MicroRNA-148a Targets DNMT1 and PPARGC1A to Regulate the Viability, Proliferation, and Milk Fat Synthesis of Ovine Mammary Epithelial Cells
Abstract
1. Introduction
2. Results
2.1. Tissue Expression of miR-148a
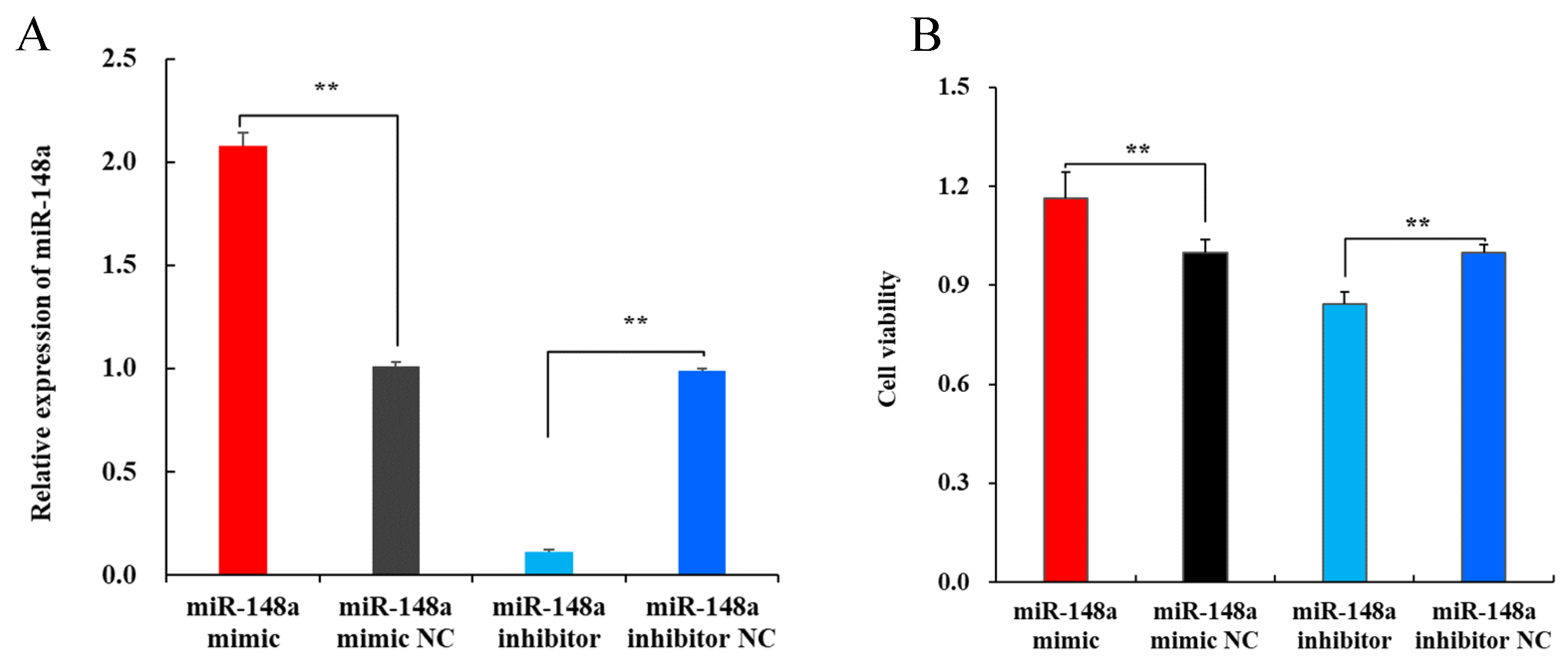
2.2. miR-148a Increases the Viability of OMECs
2.3. miR-148a Improves the Proliferation of OMECs
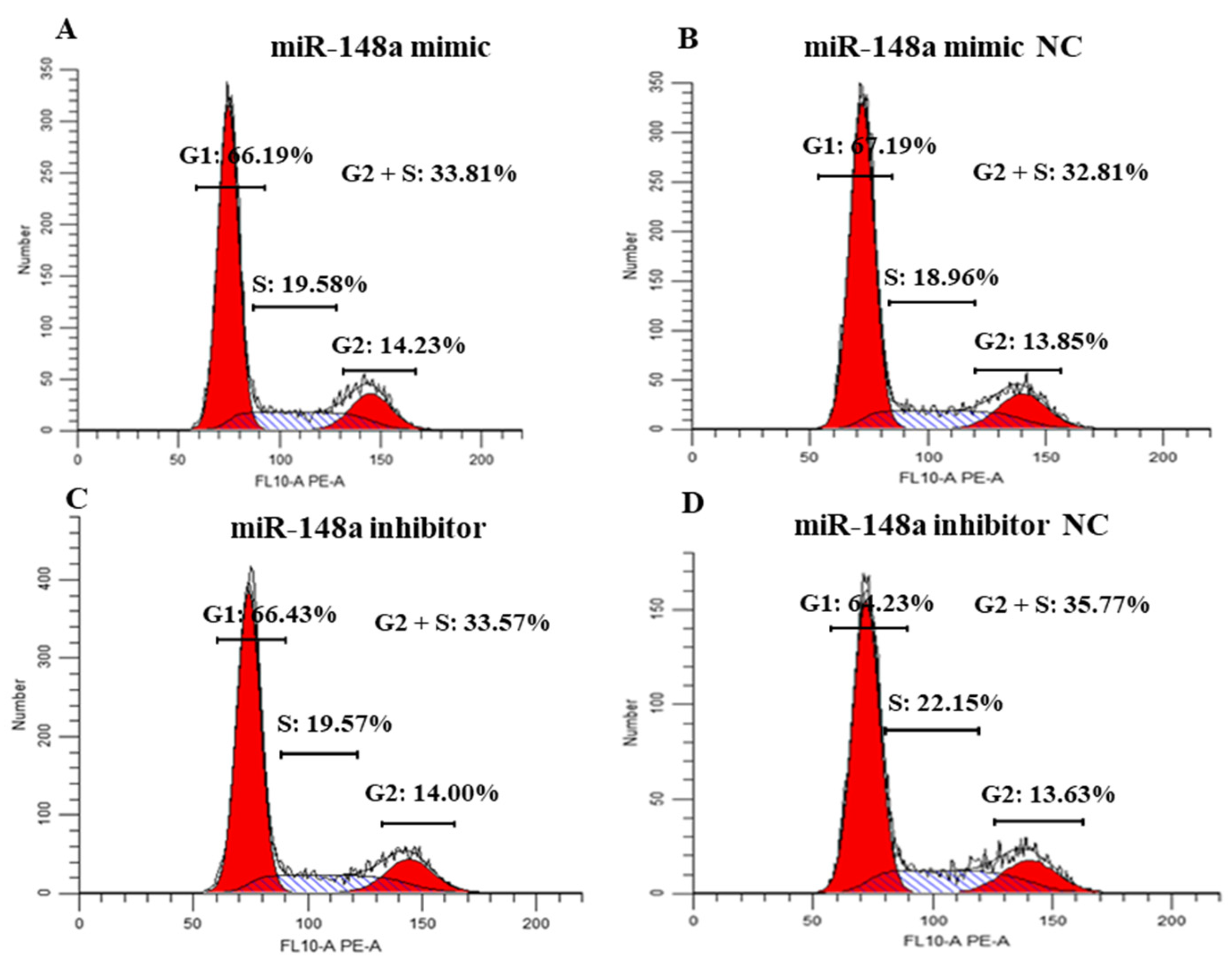
2.4. miR-148a Affects the Cycle of OMECs
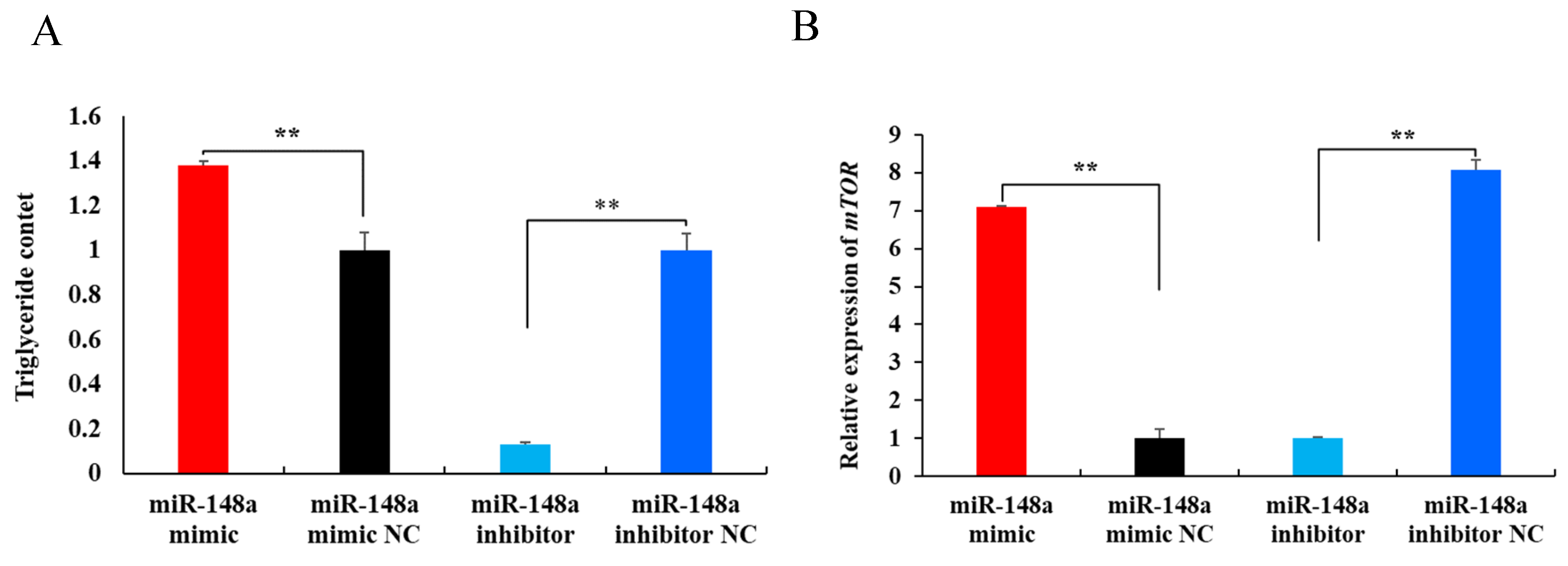
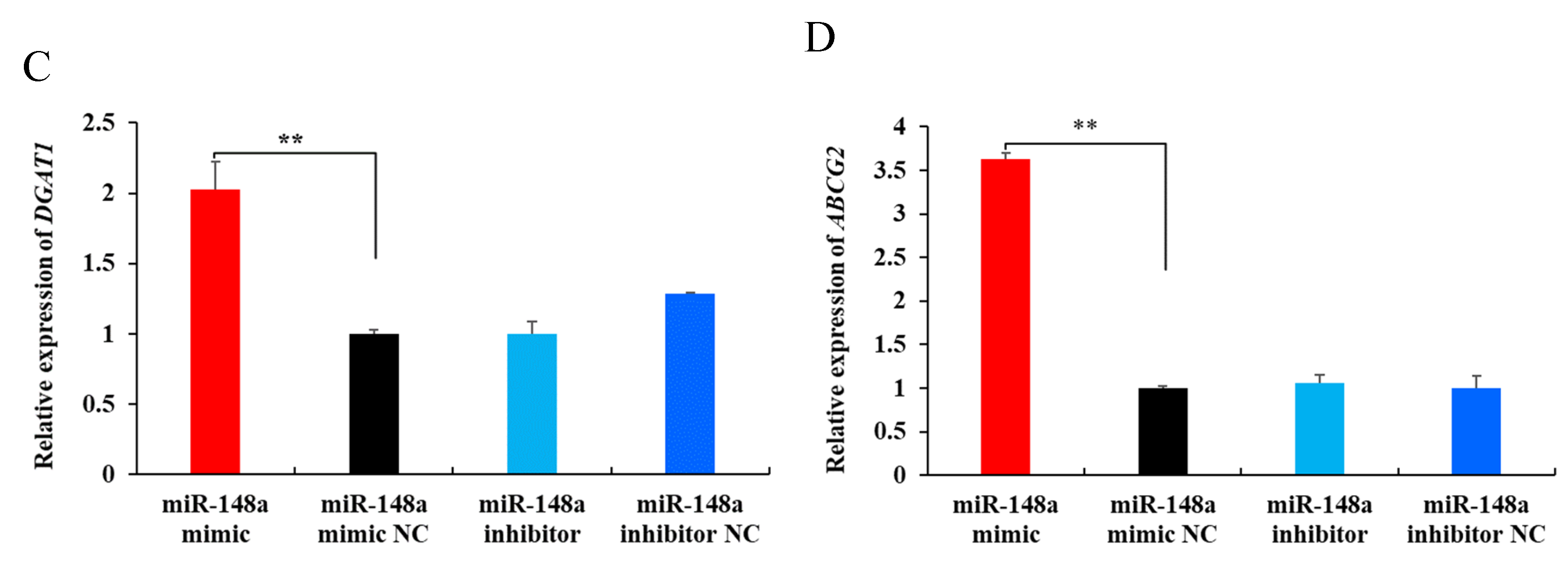
2.5. miR-148a Promotes Milk Fat Synthesis
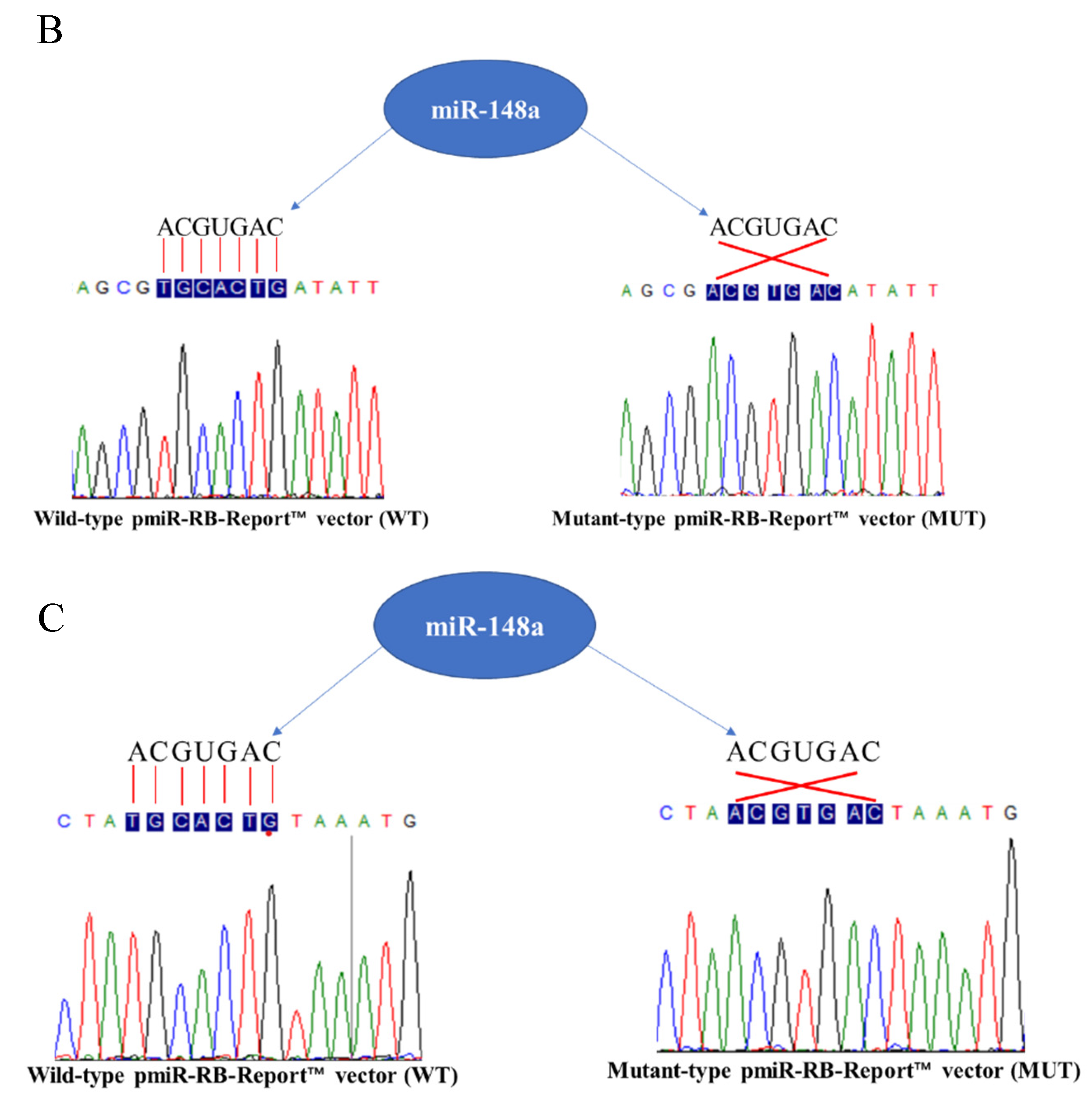
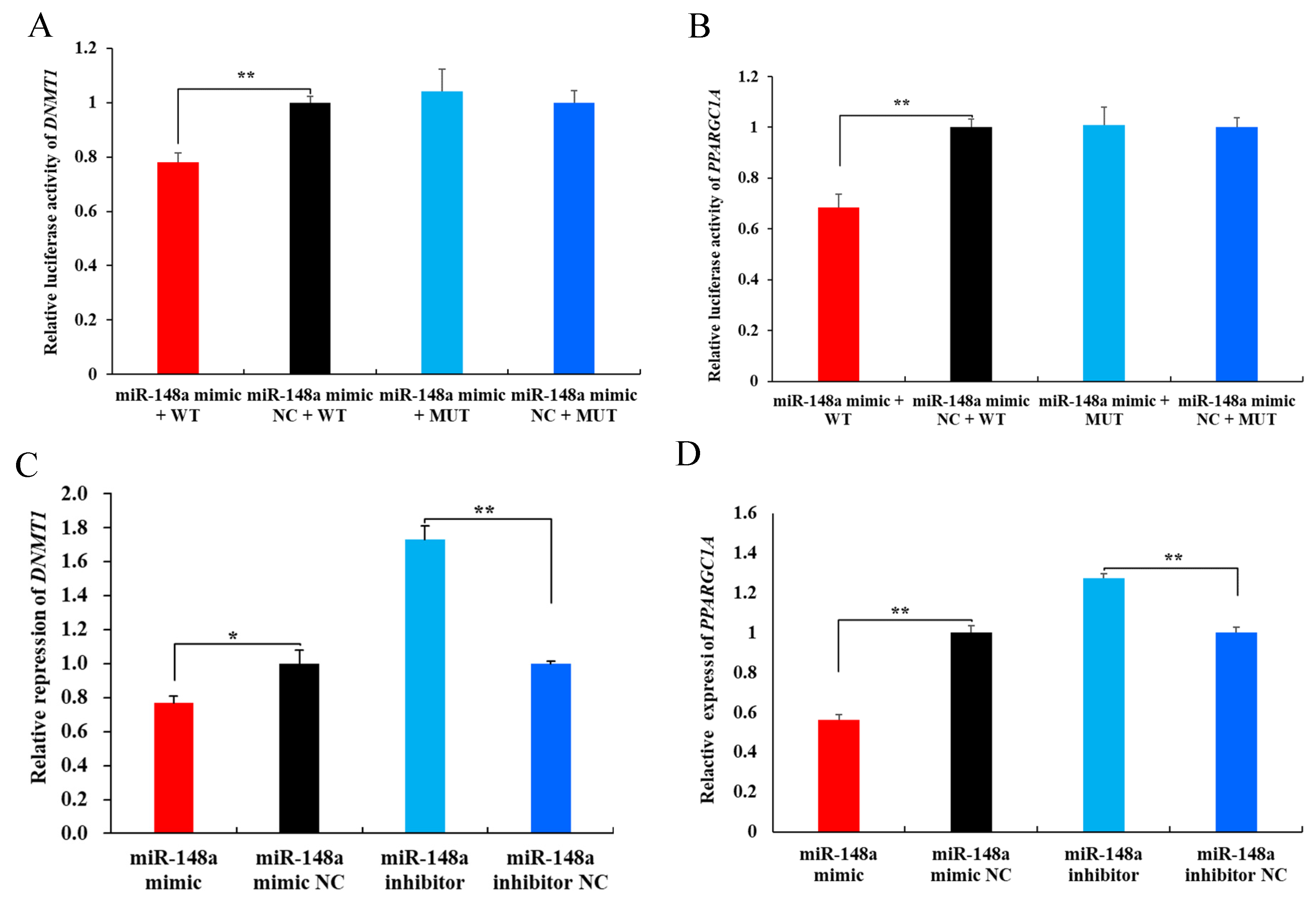
2.6. miR-148a Targets DNMT1 and PPARGC1A
2.7. Expression Levels of the Target Genes in Ovine Mammary Gland Tissue during Different Development Periods
3. Discussion
4. Materials and Methods
4.1. Ovine Tissue Sample Collection
4.2. Tissue Expression of miR-148a and Its Target Genes
4.3. Culture and Purification of OMECs
4.4. Cell Transfection and CCK8 Assay
4.5. Effect of miR-148a on the Proliferation of OMECs
4.6. Effect of miR-148a on the Milk Fat Synthesis of OMECs
4.7. Dual Luciferase Reporter Assay
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Hao, Z.; Hu, J.; Liu, X.; Li, S.; Wang, J.; Shen, J.; Song, Y.; Ke, N.; Luo, Y. Small RNA deep sequencing reveals the expressions of microRNAs in ovine mammary gland development at peak-lactation and during the non-lactating period. Genomics 2021, 113, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Arbab, A.A.I.; Zhang, H.; Yang, Y.; Lu, X.; Han, Z.; Yang, Z. MicroRNA-193a-5p regulates the synthesis of polyunsaturated fatty acids by targeting fatty acid desaturase 1 (FADS1) in bovine mammary epithelial cells. Biomolecules 2021, 11, 157. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, J.; Zhang, T.; Tian, H.; Ma, Y.; Xu, H.; Yao, D.; Loor, J.J. MicroRNA-26a/b and their host genes synergistically regulate triacylglycerol synthesis by targeting the INSIG1 gene. RNA Biol. 2016, 13, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chu, S.; Wang, X.; Fan, Y.; Zhan, T.; Arbab, A.A.I.; Li, M.; Zhang, H.; Mao, Y.; Loor, J.J.; et al. MicroRNA-106b regulates milk fat metabolism via ATP binding cassette subfamily a member 1 (ABCA1) in bovine mammary epithelial cells. J. Agric. Food Chem. 2019, 67, 3981–3990. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chu, S.; Wang, X.; Sun, Y.; Xu, T.; Mao, Y.; Loor, J.J.; Yang, Z. MiR-16a regulates milk fat metabolism by targeting large tumor suppressor kinase 1 (LATS1) in bovine mammary epithelial cells. J. Agric. Food Chem. 2019, 67, 11167–11178. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lu, Q.; Liang, Y.; Cui, X.; Wang, X.; Mao, Y.; Yang, Z. Circ11103 interacts with miR-128/PPARGC1A to regulate milk fat metabolism in dairy cows. J. Agric. Food Chem. 2021, 69, 4490–4500. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Luo, J.; Huang, L.; Tian, H.; Chen, Y.; He, Q. miR-204-5p and miR-211 synergistically downregulate the αS1-casein content and contribute to the lower allergy of goat milk. J. Agric. Food Chem. 2021, 69, 5353–5362. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Wang, J.; Luo, Y.; Hu, J.; Liu, X.; Li, S.; Li, M.; Shi, B.; Hu, L.; Liu, Y.; et al. MicroRNA-200c affects milk fat synthesis by targeting PANK3 in ovine mammary epithelial cells. Int. J. Mol. Sci. 2022, 23, 15601. [Google Scholar] [CrossRef]
- Hao, Z.; Luo, Y.; Wang, J.; Hickford, J.G.H.; Zhou, H.; Hu, J.; Liu, X.; Li, S.; Shen, J.; Ke, N.; et al. MicroRNA-432 inhibits milk fat synthesis by targeting SCD and LPL in ovine mammary epithelial cells. Food Funct. 2021, 12, 9432–9442. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Hao, Z.; Zhao, M.; Shen, J.; Ke, N.; Song, Y.; Qiao, L.; Lu, Y.; Hu, L.; Wu, X.; et al. MicroRNA-148a Regulates the proliferation and differentiation of ovine preadipocytes by targeting PTEN. Animals 2021, 11, 820. [Google Scholar] [CrossRef]
- Iqbal, A.; Yu, H.; Jiang, P.; Zhao, Z. Deciphering the key regulatory roles of KLF6 and bta-miR-148a on milk fat metabolism in bovine mammary epithelial cells. Genes 2022, 13, 1828. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, J.; Sun, S.; Cao, D.; Shi, H.; Loor, J.J. miR-148a and miR-17-5p synergistically regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells. RNA Biol. 2017, 14, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Liu, Q.; Zhang, X.; Ren, Y.; Lei, X.; Li, S.; Chen, Q.; Deng, K.; Wang, P.; Zhang, H.; et al. Identification and analysis of the expression of microRNA from lactating and nonlactating mammary glands of the Chinese swamp buffalo. J. Dairy Sci. 2017, 100, 1971–1986. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gu, X.; Lv, X.; Cao, X.; Yuan, Z.; Wang, S.; Sun, W. Non-coding transcriptomic profiles in the sheep mammary gland during different lactation periods. Front. Vet. Sci. 2022, 9, 983562. [Google Scholar] [CrossRef] [PubMed]
- Maddika, S.; Ande, S.R.; Wiechec, E.; Hansen, L.L.; Wesselborg, S.; Los, M. Akt-mediated phosphorylation of CDK2 regulates its dual role in cell cycle progression and apoptosis. J. Cell. Sci. 2008, 121, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Padmakumar, V.C.; Aleem, E.; Berthet, C.; Hilton, M.B.; Kaldis, P. Cdk2 and Cdk4 activities are dispensable for tumorigenesis caused by the loss of p53. Mol. Cell. Biol. 2009, 29, 2582–2593. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hao, Z.; Hu, L.; Qiao, L.; Luo, Y.; Hu, J.; Liu, X.; Li, S.; Zhao, F.; Shen, J.; et al. MicroRNA-199a-3p regulates proliferation and milk fat synthesis of ovine mammary epithelial cells by targeting VLDLR. Front. Vet. Sci. 2022, 9, 948873. [Google Scholar] [CrossRef] [PubMed]
- Muroya, S.; Hagi, T.; Kimura, A.; Aso, H.; Matsuzaki, M.; Nomura, M. Lactogenic hormones alter cellular and extracellular microRNA expression in bovine mammary epithelial cell culture. J. Anim. Sci. Biotechnol. 2016, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.L.; Peng, Z.; Yang, X.; Fan, K.J.; Ye, H.; Li, Z.H.; Wang, Y.; Xu, X.L.; Li, J.; Wang, Y.L.; et al. miR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int. J. Biol. Sci. 2011, 7, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Sajan, P.; Shi, J.; Shen, Y.; Wang, K.; Deng, X.; Zhou, L.; Hu, P.; Gao, L. MiR-148a increases glioma cell migration and invasion by downregulating GADD45A in human gliomas with IDH1 R132H mutations. Oncotarget 2017, 8, 25345–25361. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Nie, Y. MiR-148a participates in the growth of RPMI8226 multiple myeloma cells by regulating CDKN1B. Biomed. Pharmacother. 2016, 84, 1967–1971. [Google Scholar] [CrossRef] [PubMed]
- Bovenhuis, H.; Visker, M.H.; van Valenberg, H.J.; Buitenhuis, A.J.; van Arendonk, J.A. Effects of the DGAT1 polymorphism on test-day milk production traits throughout lactation. J. Dairy. Sci. 2015, 98, 6572–6582. [Google Scholar] [CrossRef] [PubMed]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Jiang, M.; Yang, T.; Tan, D.; Hu, G.; Zhao, G.; Zhan, K. Acetate-induced milk fat synthesis is associated with activation of the mTOR signaling pathway in bovine mammary epithelial cells. Animals 2022, 12, 2616. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Bovenhuis, H.; Visker, M.H.; van Arendonk, J.A. Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Tantia, M.S.; Vijh, R.K.; Mishra, B.P.; Mishra, B.; Kumar, S.T.; Sodhi, M. DGAT1 and ABCG2 polymorphism in Indian cattle (Bos indicus) and buffalo (Bubalus bubalis) breeds. BMC Vet. Res. 2006, 2, 32. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Zhao, Z.; Yu, X.; Lu, C.; Jiang, P.; Yu, H.; Li, X.; Yu, X.; Liu, J.; Fang, X.; et al. Integrative analysis of miRNAs and mRNAs revealed regulation of lipid metabolism in dairy cattle. Funct. Integr. Genom. 2021, 21, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Chen, Y.; Niu, Y.; Chen, W.; Wang, Q.; Xiao, S.; Li, A.; Xie, Y.; Li, J.; Zhao, X.; et al. A deep investigation into the adipogenesis mechanism: Profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genom. 2010, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, H.; Hickford, J.G.H.; Hao, Z.; Shen, J.; Luo, Y.; Hu, J.; Liu, X.; Li, S. Comparison of the transcriptome of the ovine mammary gland in lactating and non-lactating Small-Tailed Han Sheep. Front. Genet. 2020, 11, 472. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bian, Y.; Wang, Z.; Li, D.; Wang, C.; Li, Q.; Gao, X. MicroRNA-152 regulates DNA methyltransferase 1 and is involved in the development and lactation of mammary glands in dairy cows. PLoS ONE 2014, 9, e101358. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.; Zhang, S.; Yang, S.; Alim, M.A.; Zhang, Q.; Li, Y.; Liu, L. Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese Holstein cattle population based on a post genome-wide association study. BMC Genet. 2016, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Gong, F. MiR-148a promotes myocardial differentiation of human bone mesenchymal stromal cells via DNA methyltransferase 1 (DNMT1). Cell Biol. Int. 2018, 42, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Hermann, A.; Gowher, H.; Jeltsch, A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004, 61, 2571–2587. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, J.; Zhang, C.; Ma, Y.; Sun, S.; Zhang, T.; Loor, J.J. Mechanism of prolactin inhibition of miR-135b via methylation in goat mammary epithelial cells. J. Cell. Physiol. 2018, 233, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Szyf, M. The role of DNA methyltransferase 1 in growth control. Front. Biosci. 2001, 6, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Fan, X.; Zhang, Y.; Teng, X.; Miao, Y. Molecular characterization, tissue expression and polymorphisms of buffalo PPARGC1A gene. Arch. Anim. Breed. 2020, 63, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, H.; Hickford, J.G.H.; Hao, Z.; Gong, H.; Hu, J.; Liu, X.; Li, S.; Shen, J.; Ke, N.; et al. Identification and characterization of circular RNAs in mammary gland tissue from sheep at peak lactation and during the nonlactating period. J. Dairy Sci. 2021, 104, 2396–2409. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Gao, W.; Jin, C.; Wang, L.; Wang, Y.; Chen, W.; Zou, S.; Huang, S.; Li, Z.; Wang, J.; et al. Preliminary study on microR-148a and microR-10a in dermal papilla cells of Hu sheep. BMC Genet. 2019, 20, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ying, Z.Z.; Tang, Z.L.; Long, L.Q.; Li, K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012, 287, 21093–21101. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Zhou, H.; Hickford, J.G.H.; Gong, H.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Zhao, M.; Luo, Y. Transcriptome profile analysis of mammary gland tissue from two breeds of lactating sheep. Genes 2019, 10, 781. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, K.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Name | Forward (5’→3’) | Reverse (5’→3’) | Purpose of Primers |
|---|---|---|---|
| DNMT1 (WT) 1 | CCGCTCGAGCCTATCACCCATGTTTCTG | GAATGCGGCCGCGTCTTAATTTCCACTTCA | Construction of pmiR-RB-Report™ vector |
| PPARGC1A (WT) 1 | CCGCTCGAGGAGCACGGTGTTACATTA | GAATGCGGCCGCGGGCTAGGAGGTTTATTG | |
| DNMT1 (MUT) 2 | CAGCGACGTGACATATTGTTGTATTTTCACATGTCAATCG | AATATGTCAGTCGCTGGGGATCTCTGGTGG | |
| PPARGC1A (MUT) 2 | GTTACAGCTAACGTGACTAAATGCAGCCTTCTTT | GTCACGTTAGCTGTAACAAAAAAAGAACATCATTG | |
| miRNA-148a | TCAGTGCACTACAGAACTTTGT | TGGTGTCGTGGAGTCG | RT-qPCR |
| U6 | GGAACGATACAGAGAAGATTAGC | TGGAACGCTTCACGAATTTGCG | |
| CDK2 | AGAAGTGGTGGCGCTTAAAA | TCTTGAGATCCTGGTGCAGA | |
| CDK4 | GCTTTTGAGCATCCCAATGT | AGGTCTTGGTCCACATGCTC | |
| GAPDH | CACAGTCAAGGCAGAGAACG | CAGCCTTCTCCATGGTAGTG | |
| β-actin | AGCCTTCCTTCCTGGGCATGGA | GGACAGCACCGTGTTGGCGTAGA | |
| mTOR | TGCTAACTACCTTCGGAACCT | TGAAAGTGTCCCCTGCCAT | |
| DGAT1 | CCACCATTCTCTGCTTCCCA | AGCTTGAGGAAGAGGATGGT | |
| ABCG2 | AGTTACGAGGTTTCCATCCCA | CGACAAAGTAGAAAGCCAGTCT | |
| DNMT1 | TTCTCACTGCCTGACGATGT | TGACTTTAGCCAGGTAGCCC | |
| PPARGC1A | GACTCAAGTGGTGCAGTGAC | GGCAATCCGTCTTCATCCAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Ke, N.; Wu, X.; Zhen, H.; Hu, J.; Liu, X.; Li, S.; Zhao, F.; Li, M.; Shi, B.; et al. MicroRNA-148a Targets DNMT1 and PPARGC1A to Regulate the Viability, Proliferation, and Milk Fat Synthesis of Ovine Mammary Epithelial Cells. Int. J. Mol. Sci. 2024, 25, 8558. https://doi.org/10.3390/ijms25168558
Wang J, Ke N, Wu X, Zhen H, Hu J, Liu X, Li S, Zhao F, Li M, Shi B, et al. MicroRNA-148a Targets DNMT1 and PPARGC1A to Regulate the Viability, Proliferation, and Milk Fat Synthesis of Ovine Mammary Epithelial Cells. International Journal of Molecular Sciences. 2024; 25(16):8558. https://doi.org/10.3390/ijms25168558
Chicago/Turabian StyleWang, Jiqing, Na Ke, Xinmiao Wu, Huimin Zhen, Jiang Hu, Xiu Liu, Shaobin Li, Fangfang Zhao, Mingna Li, Bingang Shi, and et al. 2024. "MicroRNA-148a Targets DNMT1 and PPARGC1A to Regulate the Viability, Proliferation, and Milk Fat Synthesis of Ovine Mammary Epithelial Cells" International Journal of Molecular Sciences 25, no. 16: 8558. https://doi.org/10.3390/ijms25168558
APA StyleWang, J., Ke, N., Wu, X., Zhen, H., Hu, J., Liu, X., Li, S., Zhao, F., Li, M., Shi, B., Zhao, Z., Ren, C., & Hao, Z. (2024). MicroRNA-148a Targets DNMT1 and PPARGC1A to Regulate the Viability, Proliferation, and Milk Fat Synthesis of Ovine Mammary Epithelial Cells. International Journal of Molecular Sciences, 25(16), 8558. https://doi.org/10.3390/ijms25168558





