Surface Plasmon Resonance Immunosensor for Direct Detection of Antibodies against SARS-CoV-2 Nucleocapsid Protein
Abstract
:1. Introduction
2. Results
3. Materials and Methods
3.1. Reagents
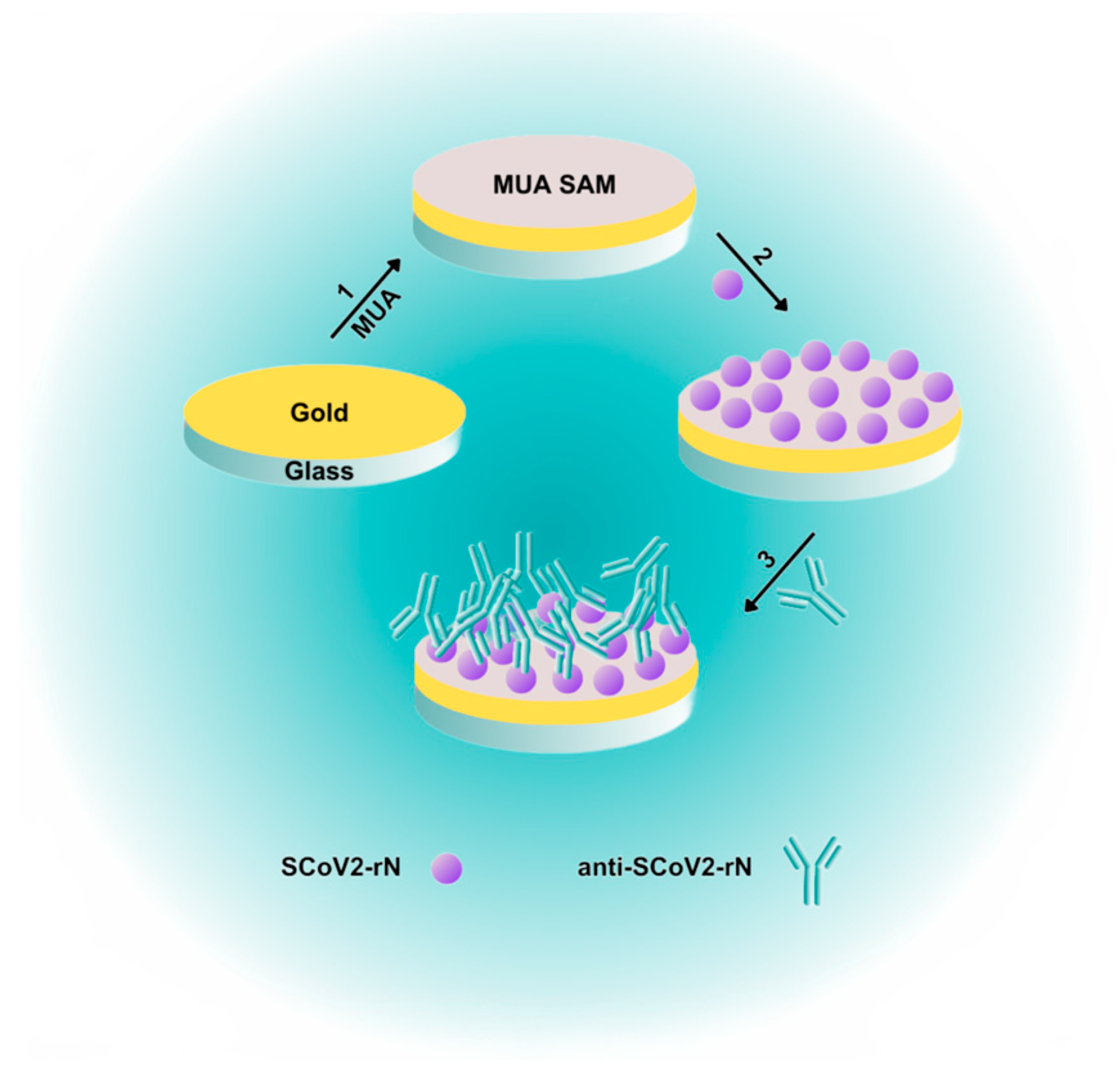
3.2. Gold-Coated Sensor Chip Preparation
3.3. Gold-Coated Sensor Chip Surface Stabilization
3.4. Nucleocapsid Protein Immobilization
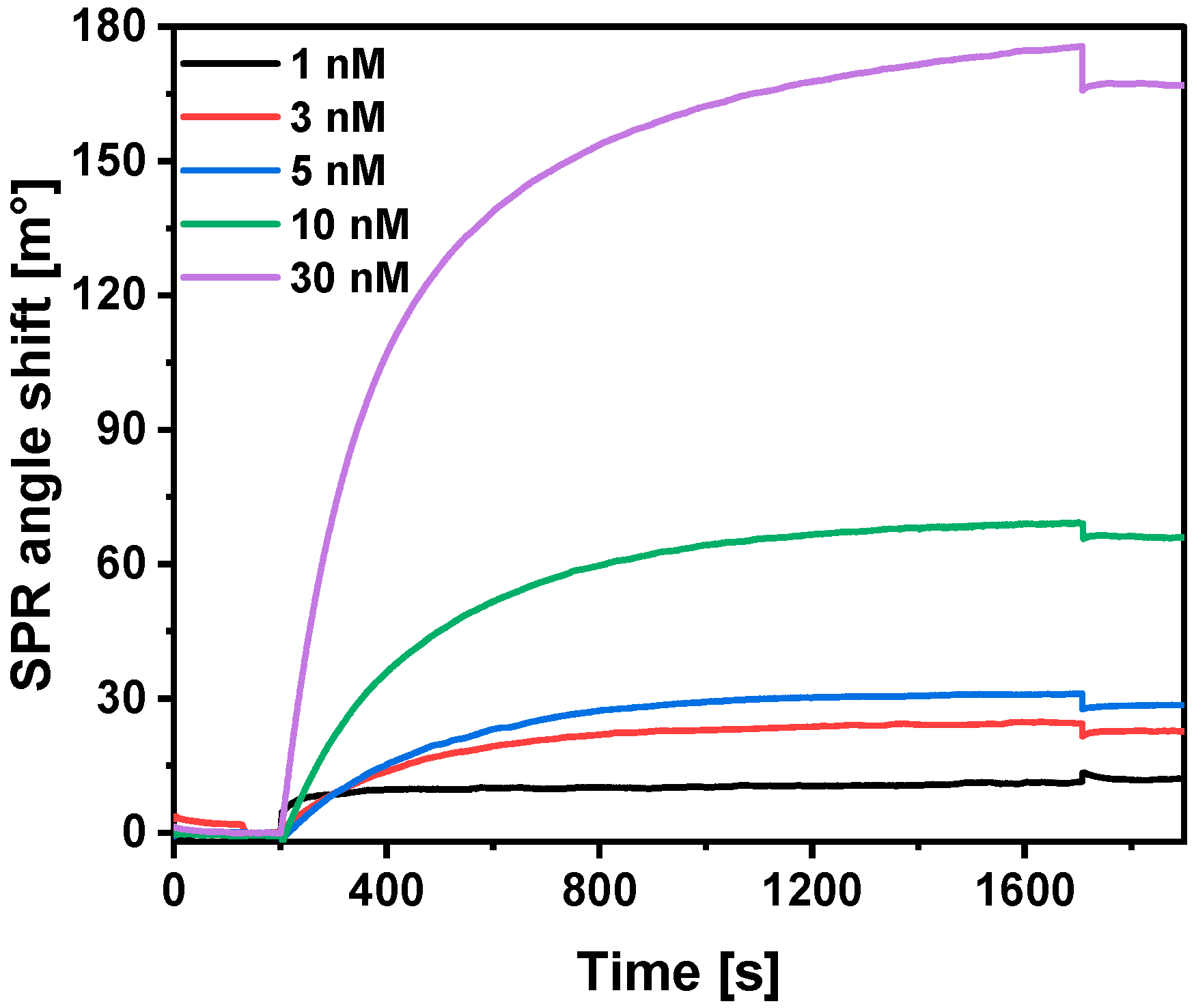
3.5. Interaction of Immobilized SCoV2-rN with Specific Antibodies
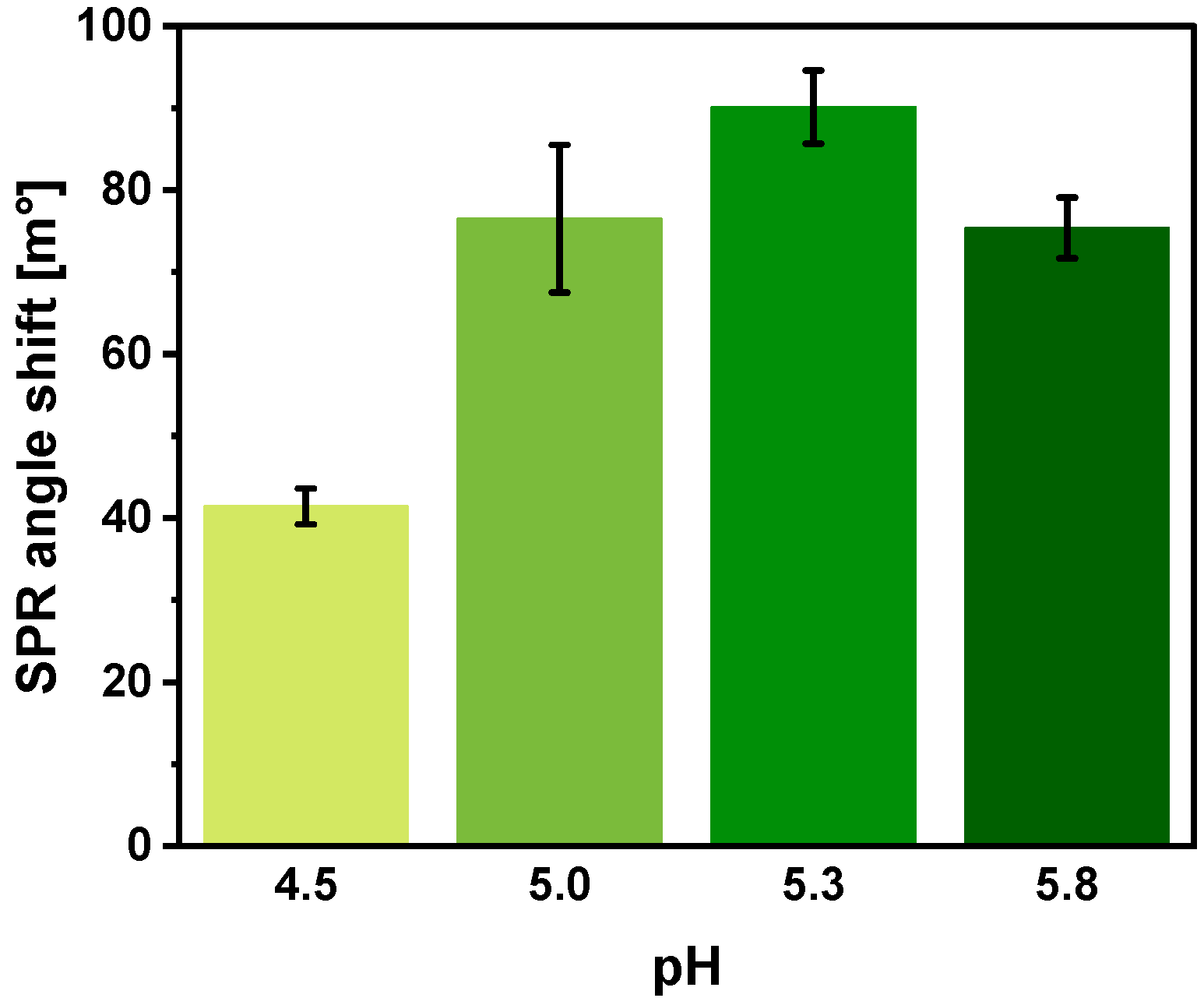
3.6. Selection of Regeneration Solution and Regeneration Duration
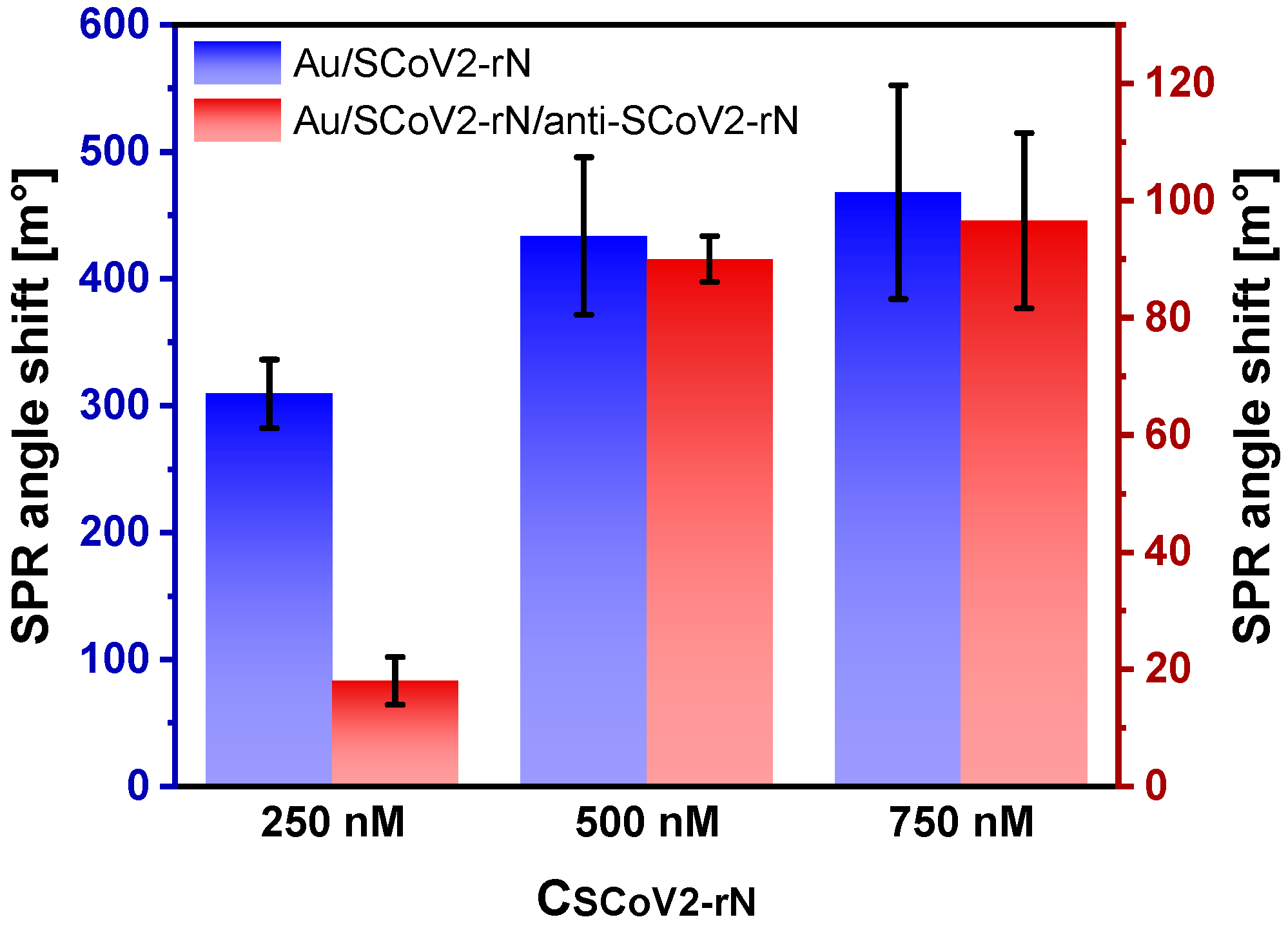
3.7. Optimization of SCoV2-rN Surface Concentration
3.8. Calculations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Szekely, J.; Mongkolprasert, J.; Jeayodae, N.; Senorit, C.; Chaimuti, P.; Swangphon, P.; Nanakorn, N.; Nualnoi, T.; Wongwitwichot, P.; Pengsakul, T. Development, Analytical, and Clinical Evaluation of Rapid Immunochromatographic Antigen Test for SARS-CoV-2 Variants Detection. Diagnostics 2022, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO COVID-19 Dashboard, World Health Organization. 2022. Available online: https://data.who.int/dashboards/covid19/cases?n=o (accessed on 18 July 2024).
- Drobysh, M.; Ramanaviciene, A.; Viter, R.; Chen, C.-F.; Samukaite-Bubniene, U.; Ratautaite, V.; Ramanavicius, A. Biosensors for the determination of SARS-CoV-2 virus and diagnosis of COVID-19 infection. Int. J. Mol. Sci. 2022, 23, 666. [Google Scholar] [CrossRef] [PubMed]
- Lutomski, C.A.; El-Baba, T.J.; Bolla, J.R.; Robinson, C.V. Multiple Roles of SARS-CoV-2 N Protein Facilitated by Proteoform-Specific Interactions with RNA, Host Proteins, and Convalescent Antibodies. JACS Au 2021, 1, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Yang, M.; Hong, Z.; Zhang, L.; Huang, Z.; Chen, X.; He, S.; Zhou, Z.; Zhou, Z.; Chen, Q.; et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 2020, 10, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Zinkernagel, R.M.; Hengartner, H. Regulation of the Immune Response by Antigen. Science 2001, 293, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, I.; Mattiuzzo, G.; Page, M.; Minor, P.; Griffiths, E.; Nuebling, M.; Moorthy, V. WHO International Standard for evaluation of the antibody response to COVID-19 vaccines: Call for urgent action by the scientific community. Lancet Microbe 2022, 3, e235–e240. [Google Scholar] [CrossRef] [PubMed]
- Plikusiene, I.; Maciulis, V.; Ramanaviciene, A.; Balevicius, Z.; Buzavaite-Verteliene, E.; Ciplys, E.; Slibinskas, R.; Simanavicius, M.; Zvirbliene, A.; Ramanavicius, A. Evaluation of kinetics and thermodynamics of interaction between immobilized SARS-CoV-2 nucleoprotein and specific antibodies by total internal reflection ellipsometry. J. Colloid Interface Sci. 2021, 594, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.F.; Vidal-Pla, M.; Moya, M.C.; Espasa, M.; Casabella, A.; Seda, M.; Calvet, J.; Gratacós, J.; Serrano, R.M.; Peña, P. SARS-CoV-2 Spike Protein Vaccine-Induced Immune Imprinting Reduces Nucleocapsid Protein Antibody Response in SARS-CoV-2 Infection. J. Immunol. Res. 2022, 2022, 8287087. [Google Scholar] [CrossRef] [PubMed]
- Soleimanian, S.; Alyasin, S.; Sepahi, N.; Ghahramani, Z.; Kanannejad, Z.; Yaghobi, R.; Karimi, M.H. An Update on Protective Effectiveness of Immune Responses after Recovery from COVID-19. Front. Immunol. 2022, 13, 884879. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, A.; Jędruchniewicz, N.; Torelli, A.; Kaczmarzyk-Radka, A.; Coluccio, R.; Kłak, M.; Konieczny, A.; Ferenc, S.; Witkiewicz, W.; Montomoli, E.; et al. Antibodies specific to SARS-CoV-2 proteins N, S and E in COVID-19 patients in the normal population and in historical samples. J. Gen. Virol. 2021, 102, 001692. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-K.; Lo, S.-C.; Wang, Y.-S.; Hou, M.-H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today 2016, 21, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.A.; Saier, M.H. The SARS-Coronavirus Infection Cycle: A Survey of Viral Membrane Proteins, Their Functional Interactions and Pathogenesis. Int. J. Mol. Sci. 2021, 22, 1308. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Hu, C.; Zhang, F.; Jahan, S.; Yuan, B.; Saleh, M.S.; Gao, S.; Panat, R. N protein-based ultrasensitive SARS-CoV-2 antibody detection in seconds via 3D nanoprinted, microarchitected array electrodes. J. Med. Virol. 2022, 94, 2067–2078. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Li, J.; Ai, Y.; He, H.; Chen, X.; Yin, M.; Li, W.; Huang, W.; Luo, M.-Y.; He, J. Current strategies for SARS-CoV-2 molecular detection. Anal. Methods 2022, 14, 4625–4642. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Brochot, E.; Demey, B.; Touze, A.; Belouzard, S.; Dubuisson, J.; Schmit, J.L. Anti-Spike anti-Nucleocapsid and neutralizing antibodies in SARS-CoV-2 hospitalized patients and asymptomatic carriers. Front. Microbiol. 2020, 11, 2468. [Google Scholar] [CrossRef] [PubMed]
- Kasetsirikul, S.; Umer, M.; Soda, N.; Sreejith, K.R.; Shiddiky, M.J.A.; Nguyen, N.-T. Detection of the SARS-CoV-2 humanized antibody with paper-based ELISA. Analyst 2020, 145, 7680–7686. [Google Scholar] [CrossRef] [PubMed]
- Damiani, V.; Pizzinato, E.; Cicalini, I.; Demattia, G.; Zucchelli, M.; Natale, L.; Palmarini, C.; Di Marzio, C.; Federici, L.; De Laurenzi, V.; et al. Development of a Method for Detection of SARS-CoV-2 Nucleocapsid Antibodies on Dried Blood Spot by DELFIA Immunoassay. Diagnostics 2023, 13, 897. [Google Scholar] [CrossRef] [PubMed]
- Sadique, M.A.; Yadav, S.; Ranjan, P.; Khan, R.; Khan, F.; Kumar, A.; Biswas, D. Highly Sensitive Electrochemical Immunosensor Platforms for Dual Detection of SARS-CoV-2 Antigen and Antibody based on Gold Nanoparticle Functionalized Graphene Oxide Nanocomposites. ACS Appl. Bio Mater. 2022, 5, 2421–2430. [Google Scholar] [CrossRef] [PubMed]
- Ke, G.; Su, D.; Li, Y.; Zhao, Y.; Wang, H.; Liu, W.; Li, M.; Yang, Z.; Xiao, F.; Yuan, Y.; et al. An accurate, high-speed, portable bifunctional electrical detector for COVID-19. Sci. China Mater. 2021, 64, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zheng, Z.; Hu, H.; Zhou, Q.; Liu, W.; Li, X.; Liu, Z.; Wang, Y.; Ma, Y. A point-of-care selenium nanoparticle-based test for the combined detection of anti-SARS-CoV-2 IgM and IgG in human serum and blood. Lab Chip 2020, 20, 4255–4261. [Google Scholar] [CrossRef] [PubMed]
- Muench, P.; Jochum, S.; Wenderoth, V.; Ofenloch-Haehnle, B.; Hombach, M.; Strobl, M.; Sadlowski, H.; Sachse, C.; Torriani, G.; Eckerle, I.; et al. Development and validation of the elecsys anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef]
- Li, Z.; Wang, A.; Zhou, J.; Chen, Y.; Liu, H.; Liu, Y.; Zhang, Y.; Ding, P.; Zhu, X.; Liang, C.; et al. A Universal Fluorescent Immunochromatography Assay Based on Quantum Dot Nanoparticles for the Rapid Detection of Specific Antibodies against SARS-CoV-2 Nucleocapsid Protein. Int. J. Mol. Sci. 2022, 23, 6225. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Lozano, O.; Sierra, M.; Soler, M.; Estévez, M.C.; Chiscano-Camón, L.; Ruiz-Sanmartin, A.; Ruiz-Rodriguez, J.C.; Ferrer, R.; González-López, J.J.; Esperalba, J.; et al. Label-Free Plasmonic Biosensor for Rapid, Quantitative, and Highly Sensitive COVID-19 Serology: Implementation and Clinical Validation. Anal. Chem. 2022, 94, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Djaileb, A.; Jodaylami, M.H.; Coutu, J.; Ricard, P.; Lamarre, M.; Rochet, L.; Cellier-Goetghebeur, S.; Macaulay, D.; Charron, B.; Lavallée, É.; et al. Cross-validation of ELISA and a portable surface plasmon resonance instrument for IgG antibody serology with SARS-CoV-2 positive individuals. Analyst 2021, 146, 4905–4917. [Google Scholar] [CrossRef] [PubMed]
- Basso, C.R.; Malossi, C.D.; Haisi, A.; Pedrosa, V.d.A.; Barbosa, A.N.; Grotto, R.T.; Junior, J.P.A. Fast and reliable detection of SARS-CoV-2 antibodies based on surface plasmon resonance. Anal. Methods 2021, 13, 3297–3306. [Google Scholar] [CrossRef] [PubMed]
- Ramanaviciene, A.; Kausaite-Minkstimiene, A.; Popov, A.; Brasiunas, B.; Ramanavicius, A. Chapter 12—Design of immunosensors for rapid and sensitive detection of biomarkers. In The Detection of Biomarkers; Ozkan, S.A., Bakirhan, N.K., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 303–333. [Google Scholar] [CrossRef]
- Capelli, D.; Scognamiglio, V.; Montanari, R. Surface plasmon resonance technology: Recent advances, applications and experimental cases. TrAC Trends Anal. Chem. 2023, 163, 117079. [Google Scholar] [CrossRef]
- Kausaite-Minkstimiene, A.; Popov, A.; Ramanaviciene, A. Ultra-sensitive SPR immunosensors: A comprehensive review of labeling and interface modification using nanostructures. TrAC Trends Anal. Chem. 2024, 170, 117468. [Google Scholar] [CrossRef]
- Fendi, F.W.S.; Mukhtar, W.M.; Abdullah, M. Surface plasmon resonance sensor for COVID-19 detection: A review on plasmonic materials. Sens. Actuators A Phys. 2023, 362, 114617. [Google Scholar] [CrossRef]
- Lv, J.; Wang, J.; Yang, L.; Liu, W.; Haihao, F.; Chu, P.K.; Liu, C. Recent advances of optical fiber biosensors based on surface plasmon resonance: Sensing principles, Structures, and Prospects. Sens. Diagn. 2024, 362, 114617. [Google Scholar] [CrossRef]
- Dixit, C.K. Surface regeneration of gold-coated chip for highly-reproducible surface plasmon resonance immunoassays. J. Biosens. Bioelectron. 2014, 5, 1. [Google Scholar] [CrossRef]
- Goode, J.A.; Rushworth, J.V.H.; Millner, P.A. Biosensor regeneration: A review of common techniques and outcomes. Langmuir 2015, 31, 6267–6276. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.J.E. Amine Coupling through EDC/NHS: A Practical Approach. In Surface Plasmon Resonance: Methods and Protocols; Mol, N.J., Fischer, M.J.E., Eds.; Humana Press: Totowa, NJ, USA, 2010; pp. 55–73. [Google Scholar] [CrossRef]
- Kausaite-Minkstimiene, A.; Popov, A.; Ramanaviciene, A. Surface plasmon resonance immunosensor with antibody-functionalized magnetoplasmonic nanoparticles for ultrasensitive quantification of the CD5 biomarker. ACS Appl. Mater. Interfaces 2022, 14, 20720–20728. [Google Scholar] [CrossRef] [PubMed]
- Scheller, C.; Krebs, F.; Minkner, R.; Astner, I.; Gil-Moles, M.; Wätzig, H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis 2020, 41, 1137–1151. [Google Scholar] [CrossRef] [PubMed]
- Schasfoort, R.B.M. Handbook of Surface Plasmon Resonance; Royal Society of Chemistry: London, UK, 2017. [Google Scholar]
- Malmqvist, M. Surface plasmon resonance for detection and measurement of antibody-antigen affinity and kinetics. Curr. Opin. Immunol. 1993, 5, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Kausaite-Minkstimiene, A.; Popov, A.; Kalvaityte, U.; Bernotiene, E.; Mobasheri, A.; Ramanaviciene, A. An ultra-sensitive SPR immunosensor for quantitative determination of human cartilage oligomeric matrix protein biomarker. Biosens. Bioelectron. 2023, 234, 115370. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zeng, W.; He, H.; Zhao, D.; Yang, Y.; Jiang, D.; Qi, Y.; He, W.; Zhao, C.; Yi, R.; et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by chemiluminescence immunoanalysis. medRxiv 2020. [Google Scholar] [CrossRef]
- Kausaite-Minkstimiene, A.; Ramanaviciene, A.; Ramanavicius, A. Surface plasmon resonance biosensor for direct detection of antibodies against human growth hormone. Analyst 2009, 134, 2051–2057. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, M.; Qi, F.; Lin, F.R.; Jen, A.K.-Y. Self-Assembled monolayers for interfacial engineering in solution-processed thin-film electronic devices: Design, fabrication, and applications. Chem. Rev. 2024, 124, 2138–2204. [Google Scholar] [CrossRef] [PubMed]
- Popov, A.; Lisyte, V.; Kausaite-Minkstimiene, A.; Bernotiene, E.; Ramanaviciene, A. Experimental evaluation of quantum dots and antibodies conjugation by surface plasmon resonance spectroscopy. Int. J. Mol. Sci. 2022, 23, 12626. [Google Scholar] [CrossRef] [PubMed]

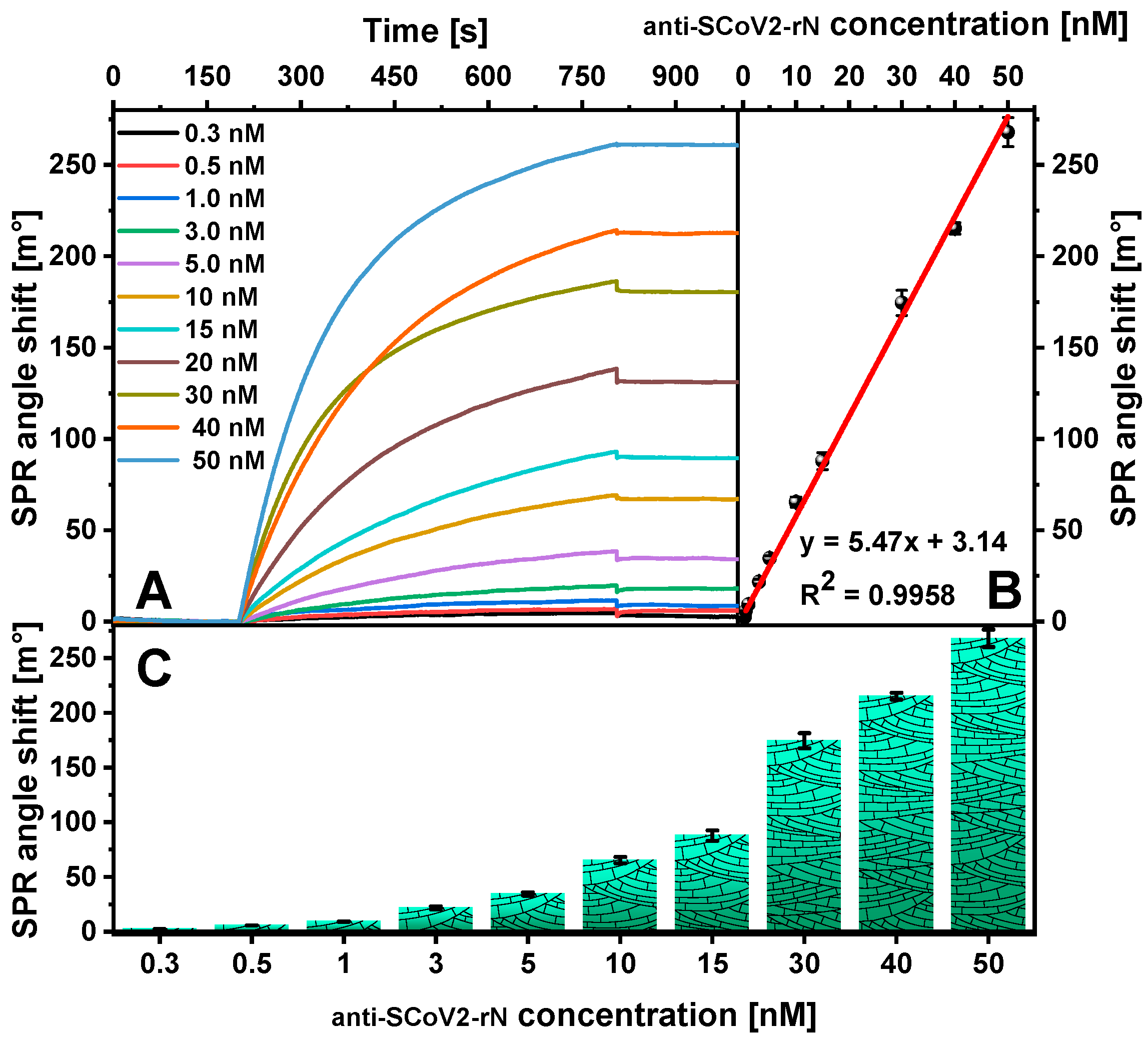
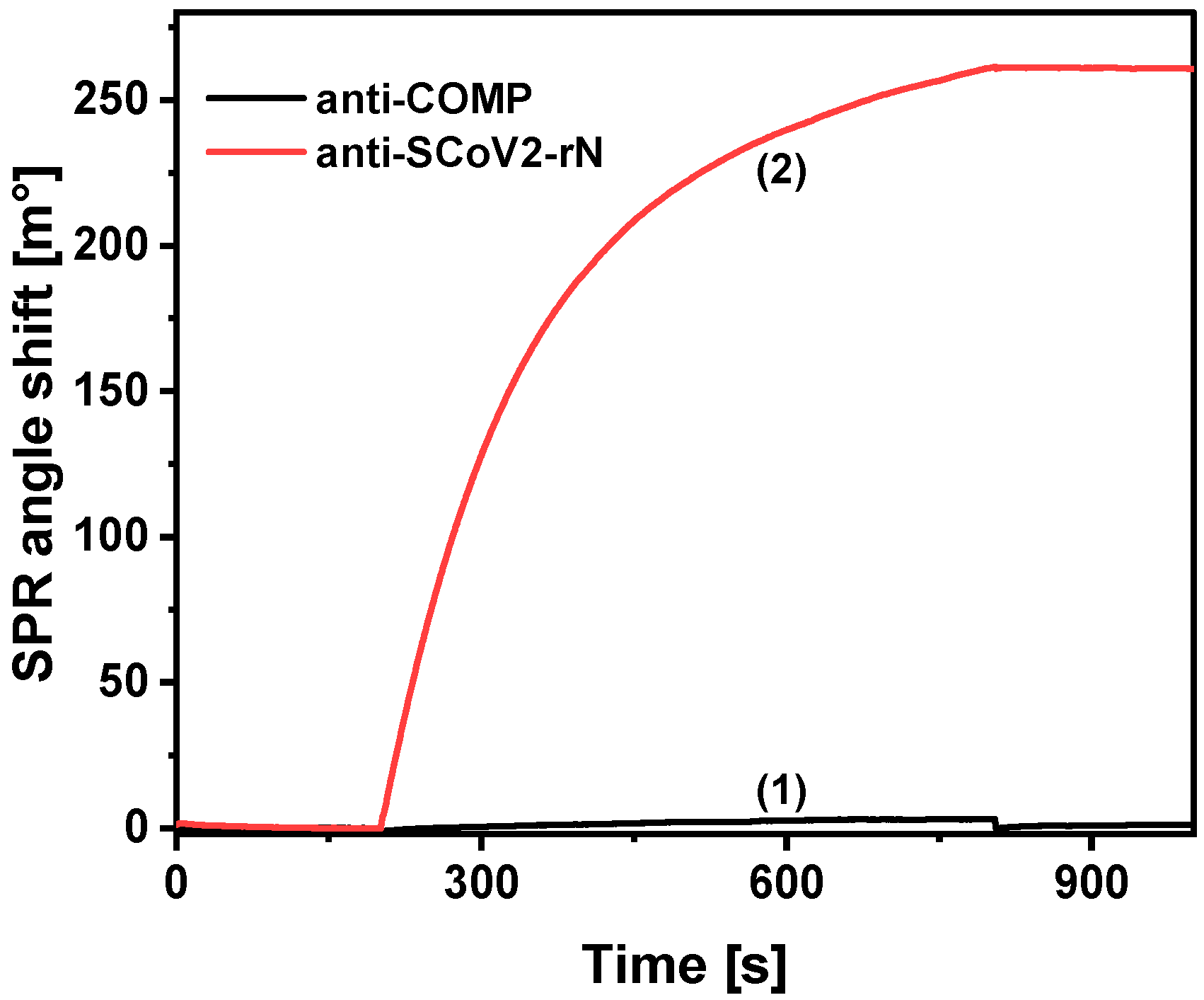
| Antibody Type | Detection Method | Linear Range [ng/mL] | LOD [ng/mL] | Reference |
|---|---|---|---|---|
| n/a | electrochemical impedance transduction | − | 0.00195 (13 fM) | [14] |
| Monoclonal humanized IgG | paper-based ELISA | 1–50 | 9 | [18] |
| Rabbit polyclonal IgG | electrochemical immunosensor | 1 × 10−6–1 | 1 × 10−6 | [20] |
| n/a | lateral flow immunoassay | − | 5 | [22] |
| Polyclonal IgG | SPR spectroscopy | 100–10,000 | 45.6 | [25] |
| Mouse monoclonal IgG | SPR spectroscopy | 75–7500 | 8.55 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lisyte, V.; Kausaite-Minkstimiene, A.; Brasiunas, B.; Popov, A.; Ramanaviciene, A. Surface Plasmon Resonance Immunosensor for Direct Detection of Antibodies against SARS-CoV-2 Nucleocapsid Protein. Int. J. Mol. Sci. 2024, 25, 8574. https://doi.org/10.3390/ijms25168574
Lisyte V, Kausaite-Minkstimiene A, Brasiunas B, Popov A, Ramanaviciene A. Surface Plasmon Resonance Immunosensor for Direct Detection of Antibodies against SARS-CoV-2 Nucleocapsid Protein. International Journal of Molecular Sciences. 2024; 25(16):8574. https://doi.org/10.3390/ijms25168574
Chicago/Turabian StyleLisyte, Viktorija, Asta Kausaite-Minkstimiene, Benediktas Brasiunas, Anton Popov, and Almira Ramanaviciene. 2024. "Surface Plasmon Resonance Immunosensor for Direct Detection of Antibodies against SARS-CoV-2 Nucleocapsid Protein" International Journal of Molecular Sciences 25, no. 16: 8574. https://doi.org/10.3390/ijms25168574








