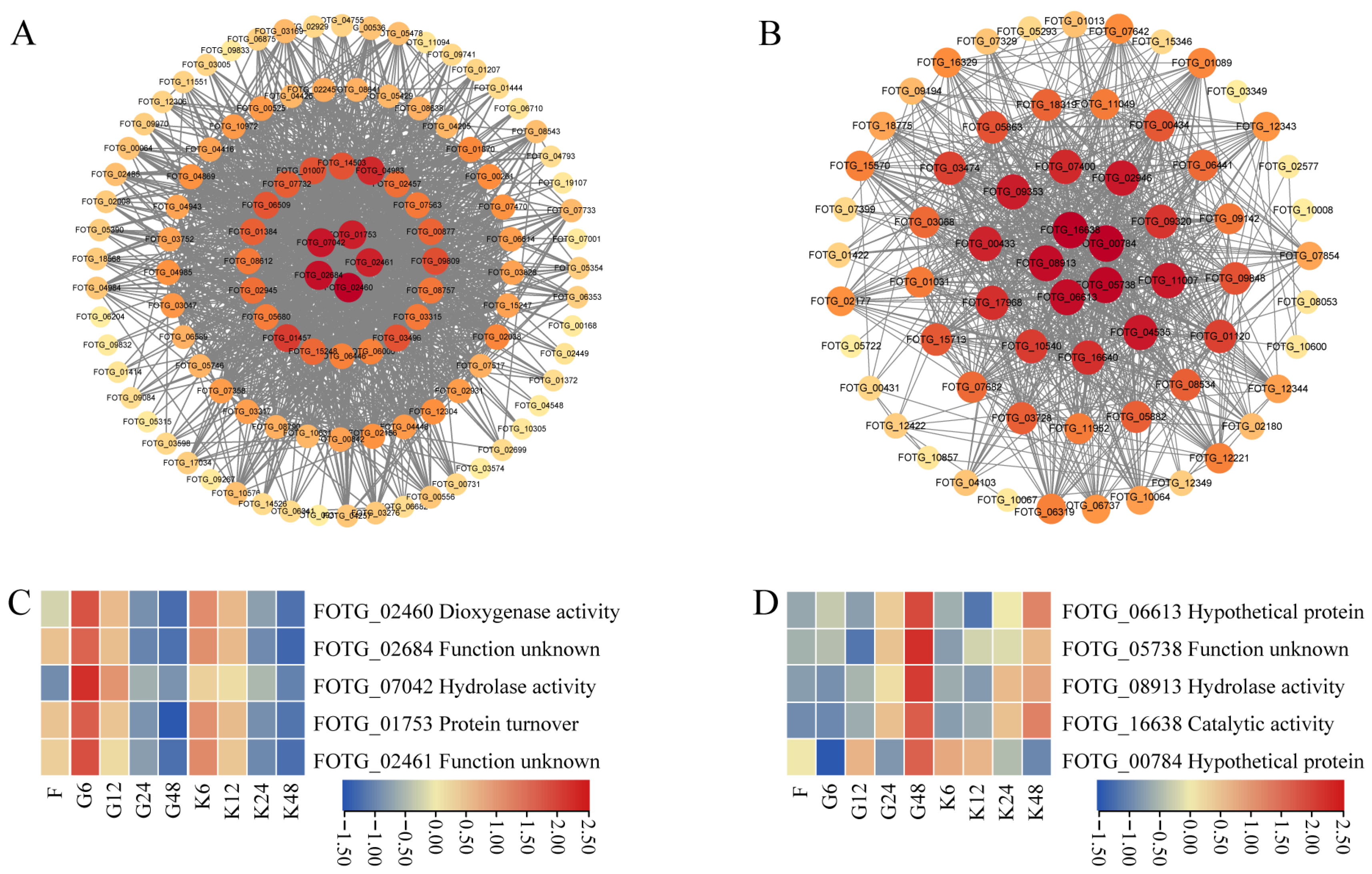
2.2. Hub-Genes with Pathogenicity of F. ox
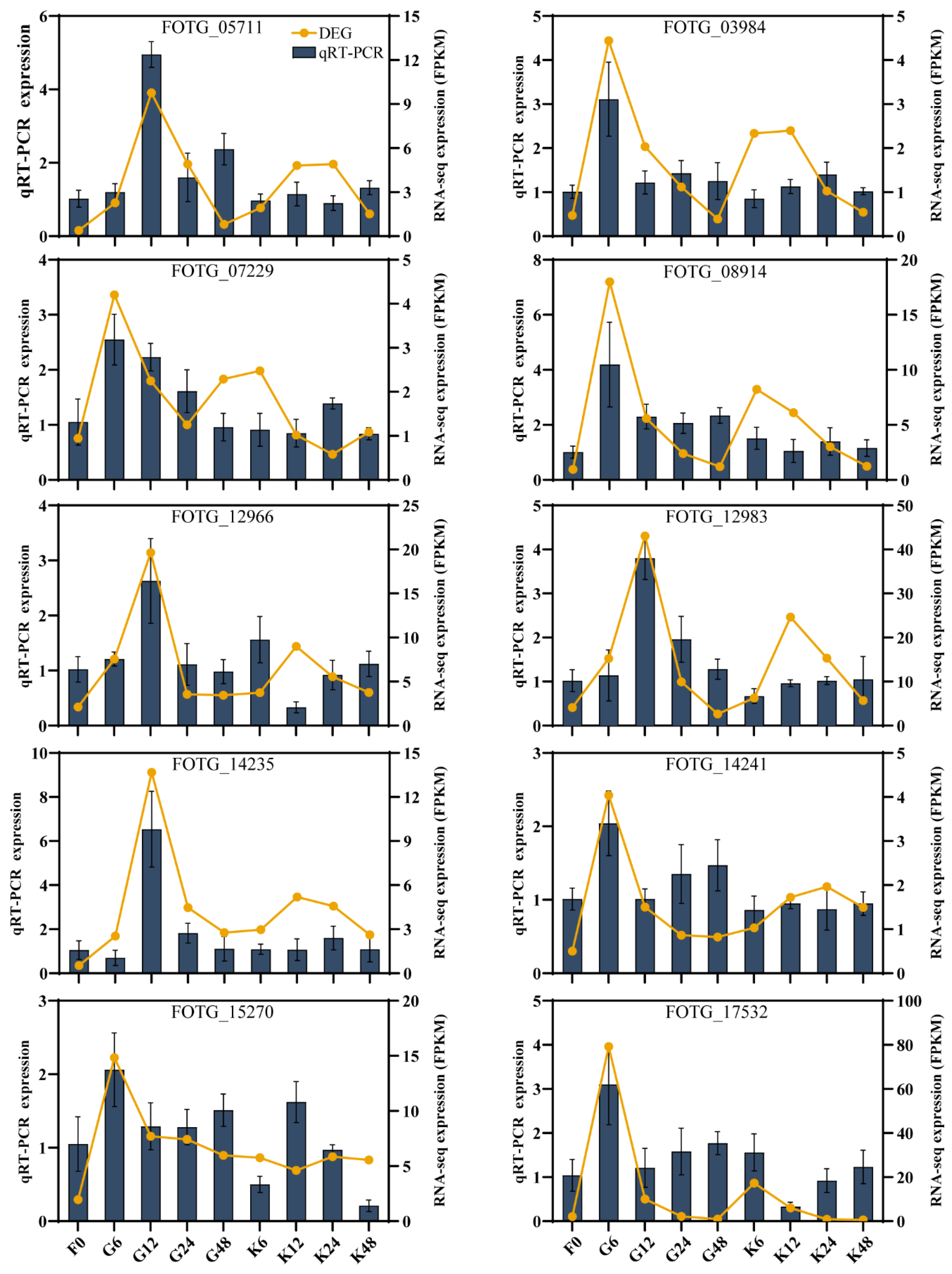
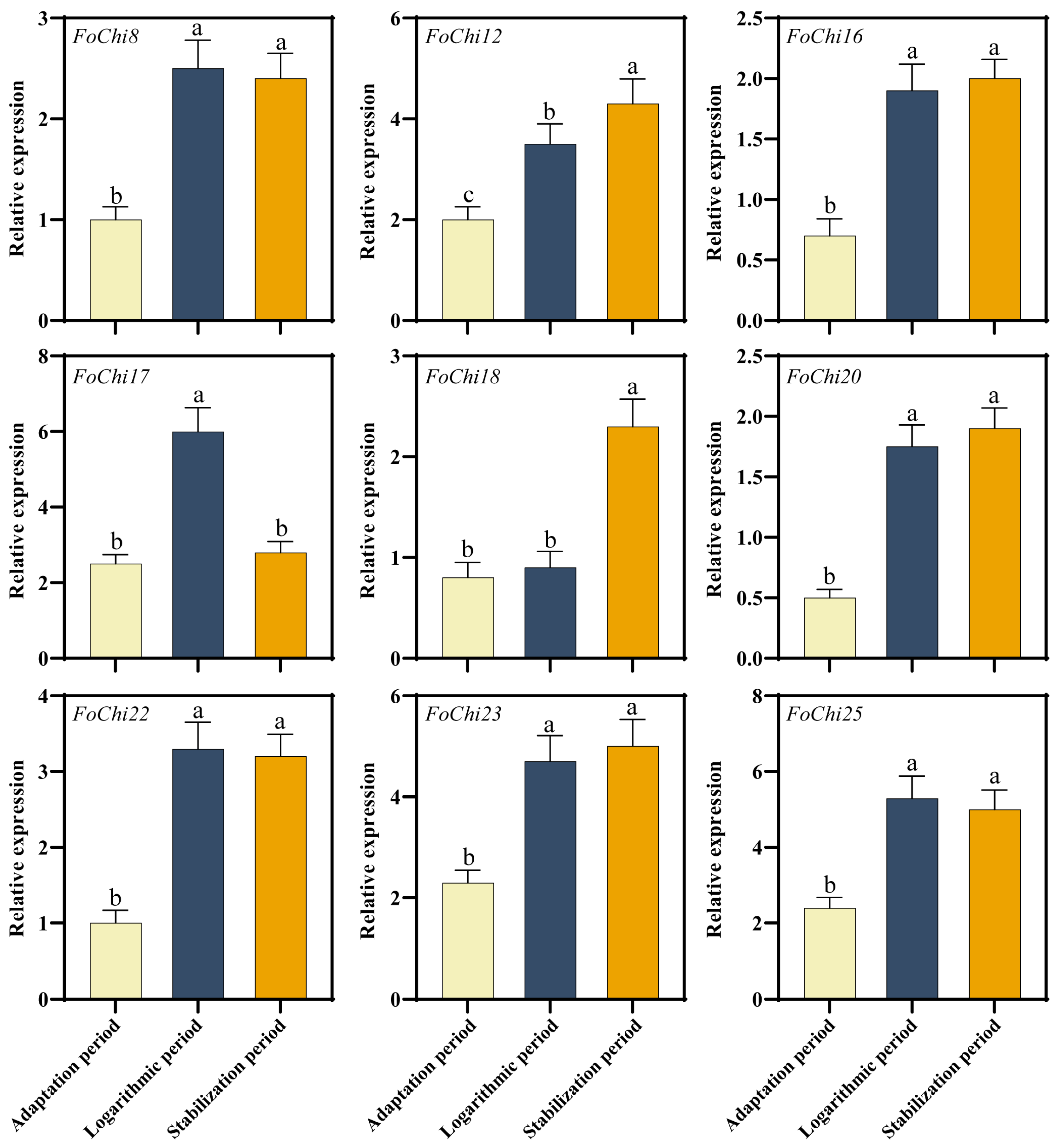
2.3. FoChi Genes in F. ox Were Involved in Resistant and Susceptible Cotton Root Secretions
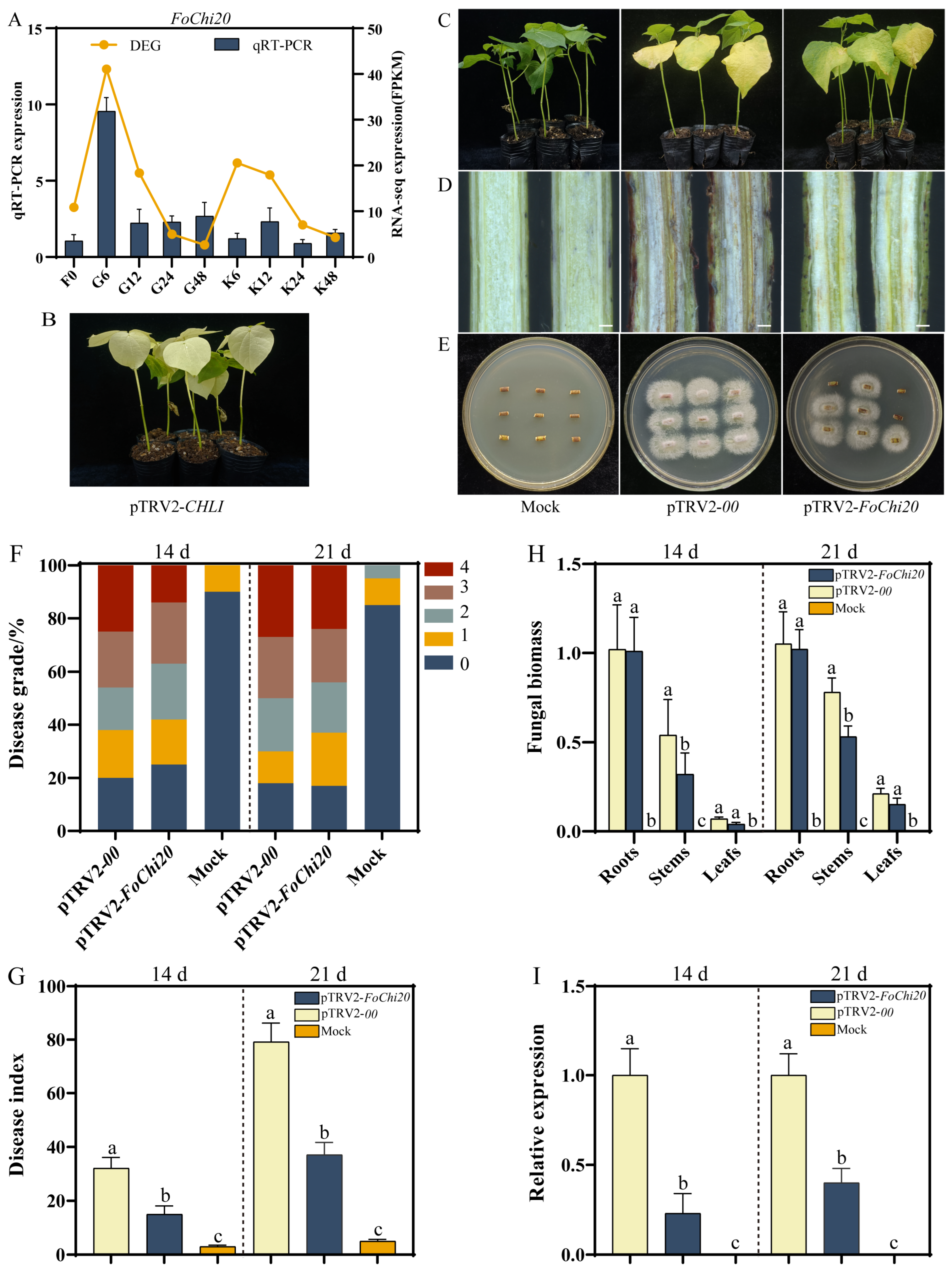
2.4. Silenced FoChi20 Gene Attenuated the Pathogenicity of F. ox and Enhanced Disease Resistance in Cotton
3. Discussion
3.1. Root Secretions Could Be Widely Used to Inhibit F. ox Growth
3.2. Pathogenicity-Related Genes in F. ox Were Predicated
3.3. Chitinase Genes Influenced the Pathogenicity of Pathogenic Bacteria
3.4. The Value of HIGS Technology in Plant Disease Resistance Research
4. Materials and Methods
4.1. Sample Collection of Cotton and Fusarium oxysporum f. sp. Vasinfectum
4.2. F. ox Morphological and Molecular Biological Characterization
4.3. RT-qPCR Experiment
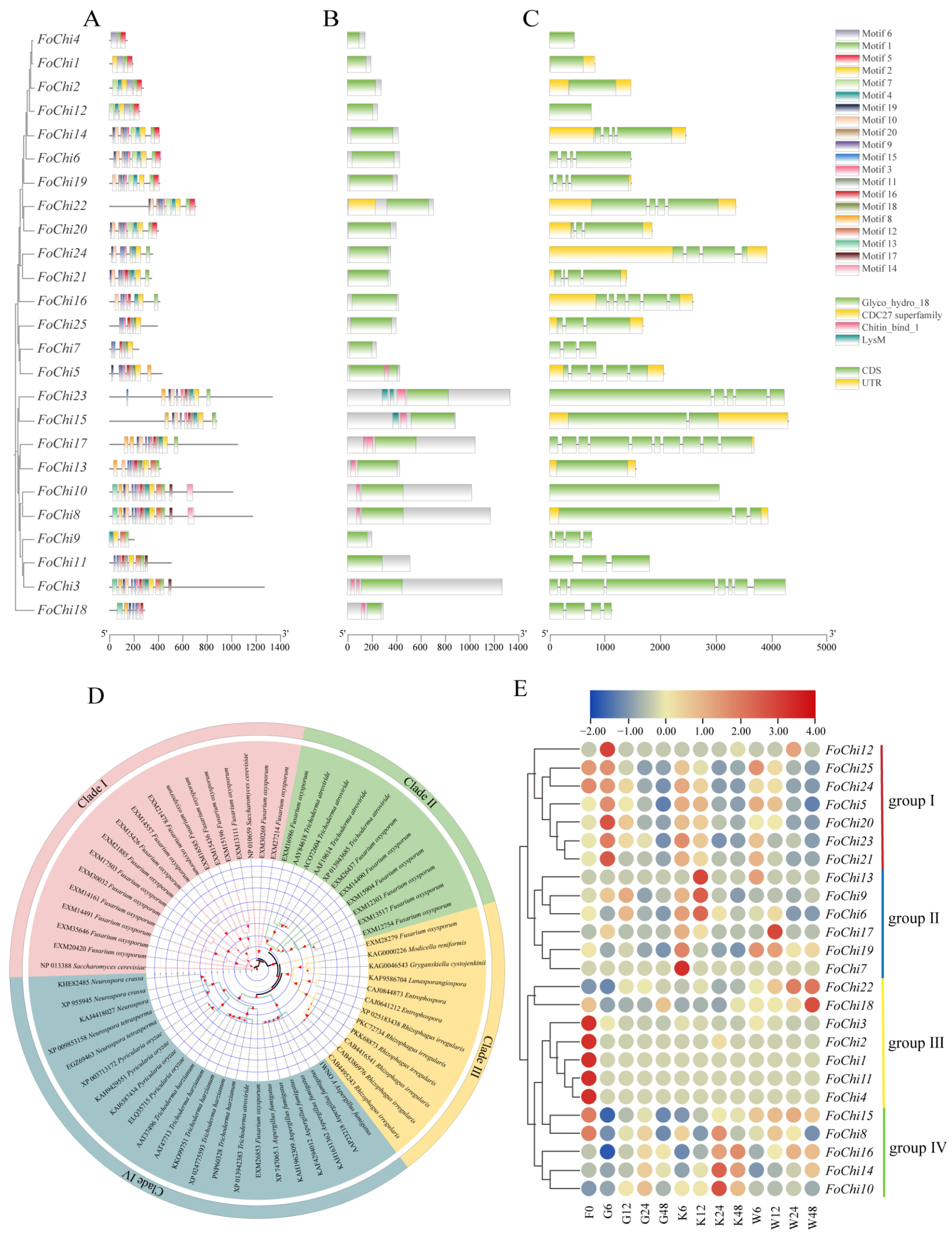
4.4. Identification of Chitinase Genes in F. ox
4.5. Evolutionary Tree and Location of the Chitinase Genes
4.6. Host-Induced FoChi20 Gene Silencing Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chávez Montes, R.A.; Ulloa, M.; Biniashvili, T.; Zackay, A.; Kfir, N.; Lopez-Arredondo, D.; Herrera-Estrella, L. Assembly and annotation of the Gossypium barbadense L.‘Pima-S6’ genome raise questions about the chromosome structure and gene content of Gossypium barbadense genomes. BMC Genom. 2023, 24, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kong, L.; Xiao, X.; Li, P.; Liu, A.; Li, J.; Gong, J.; Gong, W.; Ge, Q.; Shang, H. Genome-wide artificial introgressions of Gossypium barbadense into G. hirsutum reveal superior loci for simultaneous improvement of cotton fiber quality and yield traits. J. Adv. Res. 2023, 53, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, S.T.; Chen, H.; Hou, Z.; Li, Y.; Yang, C.L.; Wang, D.J.; Song, C.P. Screening of abiotic stress-responsive cotton genes using a cotton full-length cDNA overexpressing Arabidopsis library. J. Integr. Plant Biol. 2020, 62, 998–1016. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Liu, R.; Gong, J.; Li, P.; Li, Z.; Gong, W.; Liu, A.; Ge, Q.; Deng, X.; Li, S. Fine mapping and candidate gene analysis of qFL-A12-5: A fiber length-related QTL introgressed from Gossypium barbadense into Gossypium hirsutum. Theor. Appl. Genet. 2023, 136, 48. [Google Scholar] [CrossRef]
- Xiong, X.; Sun, C.; Chen, B.; Sun, J.; Fei, C.; Xue, F. Transcriptomic datasets of Verticillium wilt resistant and non-resistant Gossypium barbadense varieties during pathogen inoculation. Sci. Data 2024, 11, 11. [Google Scholar] [CrossRef]
- Man, M.W.; Zhu, Y.Q.; Liu, L.L.; Luo, L.; Han, X.P.; Qiu, L.; Li, F.G.; Ren, M.Z.; Xing, Y.D. Defense Mechanisms of Cotton Fusarium and Verticillium Wilt and Comparison of Pathogenic Response in Cotton and Humans. Int. J. Mol. Sci. 2022, 23, 12217. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.L.; Zhao, N.; Wang, W.R.; Wang, M.; Zhu, J.H.; Yang, J.; Lin, F.; Huang, X.L.; Zhang, Y.H.; Min, L.; et al. Glyphosate-Induced Abscisic Acid Accumulation Causes Male Sterility in Sea Island Cotton. Plants 2023, 12, 1058. [Google Scholar] [CrossRef] [PubMed]
- Su, X.J.; Zhu, G.Z.; Song, X.H.; Xu, H.J.; Li, W.X.; Ning, X.Z.; Chen, Q.J.; Guo, W.Z. Genome-wide association analysis reveals loci and candidate genes involved in fiber quality traits in sea island cotton (Gossypium barbadense). BMC Plant Biol. 2020, 20, 289. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, Y.H.; Wan, H.A.; Tang, J.; Ni, Z.Y. The sea-island cotton GbTCP4 transcription factor positively regulates drought and salt stress responses. Plant Sci. 2022, 322, 111329. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.H.; Wan, H.N.; Ni, Z.Y. Sea-island cotton (Gossypium barbadense L.) GbTCP5 improves plant adaptation to drought and salt stress by directly activating GbERD7, GbUBC19, and GbGOLS2 expression. Ind. Crops Prod. 2023, 203, 117209. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Zhu, Y.; Zeng, L.; Stetina, S.; Zhang, J. A genome-wide association study for resistance to Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) race 4 in diploid cotton (Gossypium arboreum) and resistance transfer to tetraploid Gossypium hirsutum. Mol. Genet. Genom. 2024, 299, 30. [Google Scholar] [CrossRef]
- Bustamante, M.I.; Todd, C.; Elfar, K.; Hamid, M.I.; Garcia, J.F.; Cantu, D.; Rolshausen, P.E.; Eskalen, A. Identification and Pathogenicity of Fusarium Species Associated with Young Vine Decline in California. Plant Dis. 2024, 108, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Zu, Q.; Deng, X.; Qu, Y.; Chen, X.; Cai, Y.; Wang, C.; Li, Y.; Chen, Q.; Zheng, K.; Liu, X. Genetic Channelization Mechanism of Four Chalcone Isomerase Homologous Genes for Synergistic Resistance to Fusarium wilt in Gossypium barbadense L. Int. J. Mol. Sci. 2023, 24, 14775. [Google Scholar] [CrossRef] [PubMed]
- Babilonia, K.; Wang, P.; Liu, Z.Y.; Jamieson, P.; Mormile, B.; Rodrigues, O.; Zhang, L.; Lin, W.W.; Clement, C.D.; de Moura, S.M.; et al. A nonproteinaceous Fusarium cell wall extract triggers receptor-like protein-dependent immune responses in Arabidopsis and cotton. New Phytol. 2021, 230, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Guo, Y.; Xu, J.; Zhang, H.; Ma, Z.; Wu, H. Isolation of anthraquinone derivatives from Rubia cordifolia (Rubiaceae) and their bioactivities against plant pathogenic microorganisms. Pest Manag. Sci. 2024. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Elkins-Arce, H.; Wheeler, T.A.; Dever, J.; Whitelock, D.; Hake, K.; Wedegaertner, T.; Zhang, J.F. Effect of growth stage, cultivar, and root wounding on disease development in cotton caused by Fusarium wilt race 4 (Fusarium oxysporum f. sp. vasinfectum). Crop Sci. 2023, 63, 101–114. [Google Scholar] [CrossRef]
- Zhu, Y.; Lujan, P.A.; Wedegaertner, T.; Nichols, R.; Abdelraheem, A.; Zhang, J.F.; Sanogo, S. First Report of Fusarium oxysporum f. sp. vasinfectum Race 4 Causing Fusarium Wilt of Cotton in New Mexico, USA. Plant Dis. 2020, 104, 588. [Google Scholar] [CrossRef]
- Sakane, K.; Akiyama, M.; Ando, A.; Shigyo, M.; Ito, S.-i.; Sasaki, K.J.P.; Pathology, M.P. Identification of a novel effector gene and its functional tradeoff in Fusarium oxysporum f. sp. cepae that infects Welsh onion. Physiol. Mol. Plant Pathol. 2023, 123, 101939. [Google Scholar] [CrossRef]
- Srivastava, V.; Patra, K.; Pai, H.; Aguilar-Pontes, M.V.; Berasategui, A.; Kamble, A.; Di Pietro, A.; Redkar, A. Molecular Dialogue During Host Manipulation by the Vascular Wilt Fungus Fusarium oxysporum. Annu. Rev. Phytopathol. 2024, 62. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, X.; Ming, Y.; Liu, C.; Zhang, X.; Liu, S.; Zhu, L. Characterization of the High-Quality Genome Sequence and Virulence Factors of Fusarium oxysporum f. sp. vasinfectum Race 7. J. Fungi 2024, 10, 242. [Google Scholar] [CrossRef]
- Zhu, Y.; Abdelraheem, A.; Cooke, P.; Wheeler, T.; Dever, J.K.; Hake, K.; Bissonnette, K.; Zhang, J. Comparative analysis of infection process in Upland cottons differing in resistance to Fusarium wilt caused by Fusarium oxysporum f. sp. vasinfectum race 4. Crop Sci. 2023, 63, 1330–1343. [Google Scholar] [CrossRef]
- Maqsood, A.; Aslam, M.N.; Khaliq, H.; Shakeel, M.T.; Wu, H.; Fahad, S. Endophytic Bacillus spp. Mediated Plant Growth Promotion of Tomato Seedlings and Suppression of Meloidogyne incognita and Fusarium oxysporum Disease Complex. J. Plant Growth Regul. 2024, 43, 2454–2469. [Google Scholar] [CrossRef]
- Mokhtari, S.; Chavez, M.; Ali, A. First Report of Fusarium oxysporum f. sp. vasinfectum Causing Fusarium Wilt of Cotton in Kansas, USA. Plant Dis. 2023, 107, 1239. [Google Scholar] [CrossRef] [PubMed]
- Cen, Z.X.; Hu, B.J.; Yang, S.Y.; Ma, G.L.; Zheng, Y.R.; Dong, Y. Mechanism of benzoxazinoids affecting the growth and development of Fusarium oxysporum f. sp. fabae. Plant Mol. Biol. 2024, 114, 42. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Ding, Z.X.; Thomashow, L.S.; Hale, L.; Yang, S.D.; Xie, P.H.; Liu, X.Y.; Wang, H.Q.; Shen, Q.R.; Yuan, J. Deciphering the mechanism of fungal pathogen-induced disease-suppressive soil. New Phytol. 2023, 238, 2634–2650. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhuo, Y.H.; Lin, Y.Q.; Huang, M.M.; Tang, W.; Zheng, W.H.; Lu, G.D.; Wang, Z.H.; Yun, Y.Z. The Signal Peptidase FoSpc2 Is Required for Normal Growth, Conidiation, Virulence, Stress Response, and Regulation of Light Sensitivity in Fusarium odoratissimum. Microbiol. Spectr. 2023, 11, e04403-22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Lv, F.J.; Qiu, F.H.; Han, D.H.; Xu, Y.; Liang, W.X. Pathogenic fungi neutralize plant-derived ROS via Srpk1 deacetylation. EMBO J. 2023, 42, e112634. [Google Scholar] [CrossRef]
- Bani, M.; Cimmino, A.; Evidente, A.; Rubiales, D.; Rispail, N. Pisatin involvement in the variation of inhibition of Fusarium oxysporum f. sp pisi spore germination by root exudates of Pisum spp. germplasm. Plant Pathol. 2018, 67, 1046–1054. [Google Scholar] [CrossRef]
- Xu, W.H.; Wang, Z.G.; Wu, F.Z. Companion cropping with wheat increases resistance to Fusarium wilt in watermelon and the roles of root exudates in watermelon root growth. Physiol. Mol. Plant Pathol. 2015, 90, 12–20. [Google Scholar] [CrossRef]
- Wu, F.Z.; Liu, B.; Zhou, X.G. Effects of root exudates of watermelon cultivars differing in resistance to Fusarium wilt on the growth and development of Fusarium oxysporum f. sp. niveum. Allelopath. J. 2010, 25, 403–413. [Google Scholar]
- Yang, S.Y.; Zheng, Y.R.; Guo, Y.T.; Yang, Z.X.; Dong, Y. Faba bean-wheat intercropping and appropriate nitrogen supply control fusarium wilt in faba bean via altering specific amino acids in the root exudate of faba bean. Physiol. Mol. Plant Pathol. 2023, 124, 101961. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, F.Z.; Liu, S.W. Allelopathic effects of root exudates of Chinese onion accessions on cucumber yield and Fusarium oxysporum f. sp. cucumerinum. Allelopath. J. 2011, 27, 75–85. [Google Scholar]
- Aro, N.; Pakula, T.; Penttilä, M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 719–739. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, L.; Pinar, M.; Le Borgne, R.; Durieu, C.; Penalva, M.A.; Boudaoud, A.; Minc, N. Cell wall dynamics stabilize tip growth in a filamentous fungus. PLoS Biol. 2023, 21, e3001981. [Google Scholar] [CrossRef] [PubMed]
- Pate, P.K.; Free, S.J. The Genetics and Biochemistry of Cell Wall Structure and Synthesis in Neurospora crassa, a Model Filamentous Fungus. Front. Microbiol. 2019, 10, 2294. [Google Scholar]
- Yoshimi, A.; Miyazawa, K.; Kawauchi, M.; Abe, K. Cell Wall Integrity and Its Industrial Applications in Filamentous Fungi. J. Fungi 2022, 8, 435. [Google Scholar] [CrossRef]
- Z hao, S.; Zhang, T.; Hasunuma, T.; Kondo, A.; Zhao, X.Q.; Feng, J.X. Every road leads to Rome: Diverse biosynthetic regulation of plant cell wall-degrading enzymes in filamentous fungi Penicillium oxalicum and Trichoderma reesei. Crit. Rev. Biotechnol. 2023, 1–21. [Google Scholar] [CrossRef]
- Zhou, J.S.; Kang, L.Q.; Liu, C.C.; Niu, X.; Wang, X.J.; Liu, H.L.; Zhang, W.M.; Liu, Z.H.; Latge, J.P.; Yuan, S. Chitinases Play a Key Role in Stipe Cell Wall Extension in the Mushroom Coprinopsis cinerea. Appl. Environ. Microbiol. 2019, 85, e00532-19. [Google Scholar] [CrossRef] [PubMed]
- El-Katatny, M.H.; Somitsch, W.; Robra, K.H.; El-Katatny, M.S.; Gubitz, G.M. Production of chitinase and beta-1,3-glucanase by Trichoderma harzianum for control of the phytopathogenic fungus Sclerotium rolfsii. Food Technol. Biotechnol. 2000, 38, 173–180. [Google Scholar]
- Muñoz, W.R.; Ramírez, M.I.; Molina, F.R.; Crespo, S.G.; Rodríguez-Medina, J.R. Myosin II is important for maintaining regulated secretion and asymmetric localization of chitinase 1 in the budding yeast Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2003, 409, 411–413. [Google Scholar] [CrossRef]
- Pocsi, I.; Leiter, E.; Kwon, N.J.; Shin, K.S.; Kwon, G.S.; Pusztahelyi, T.; Emri, T.; Abuknesha, R.A.; Price, R.G.; Yu, J.H. Asexual sporulation signalling regulates autolysis of Aspergillus nidulans via modulating the chitinase ChiB production. J. Appl. Microbiol. 2009, 107, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Mou, Y.; Li, Y.X.; Yang, Q.; Wang, X.; Lin, H.C.; Kang, Z.S.; Guo, J. Silencing a Chitinase Gene, PstChia1, Reduces Virulence of Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2023, 24, 8215. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.J.; Song, L.L.; Peng, C.L.; Liu, X.; Liu, L.H.; Zhang, Y.H.; Wang, W.Z.; Zhou, J.; Wang, S.H.; Ebbole, D.; et al. A Magnaporthe Chitinase Interacts with a Rice Jacalin-Related Lectin to Promote Host Colonization. Plant Physiol. 2019, 179, 1416–1430. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Li, C.; Chen, X.F.; Cheng, J.S.; Xie, J.T.; Lin, Y.; Fu, Y.P.; Jiang, D.H.; Chen, T. Overexpression of chitinase PbChia1 from Plasmodiophora brassicae improves broad-spectrum disease resistance of Arabidopsis. Virulence 2023, 14, 2233147. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.Y.; Liu, M.X.; Wang, Y.; Zou, Y.B.; You, Y.M.; Yang, L.N.; Hu, J.X.; Zhang, H.F.; Zheng, X.B.; et al. Magnaporthe oryzae Auxiliary Activity Protein MoAa91 Functions as Chitin-Binding Protein to Induce Appressorium Formation on Artificial Inductive Surfaces and Suppress Plant Immunity. Mbio 2020, 11, e03304-19. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Zhu, J.C.; Yang, Y.; Zhang, W. Effects of root secretions of resistant and susceptible varieties of cotton on the growth and gene expression of Fusarium spinosum. Biotechnol. Bull. 2023, 39, 156–167. [Google Scholar]
- Sun, J.; Yang, J.; Zhao, S.Y.; Yu, Q.; Weng, L.L.; Xiao, C.P. Root exudates influence rhizosphere fungi and thereby synergistically regulate Panax ginseng yield and quality. Front. Microbiol. 2023, 14, 1194224. [Google Scholar] [CrossRef] [PubMed]
- Li, X.G.; Wei, Q.; Liu, B.; Alam, M.S.; Wang, X.X.; Shen, W.J.; Han, Z.M. Root exudates of transgenic cotton and their effects on Fusarium oxysporum. Front. Biosci. Landmark 2013, 18, 725–733. [Google Scholar] [CrossRef][Green Version]
- Were, E.; Schone, J.; Viljoen, A.; Rasche, F. Phenolics mediate suppression of Fusarium oxysporum f. sp. cubense TR4 by legume root exudates. Rhizosphere 2022, 21, 100459. [Google Scholar] [CrossRef]
- Tang, W.X.; Wang, G.S.; Chen, R.; Liu, X.; Chen, X.S.; Shen, X.; Yin, C.M.; Mao, Z.Q. Allium fistulosum L. Alleviates Apple Replant Disease by Suppressing Fusarium solani. J. Fungi 2022, 8, 1071. [Google Scholar] [CrossRef]
- Wu, F.Z.; Han, X.; Wang, X.Z. Allelopathic effect of root exudates of cucumber cultivars on Fusarium oxysporum. Allelopath. J. 2006, 18, 163–172. [Google Scholar]
- Wang, R.Q.; Wang, Y.T.; Yao, W.J.; Ge, W.G.; Jiang, T.B.; Zhou, B.R. Transcriptome Sequencing and WGCNA Reveal Key Genes in Response to Leaf Blight in Poplar. Int. J. Mol. Sci. 2023, 24, 10047. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.M.; Wang, X.H.; Long, J.H.; Bai, S.Y.; Cui, Y.Y.; Wang, Z.Y.; Liu, C.X.; Liu, F.L.; Wang, Z.J.; Li, Q.F. Identification of Key Modules and Candidate Genes for Powdery Mildew Resistance of Wheat-Agropyron cristatum Translocation Line WAT-2020-17-6 by WGCNA. Plants 2023, 12, 335. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.J.; Han, S.Y.; Wang, D.; Haider, M.S.; Guo, J.J.; Zhao, Q.; Du, P.; Sun, Z.Q.; Qi, F.Y.; Zheng, Z.; et al. Gene Co-expression Network Analysis of the Comparative Transcriptome Identifies Hub Genes Associated with Resistance to Aspergillus flavus L. in Cultivated Peanut (Arachis hypogaea L.). Front. Plant Sci. 2022, 13, 899177. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.B.; Pan, Y.B.; Su, Y.C.; Zou, W.H.; Xu, F.; Sun, T.T.; Grisham, M.P.; Yang, S.L.; Xu, L.P.; Que, Y.X. WGCNA Identifies a Comprehensive and Dynamic Gene Co-Expression Network That Associates with Smut Resistance in Sugarcane. Int. J. Mol. Sci. 2022, 23, 10770. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sossah, F.L.; Li, Z.; Hyde, K.D.; Li, D.; Xiao, S.J.; Fu, Y.P.; Yuan, X.H.; Li, Y. Genome-Wide Identification and Analysis of Chitinase GH18 Gene Family in Mycogone perniciosa. Front. Microbiol. 2021, 11, 596719. [Google Scholar] [CrossRef] [PubMed]
- McCreath, K.J.; Specht, C.A.; Liu, Y.L.; Robbins, P.W. Molecular cloning of a third chitinase gene (CHT1) from Candida albicans. Yeast 1996, 12, 501–504. [Google Scholar] [CrossRef]
- Yang, C.; Yu, Y.Q.; Huang, J.K.; Meng, F.W.; Pang, J.H.; Zhao, Q.Q.; Islam, M.A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae Chitinase MoChia1 by a Rice Tetratricopeptide Repeat Protein Allows Free Chitin to Trigger Immune Responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef]
- Tzelepis, G.D.; Melin, P.; Jensen, D.F.; Stenlid, J.; Karlsson, M. Functional analysis of glycoside hydrolase family 18 and 20 genes in Neurospora crassa. Fungal Genet. Biol. 2012, 49, 717–730. [Google Scholar] [CrossRef]
- Koch, A.; Wassenegger, M. Host-induced gene silencing—Mechanisms and applications. New Phytol. 2021, 231, 54–59. [Google Scholar] [CrossRef]
- Wang, Z.W.; Gao, X.; Zhong, S.; Li, Y.; Shi, M.R.; Zhang, B.R.; Zhang, S.C.; Shen, H.L.; Liu, X.L. Host-induced gene silencing of PcCesA3 and PcOSBP1 confers resistance to Phytophthora capsici in Nicotiana benthamiana through NbDCL3 and NbDCL4 processed small interfering RNAs. Int. J. Biol. Macromol. 2022, 222, 1665–1675. [Google Scholar] [CrossRef] [PubMed]
- Ghag, S.B.; Shekhawat, U.K.S.; Ganapathi, T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014, 12, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Tian, J.; Feng, Z.D.; Wang, H.; Sun, J.; Kong, Z.S. Chitin synthases containing myosin motor-like domain are required for cell wall integrity and virulence of vascular wilt pathogen Verticillium dahliae. Phytopathol. Res. 2023, 5, 21. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, H.; Zhu, J.; Zhao, Z.; Han, Z.; Zhang, W. Chitinase Gene FoChi20 in Fusarium oxysporum Reduces Its Pathogenicity and Improves Disease Resistance in Cotton. Int. J. Mol. Sci. 2024, 25, 8517. https://doi.org/10.3390/ijms25158517
Lou H, Zhu J, Zhao Z, Han Z, Zhang W. Chitinase Gene FoChi20 in Fusarium oxysporum Reduces Its Pathogenicity and Improves Disease Resistance in Cotton. International Journal of Molecular Sciences. 2024; 25(15):8517. https://doi.org/10.3390/ijms25158517
Chicago/Turabian StyleLou, Hui, Jincheng Zhu, Zengqiang Zhao, Zegang Han, and Wei Zhang. 2024. "Chitinase Gene FoChi20 in Fusarium oxysporum Reduces Its Pathogenicity and Improves Disease Resistance in Cotton" International Journal of Molecular Sciences 25, no. 15: 8517. https://doi.org/10.3390/ijms25158517
APA StyleLou, H., Zhu, J., Zhao, Z., Han, Z., & Zhang, W. (2024). Chitinase Gene FoChi20 in Fusarium oxysporum Reduces Its Pathogenicity and Improves Disease Resistance in Cotton. International Journal of Molecular Sciences, 25(15), 8517. https://doi.org/10.3390/ijms25158517






