Integrating 16S rRNA Sequencing, Microflora Metabolism, and Network Pharmacology to Investigate the Mechanism of SBL in Alleviating HDM-Induced Allergic Rhinitis
Abstract
1. Introduction
2. Results
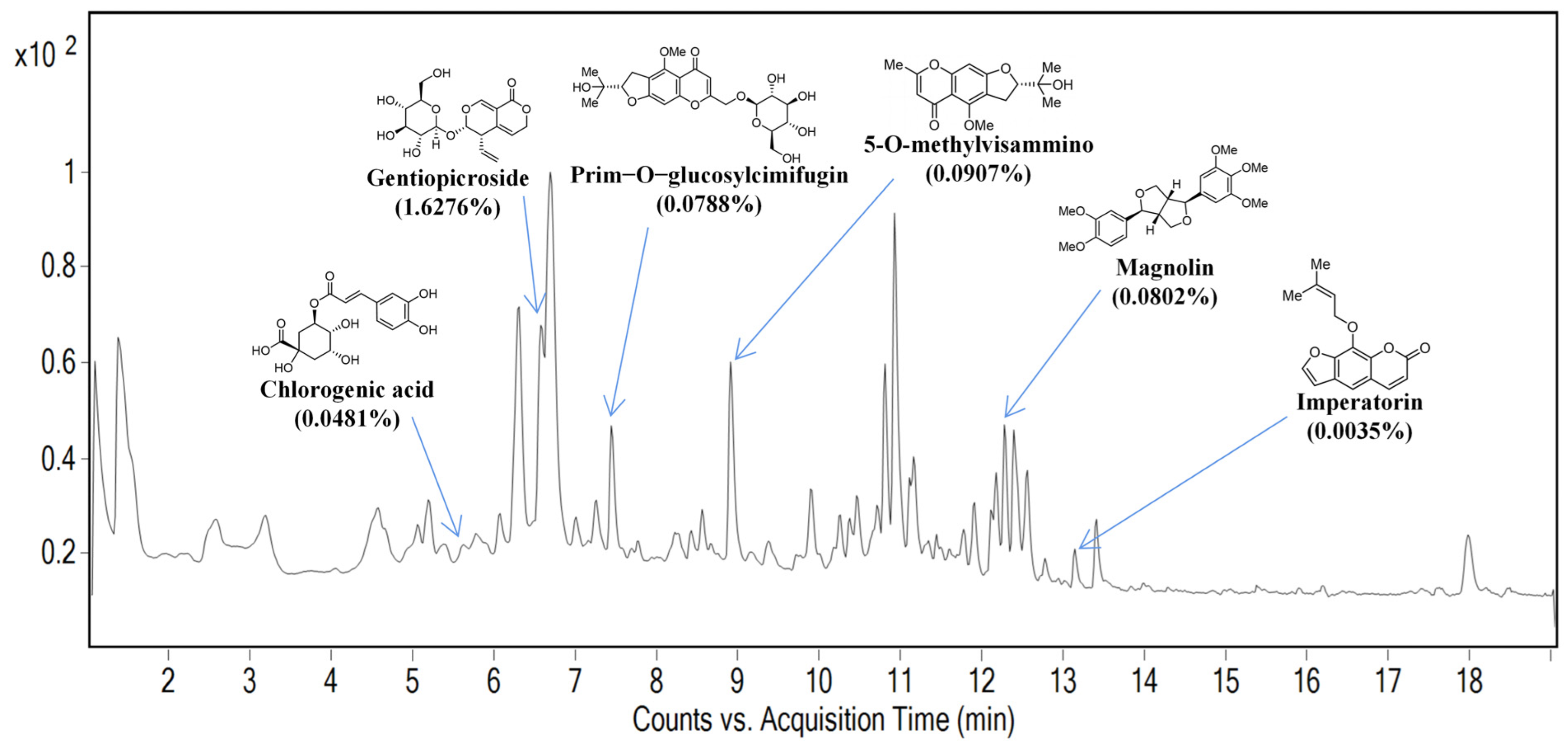
2.1. Identification of Marker Compounds of SBL by UPLC Analysis
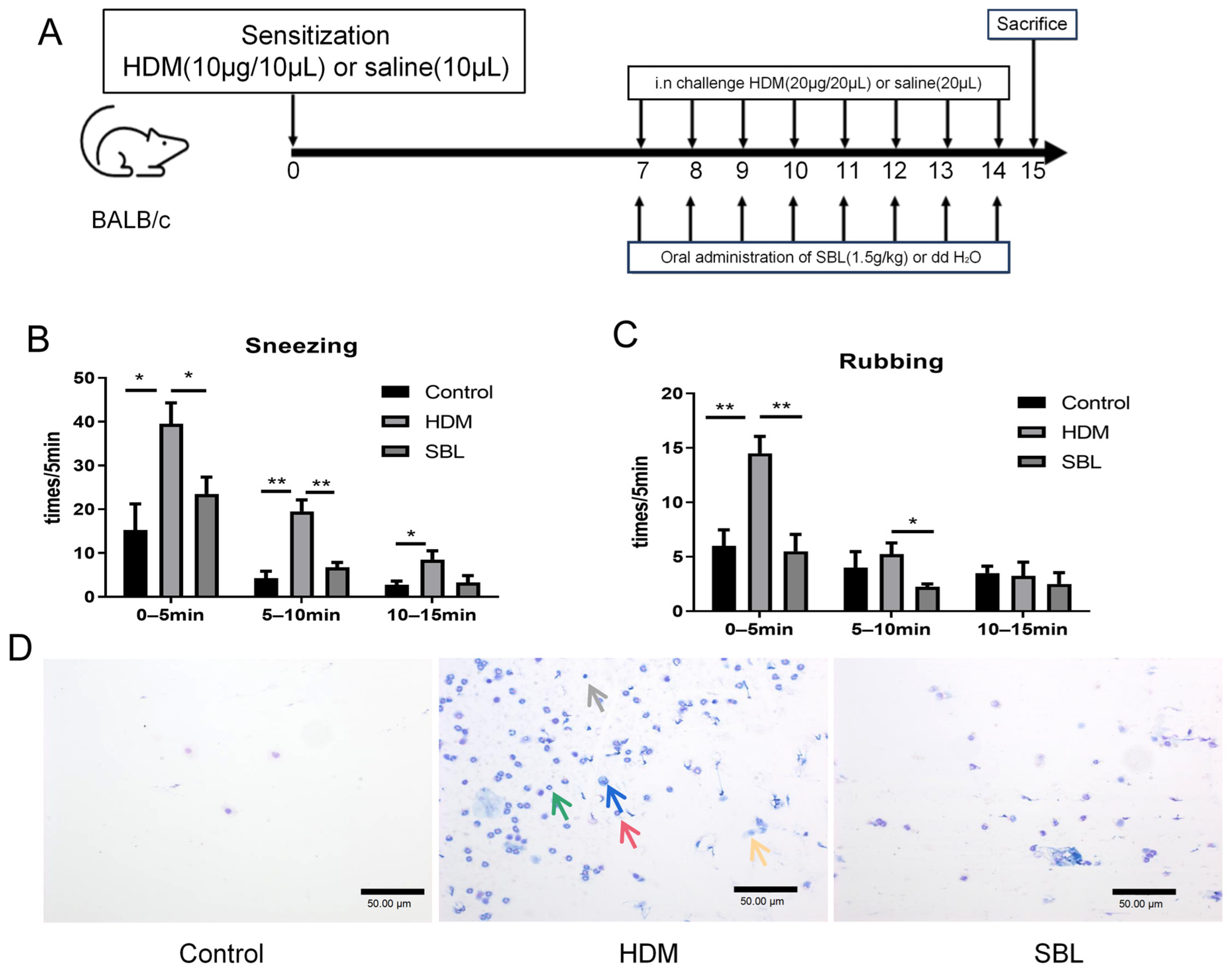
2.2. SBL Treatment Alleviated Nasal Symptoms in HDM-Induced AR Mouse Model
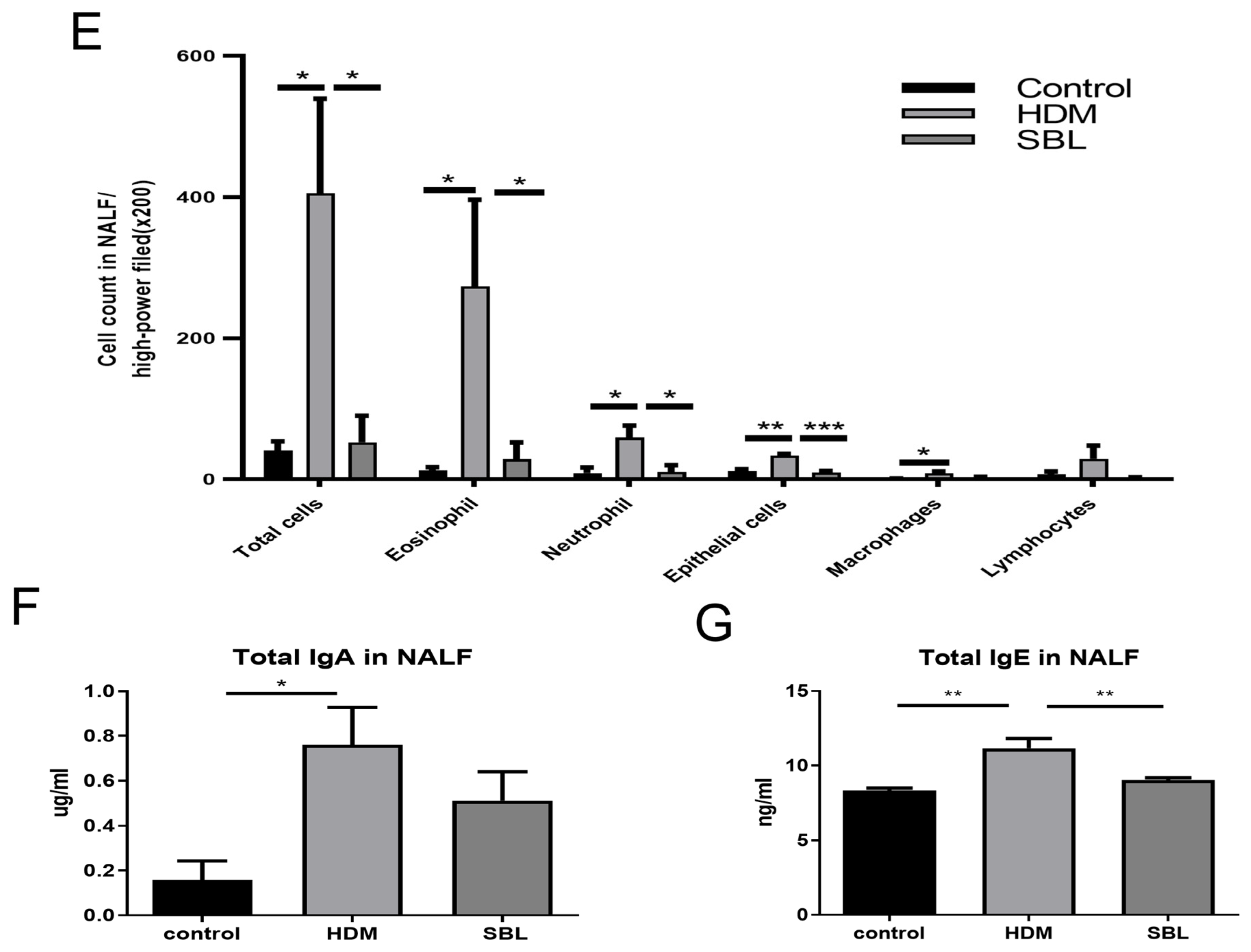
2.3. Oral Treatment of SBL Decreases the Infiltration of Different Inflammatory Cells, Nasal Epithelial Cells, and Immunoglobulin in the Nasal Lavage Fluid (NALF)
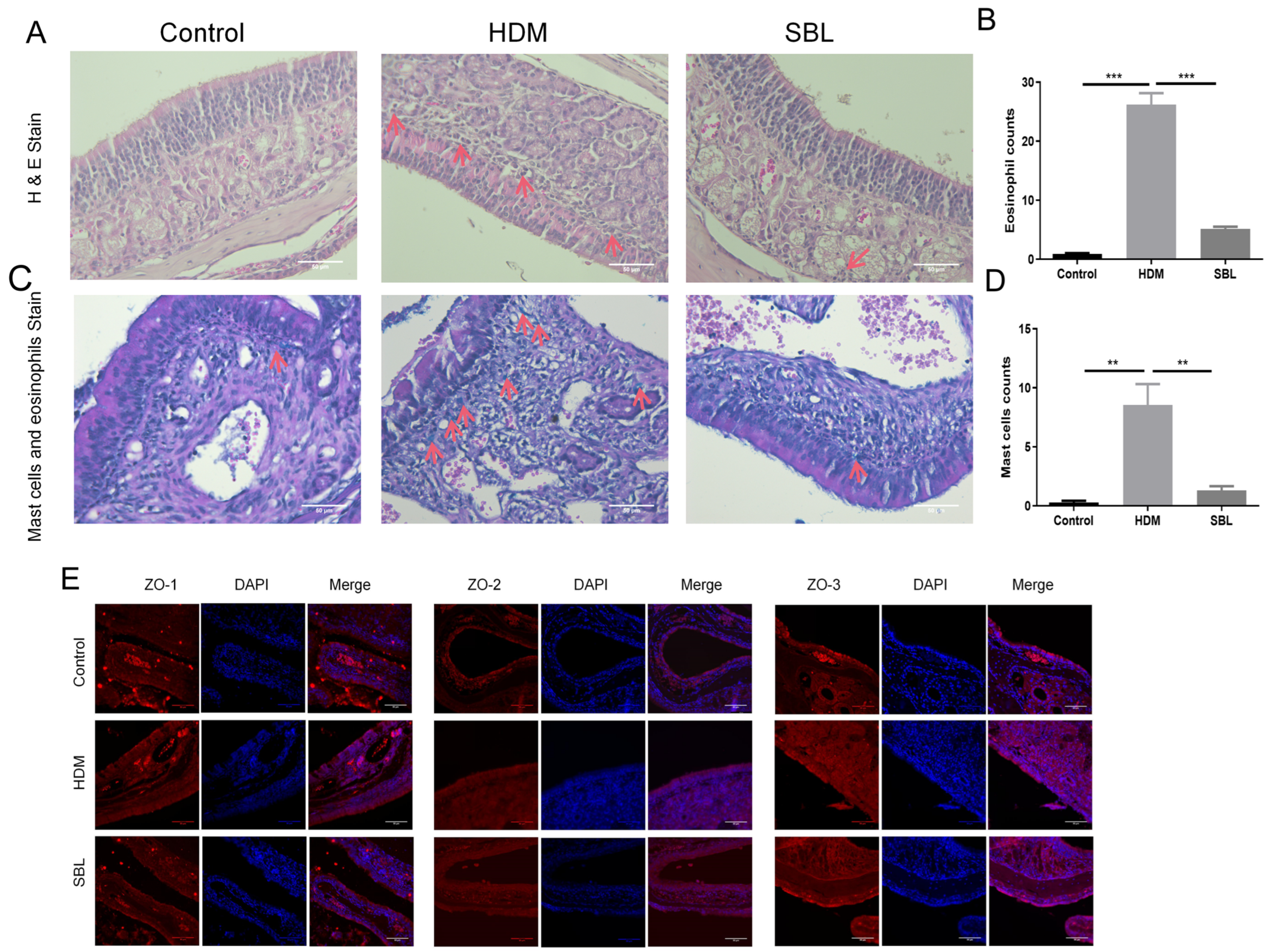
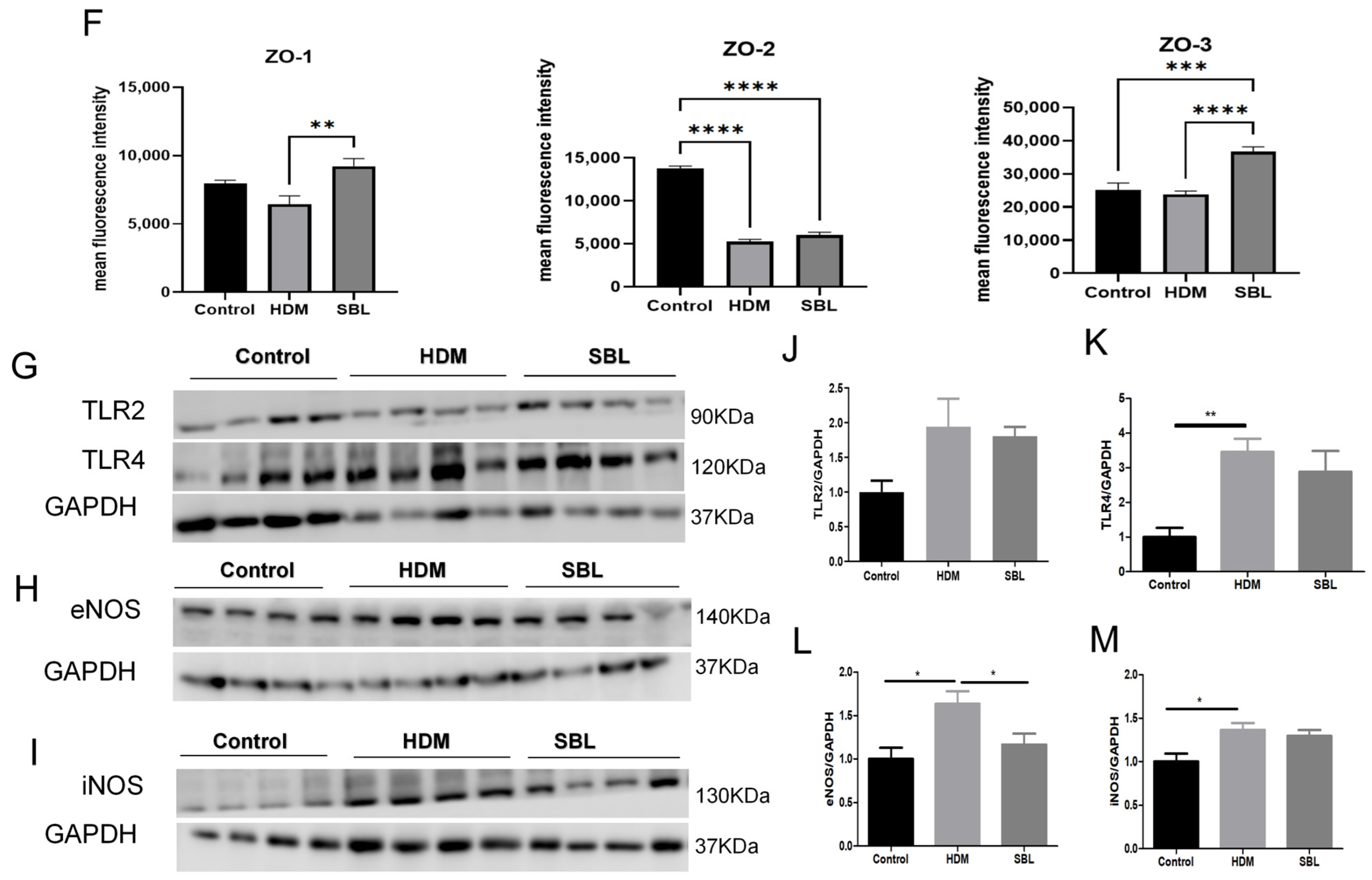
2.4. SBL Reduced Nasal Mucosa Histopathological Injuries and Nitric Oxide Protein in the HDM-Induced AR Mouse Model
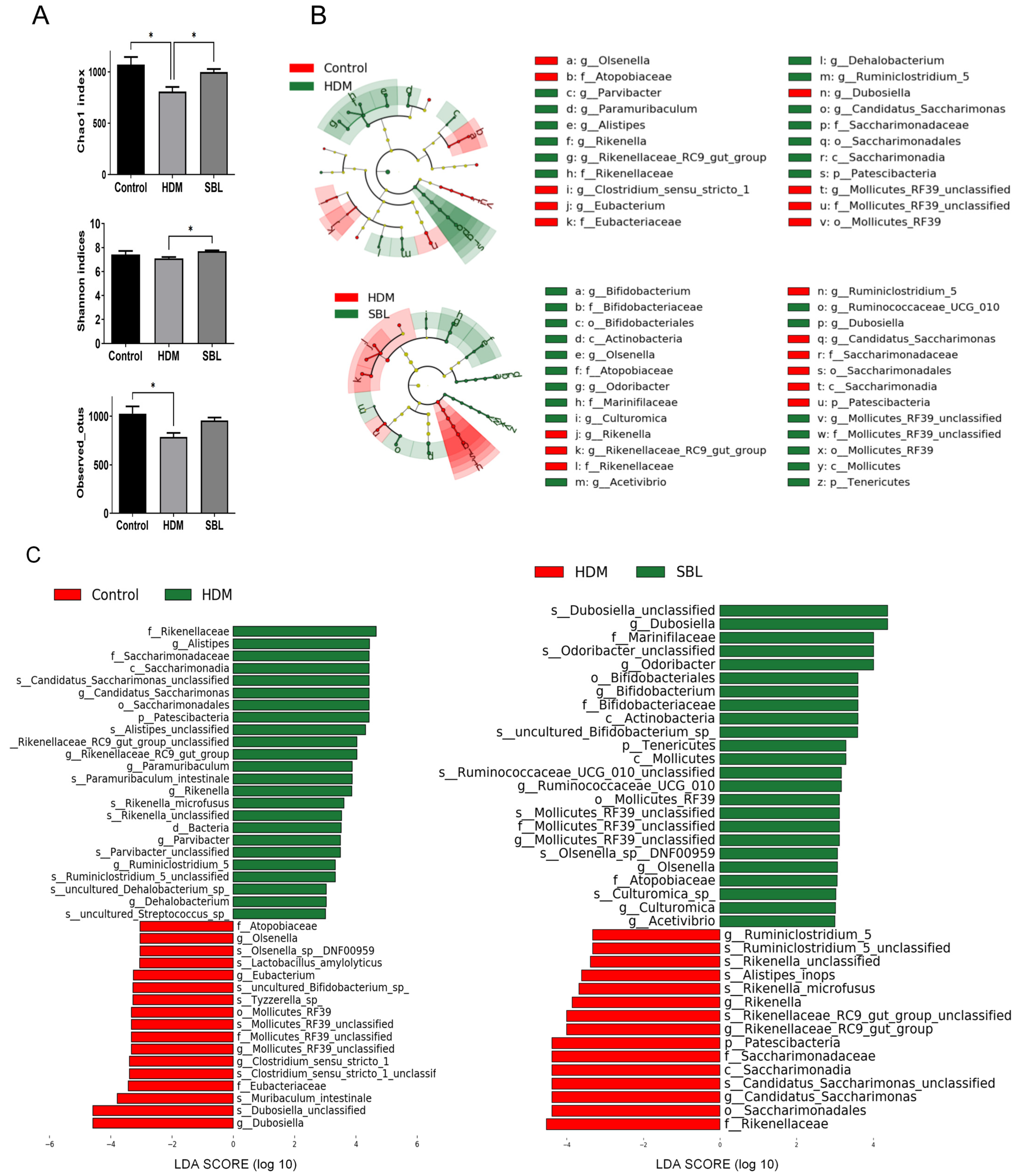
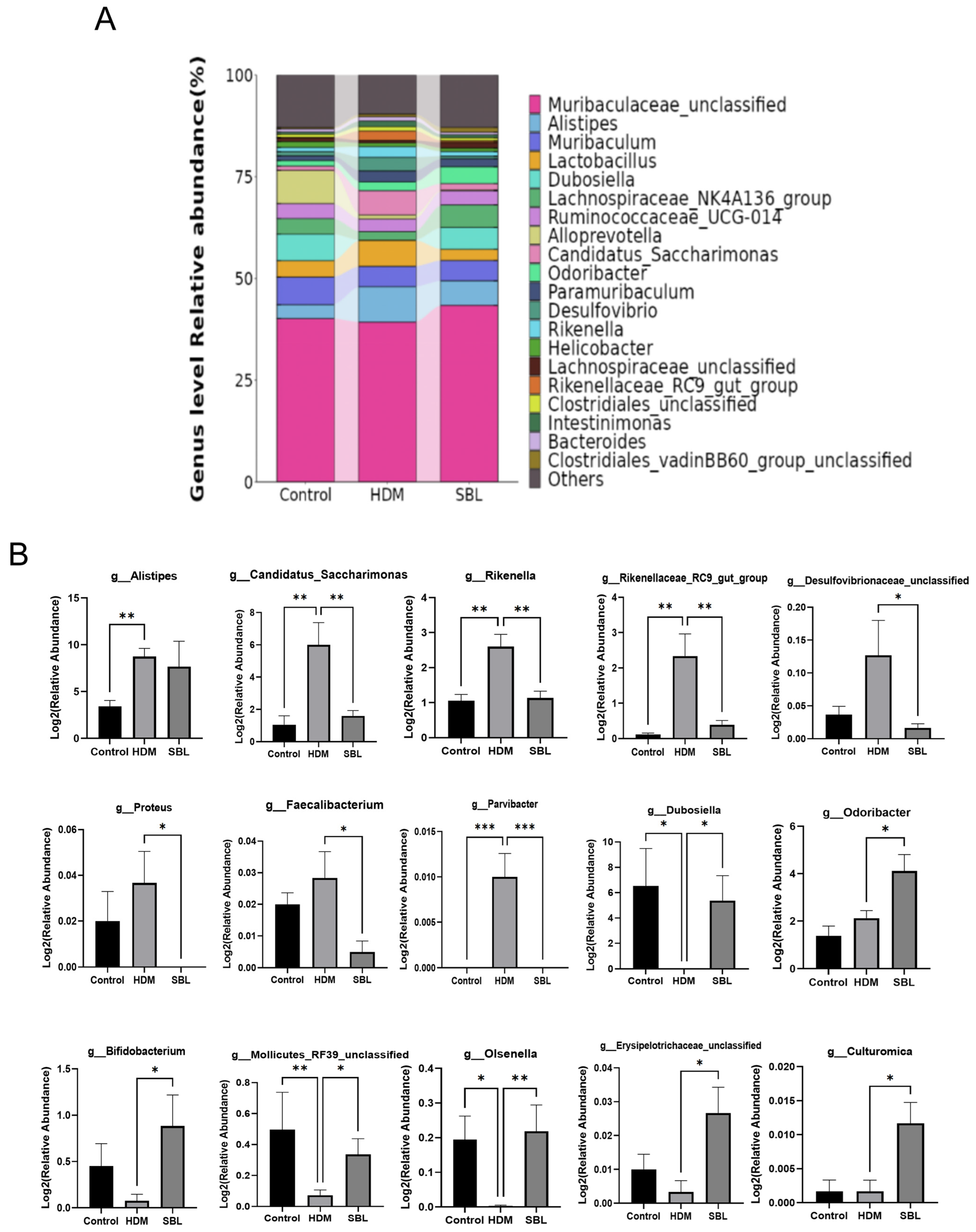
2.5. Effect of SBL on Gut Microbiota Profiles in HDM-Induced AR Mice
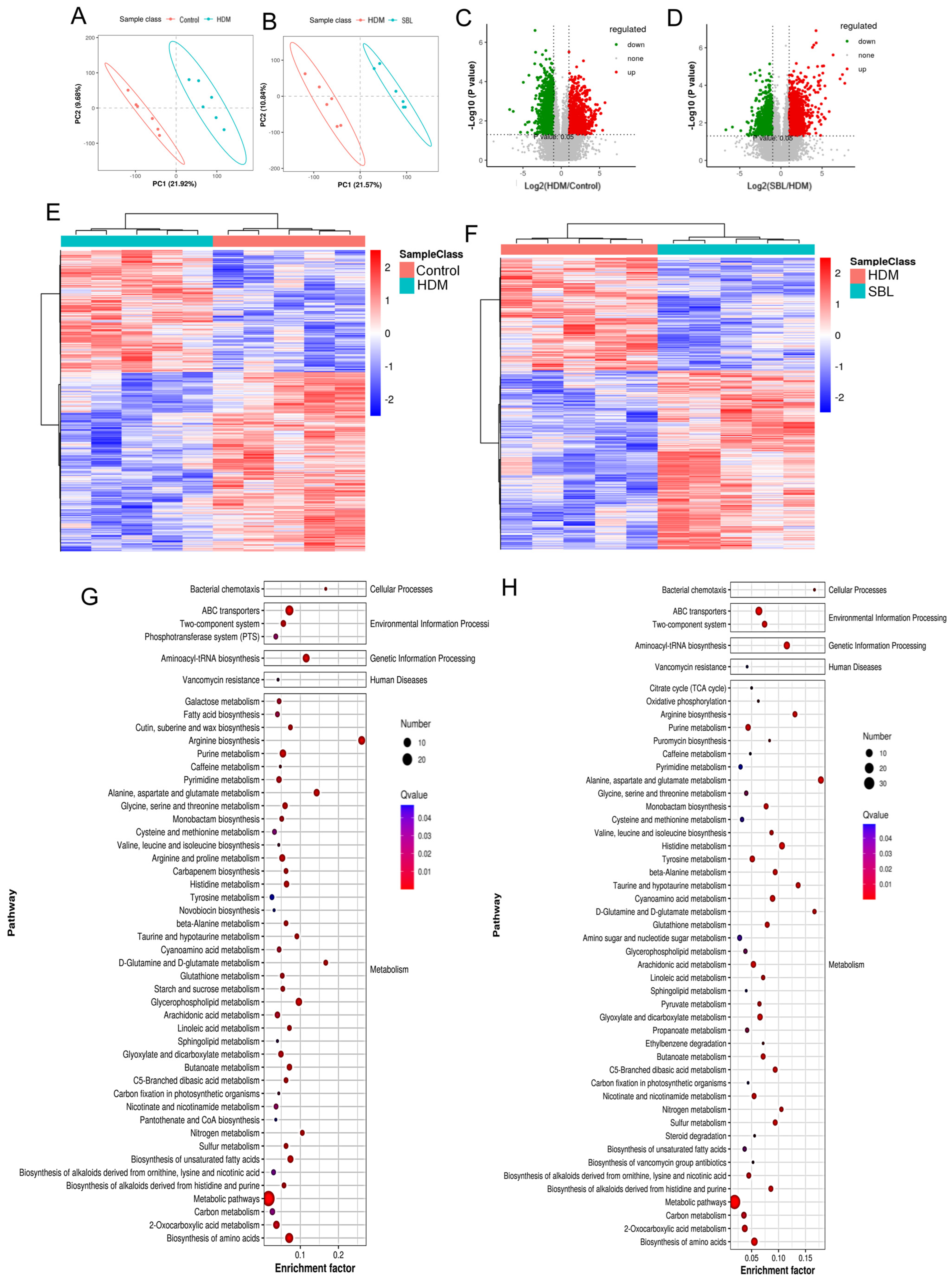
2.6. Oral Treatment of SBL Altered the Gut Metabolites in HDM-Induced AR Mice
2.7. Network Pharmacology Analysis
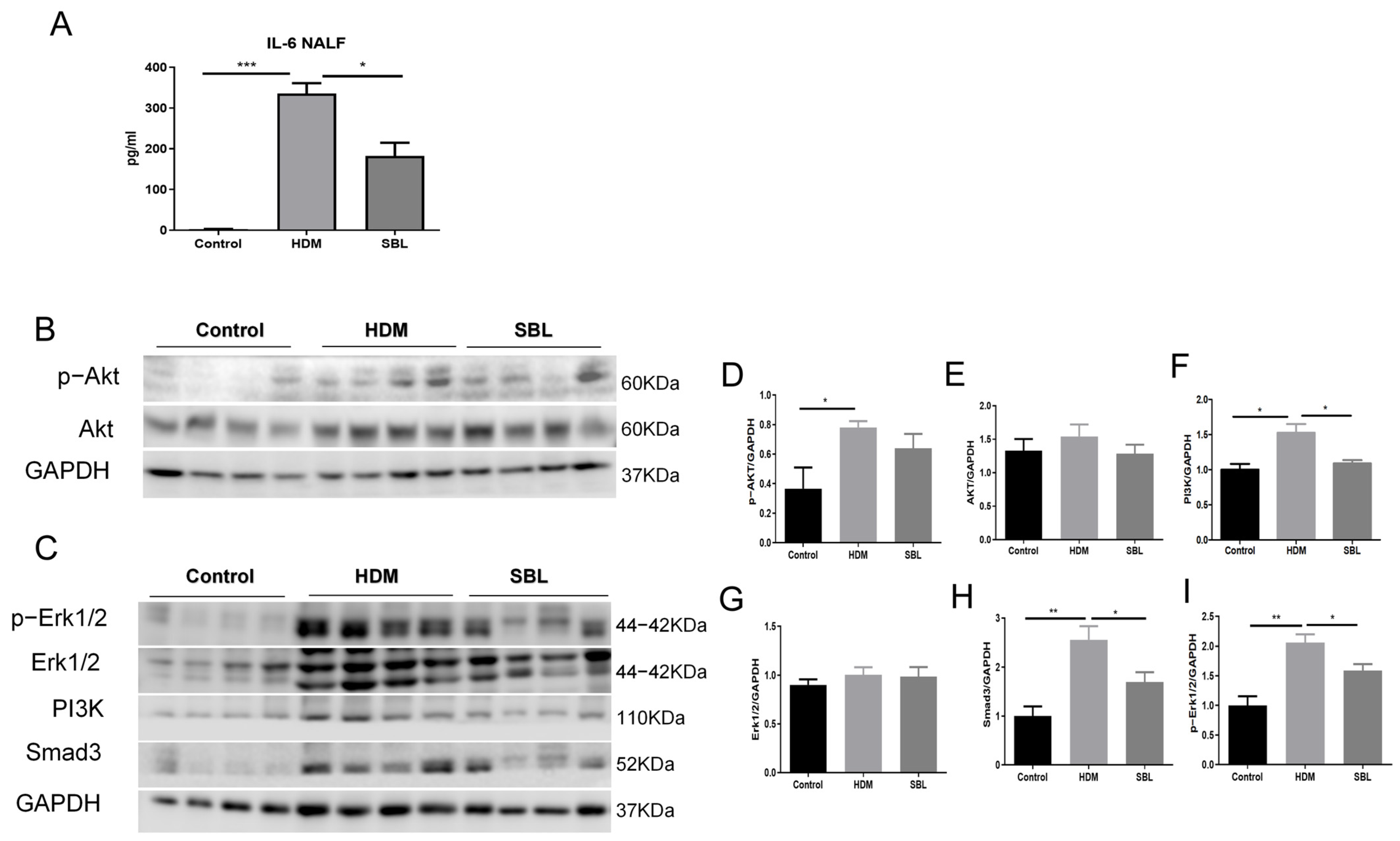
2.8. Alleviation of IL-6 Level in NALF and Suppression of the Erk and PI3K/AKT Signaling in Nasal Mucosa Upon SBL Treatment
3. Discussion
4. Materials and Methods
4.1. Preparations of Shi-Bi-Li (SBL)
4.2. Animal Experiment
4.3. Evaluation of Nasal Allergic Symptoms
4.4. Collection of Nasal Lavage Fluid (NALF) and Quantification of Immunoglobulin and Cytokines
4.5. Determination of Nasal Epithelial Cells and Immune Cells in NALF
4.6. Histological Examination and Immunofluorescence (IF) Study
4.7. Western Blot
4.8. 16S rRNA Sequence
4.9. Metabolism Studies
4.10. Network Pharmacology
4.10.1. Screening the Potential Active Ingredients and Prediction of AR-Related Targets
4.10.2. SBL-AR-Related Target Screening and PPI Network Construction
4.10.3. Molecular Docking
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Yan, Z.; Liu, L.; Yuan, J.; Jiao, L.; Zhou, M.; Liu, J.; Wen, X.; Liu, S.; Hao, P.; Liu, J.; et al. Yiqi Jiemin Decoction Alleviates Allergic Rhinitis in a Guinea Pig Model. Aging 2021, 13, 18423–18441. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Wang, C.; Zhang, L. Recent developments and highlights in allergic rhinitis. Allergy 2019, 74, 2320–2328. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, L.M.; Togias, A. Clinical practice. Allergic rhinitis. N. Engl. J. Med. 2015, 372, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Nur Husna, S.M.; Tan, H.T.; Md Shukri, N.; Mohd Ashari, N.S.; Wong, K.K. Nasal Epithelial Barrier Integrity and Tight Junctions Disruption in Allergic Rhinitis: Overview and Pathogenic Insights. Front. Immunol. 2021, 12, 663626. [Google Scholar] [CrossRef] [PubMed]
- Kan, L.L.-Y.; Li, P.; Hon, S.S.-M.; Lai, A.Y.-T.; Li, A.; Wong, K.C.-Y.; Huang, D.; Wong, C.-K. Deciphering the Interplay between the Epithelial Barrier, Immune Cells, and Metabolic Mediators in Allergic Disease. Int. J. Mol. Sci. 2024, 25, 6913. [Google Scholar] [CrossRef] [PubMed]
- Georas, S.N.; Rezaee, F. Epithelial barrier function: At the front line of asthma immunology and allergic airway inflammation. J. Allergy Clin. Immunol. 2014, 134, 509–520. [Google Scholar] [CrossRef]
- Schleimer, R.P.; Kato, A.; Kern, R.; Kuperman, D.; Avila, P.C. Epithelium: At the interface of innate and adaptive immune responses. J. Allergy Clin. Immunol. 2007, 120, 1279–1284. [Google Scholar] [CrossRef] [PubMed]
- Steelant, B.; Farre, R.; Wawrzyniak, P.; Belmans, J.; Dekimpe, E.; Vanheel, H.; Van Gerven, L.; Kortekaas Krohn, I.; Bullens, D.M.A.; Ceuppens, J.L.; et al. Impaired barrier function in patients with house dust mite-induced allergic rhinitis is accompanied by decreased occludin and zonula occludens-1 expression. J. Allergy Clin. Immunol. 2016, 137, 1043–1053. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Piao, C.H.; Hyeon, E.; Jung, S.Y.; Eom, J.E.; Shin, H.S.; Song, C.H.; Chai, O.H. Gallic acid alleviates nasal inflammation via activation of Th1 and inhibition of Th2 and Th17 in a mouse model of allergic rhinitis. Int. Immunopharmacol. 2019, 70, 512–519. [Google Scholar] [CrossRef]
- Moote, W.; Kim, H.; Ellis, A.K. Allergen-Specific immunotherapy. Allergy Asthma Clin. Immunol. 2018, 14, 53. [Google Scholar] [CrossRef]
- Eng, P.A.; Borer-Reinhold, M.; Heijnen, I.A.; Gnehm, H.P. Twelve-Year follow-up after discontinuation of preseasonal grass pollen immunotherapy in childhood. Allergy 2006, 61, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Tsang, M.S.; Kan, L.L.; Hou, T.; Hon, S.S.; Chan, B.C.; Chu, I.M.; Lam, C.W.; Leung, P.C.; Wong, C.K. The Immuno-Modulatory Activities of Pentaherbs Formula on Ovalbumin-Induced Allergic Rhinitis Mice via the Activation of Th1 and Treg Cells and Inhibition of Th2 and Th17 Cells. Molecules 2021, 27, 239. [Google Scholar] [CrossRef]
- Zhao, Y.; van Hasselt Charlie, A.; Woo, J.K.; Chen, G.G.; Wong, Y.O.; Wang, L.H.; Leung, P.C. Effect of a Chinese herbal formula, Shi-Bi-Lin, on an experimental model of allergic rhinitis. Ann. Allergy Asthma Immunol. 2006, 96, 844–850. [Google Scholar] [CrossRef]
- Kong, Y.; Hao, M.; Chen, A.; Yi, T.; Yang, K.; Li, P.; Wang, Y.; Li, P.; Jia, X.; Qin, H.; et al. SymMap database and TMNP algorithm reveal Huanggui Tongqiao granules for Allergic rhinitis through IFN-mediated neuroimmuno-modulation. Pharmacol. Res. 2022, 185, 106483. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; van Hasselt, C.A.; Woo, J.K.; Chen, G.G.; Wong, Y.O.; Wang, L.H.; Leung, P.C. Effects of the Chinese herbal formula Shi-Bi-Lin on cytokine release from the human mast cell line. Ann. Allergy Asthma Immunol. 2005, 95, 79–85. [Google Scholar] [CrossRef]
- Zhao, Y.; Woo, K.S.; Ma, K.H.; Van Hansselt, C.A.; Wong, K.C.; Cheng, K.F.; Lam, C.W.; Leung, P.C. Treatment of perennial allergic rhinitis using Shi-Bi-Lin, a Chinese herbal formula. J. Ethnopharmacol. 2009, 122, 100–105. [Google Scholar] [CrossRef]
- Albenberg, L.G.; Wu, G.D. Diet and the Intestinal Microbiome: Associations, Functions, and Implications for Health and Disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, Z.; Lin, X.; Yao, S.; Xian, M.; Ning, X.; Fu, W.; Jiang, M.; Li, N.; Xiao, X.; et al. Rural environment reduces allergic inflammation by modulating the gut microbiota. Gut Microbes 2022, 14, 2125733. [Google Scholar] [CrossRef]
- Duan, F.P.; Li, Y.S.; Hu, T.Y.; Pan, X.Q.; Ma, F.; Feng, Y.; Qiu, S.Q.; Zheng, Y.Q. Dendrobium nobile protects against ovalbumin-induced allergic rhinitis by regulating intestinal flora and suppressing lung inflammation. Chin. J. Nat. Med. 2022, 20, 443–457. [Google Scholar] [CrossRef]
- Zhao, N.; Wang, Y.; Ma, Y.; Liang, X.; Zhang, X.; Gao, Y.; Dong, Y.; Bai, D.; Hu, J. Jia-Wei-Si-Miao-Yong-An decoction modulates intestinal flora and metabolites in acute coronary syndrome model. Front. Cardiovasc. Med. 2022, 9, 1038273. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Ao, H.; Peng, C.; Yan, D. Gut microbiota, a new frontier to understand traditional Chinese medicines. Pharmacol. Res. 2019, 142, 176–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, L.; Liu, D.; Chen, H.; Tang, D.D.; Zhao, Y.Y. Metabolomics highlights pharmacological bioactivity and biochemical mechanism of traditional Chinese medicine. Chem. Biol. Interact. 2017, 273, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.-Y.; Nakajima-Adach, H.; Kaminogawa, S.; Kumagai, S.; Sugita-Konishi, Y. A Novel Effect of Nivalenol (NIV) on the Immune Response. In Animal Cell Technology: Challenges for the 21st Century; Springer: Dordrecht, The Netherlands, 2002; pp. 155–160. [Google Scholar] [CrossRef]
- Tu, Y.; Li, X.; Fu, Y.; Chen, Y.; Fang, H.; Li, Y.; Gu, Y.; Zhang, J. Isocorydine Ameliorates IL-6 Expression in Bone Marrow-Derived Macrophages and Acute Lung Injury Induced by Lipopolysaccharide. Int. J. Mol. Sci. 2023, 24, 4629. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wang, N.; Hua, L.; Deng, F.; Liu, D.; Zhou, J.; Yuan, Y.; Ouyang, F.; Chen, X.; Long, S.; et al. The Anti-Sepsis Effect of Isocorydine Screened from Guizhou Ethnic Medicine is Closely Related to Upregulation of Vitamin D Receptor Expression and Inhibition of NFkappaB p65 Translocation into the Nucleus. J. Inflamm. Res. 2022, 15, 5649–5664. [Google Scholar] [CrossRef] [PubMed]
- Shifrin, H.; Nadler-Milbauer, M.; Shoham, S.; Weinstock, M. Rivastigmine alleviates experimentally induced colitis in mice and rats by acting at central and peripheral sites to modulate immune responses. PLoS ONE 2013, 8, e57668. [Google Scholar] [CrossRef] [PubMed]
- Aboulaghras, S.; Sahib, N.; Bakrim, S.; Benali, T.; Charfi, S.; Guaouguaou, F.E.; Omari, N.E.; Gallo, M.; Montesano, D.; Zengin, G.; et al. Health Benefits and Pharmacological Aspects of Chrysoeriol. Pharmaceuticals 2022, 15, 973. [Google Scholar] [CrossRef]
- Cheng, Q.; Wang, Z.; Ma, R.; Chen, Y.; Yan, Y.; Miao, S.; Jiao, J.; Cheng, X.; Kong, L.; Ye, D. Lipoxin A4 protects against lipopolysaccharide-induced sepsis by promoting innate response activator B cells generation. Int. Immunopharmacol. 2016, 39, 229–235. [Google Scholar] [CrossRef]
- Yoon, H.S.; Park, C.M. Chrysoeriol ameliorates COX-2 expression through NF-kappaB, AP-1 and MAPK regulation via the TLR4/MyD88 signaling pathway in LPS-stimulated murine macrophages. Exp. Ther. Med. 2021, 22, 718. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, X.; Liu, B.; Xia, Y.; Xin, Z.; Deng, B.; He, L.; Deng, J.; Ren, W. Aspartate Metabolism Facilitates IL-1beta Production in Inflammatory Macrophages. Front. Immunol. 2021, 12, 753092. [Google Scholar]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Husain, S.; Zahedi, F.D.; Mohamad, S.; Abdullah, B. House Dust Mite-Induced Allergic Rhinitis: Is Prevention an Option? Curr. Treat. Options Allergy 2019, 6, 338–349. [Google Scholar] [CrossRef]
- Jung, M.A.; Song, H.K.; Jo, K.; Lee, A.; Hwang, Y.H.; Ji, K.Y.; Jung, D.H.; Cai, M.; Lee, J.Y.; Pyun, B.J.; et al. Gleditsia sinensis Lam. aqueous extract attenuates nasal inflammation in allergic rhinitis by inhibiting MUC5AC production through suppression of the STAT3/STAT6 pathway. Biomed. Pharmacother. 2023, 161, 114482. [Google Scholar] [CrossRef] [PubMed]
- Sanchez Montalvo, A.; Gohy, S.; Rombaux, P.; Pilette, C.; Hox, V. The Role of IgA in Chronic Upper Airway Disease: Friend or Foe? Front. Allergy 2022, 3, 852546. [Google Scholar] [CrossRef] [PubMed]
- Zoabi, Y.; Levi-Schaffer, F.; Eliashar, R. Allergic Rhinitis: Pathophysiology and Treatment Focusing on Mast Cells. Biomedicines 2022, 10, 2486. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, A.; Bolli, E.; Tarone, L.; Cavallo, F.; Conti, L. Toll-Like Receptor 2 at the Crossroad between Cancer Cells, the Immune System, and the Microbiota. Int. J. Mol. Sci. 2020, 21, 9418. [Google Scholar] [CrossRef]
- Yuan, S.; Li, Y.; Li, J.; Xue, J.-C.; Wang, Q.; Hou, X.-T.; Meng, H.; Nan, J.-X.; Zhang, Q.-G. Traditional Chinese Medicine and Natural Products: Potential Approaches for Inflammatory Bowel Disease. Front. Pharmacol. 2022, 13, 892790. [Google Scholar] [CrossRef]
- Iglesias, D.E.; Cremonini, E.; Hester, S.N.; Wood, S.M.; Bartlett, M.; Fraga, C.G.; Oteiza, P.I. Cyanidin and delphinidin restore colon physiology in high fat diet-fed mice: Involvement of TLR-4 and redox-regulated signaling. Free. Radic. Biol. Med. 2022, 188, 71–82. [Google Scholar] [CrossRef]
- Li, C.J.; Elsasser, T.H.; Kahl, S. AKT/eNOS signaling module functions as a potential feedback loop in the growth hormone signaling pathway. J. Mol. Signal 2009, 4, 1–13. [Google Scholar] [CrossRef][Green Version]
- Kawai, T.; Akira, S. TLR signaling. Semin Immunol. 2007, 19, 24–32. [Google Scholar] [CrossRef]
- Pantazi, A.C.; Mihai, C.M.; Balasa, A.L.; Chisnoiu, T.; Lupu, A.; Frecus, C.E.; Mihai, L.; Ungureanu, A.; Kassim, M.A.K.; Andrusca, A.; et al. Relationship between Gut Microbiota and Allergies in Children: A Literature Review. Nutrients 2023, 15, 2529. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.L.; Lu, C.C.; Lai, W.F.; Wu, T.S.; Lu, J.J.; Chen, Y.M.; Tzeng, C.M.; Liu, H.T.; Wei, H.; Lai, H.C. Role of gut microbiota in identification of novel TCM-derived active metabolites. Protein Cell 2021, 12, 394–410. [Google Scholar] [CrossRef]
- Mao, D.; Tao, B.; Sheng, S.; Jin, H.; Chen, W.; Gao, H.; Deng, J.; Li, Z.; Chen, F.; Chan, S.; et al. Causal Effects of Gut Microbiota on Age-Related Macular Degeneration: A Mendelian Randomization Study. Investig. Ophthalmol. Vis. Sci. 2023, 64, 32. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yin, Q.; Xie, Q.; Jiang, C.; Zhou, L.; Liu, J.; Li, B.; Jiang, S. 2′-Fucosyllactose Promotes the Production of Short-Chain Fatty Acids and Improves Immune Function in Human-Microbiota—Associated Mice by Regulating Gut Microbiota. J. Agric. Food Chem. 2022, 70, 13615–13625. [Google Scholar] [CrossRef]
- Gavzya, S.J.; Kensiskia, A.; Lee, Z.L.; Mongodin, E.F.; Ma, B.; Bromberg, J.S.; Mongodinc, E.F.; Bing, M.; Bromberg, J.S. Bifidobacterium mechanisms of immune modulation and tolerance. Gut Microbes 2023, 15, 2291164. [Google Scholar] [CrossRef]
- Liu, T.H.; Wang, J.; Zhang, C.Y.; Zhao, L.; Sheng, Y.Y.; Tao, G.S.; Xue, Y.Z. Gut microbial characteristical comparison reveals potential anti-aging function of Dubosiella newyorkensis in mice. Front. Endocrinol. 2023, 14, 1133167. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, Y.; Lai, Z.; Huang, J.; Li, C.; Zhang, Y.; Gong, X.; Deng, J.; Ye, X.; Li, X. An integrated network pharmacology and transcriptomic method to explore the mechanism of the total Rhizoma Coptidis alkaloids in improving diabetic nephropathy. J. Ethnopharmacol. 2021, 270, 113806. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Oh, S.; Shin, H.W.; Lee, Y.; Lee, H.; Seok, S.H. Matrix Stiffening Enhances DNCB-Induced IL-6 Secretion in Keratinocytes Through Activation of ERK and PI3K/Akt Pathway. Front. Immunol. 2021, 12, 759992. [Google Scholar] [CrossRef]
- Li, S.; Tian, J.; Zhang, H.; Zhou, S.; Wang, X.; Zhang, L.; Yang, J.; Zhang, Z.; Ji, Z. Down-regulating IL-6/GP130 targets improved the anti-tumor effects of 5-fluorouracil in colon cancer. Apoptosis 2018, 23, 356–374. [Google Scholar] [CrossRef]
- Esnault, S.; Hosravi, M.; Kelly, E.A.; Liu, L.Y.; Bochkov, Y.A.; Tattersall, M.C.; Jarjour, N.N. Increased IL-6 and Potential IL-6 trans-signalling in the airways after an allergen challenge. Clin. Exp. Allergy 2021, 51, 564–573. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Hon, S.S.-M.; Tsang, M.S.-M.; Kan, L.L.-Y.; Lai, A.Y.-T.; Chan, B.C.-L.; Leung, P.-C.; Wong, C.-K. Integrating 16S rRNA Sequencing, Microflora Metabolism, and Network Pharmacology to Investigate the Mechanism of SBL in Alleviating HDM-Induced Allergic Rhinitis. Int. J. Mol. Sci. 2024, 25, 8655. https://doi.org/10.3390/ijms25168655
Li P, Hon SS-M, Tsang MS-M, Kan LL-Y, Lai AY-T, Chan BC-L, Leung P-C, Wong C-K. Integrating 16S rRNA Sequencing, Microflora Metabolism, and Network Pharmacology to Investigate the Mechanism of SBL in Alleviating HDM-Induced Allergic Rhinitis. International Journal of Molecular Sciences. 2024; 25(16):8655. https://doi.org/10.3390/ijms25168655
Chicago/Turabian StyleLi, Peiting, Sharon Sze-Man Hon, Miranda Sin-Man Tsang, Lea Ling-Yu Kan, Andrea Yin-Tung Lai, Ben Chung-Lap Chan, Ping-Chung Leung, and Chun-Kwok Wong. 2024. "Integrating 16S rRNA Sequencing, Microflora Metabolism, and Network Pharmacology to Investigate the Mechanism of SBL in Alleviating HDM-Induced Allergic Rhinitis" International Journal of Molecular Sciences 25, no. 16: 8655. https://doi.org/10.3390/ijms25168655
APA StyleLi, P., Hon, S. S.-M., Tsang, M. S.-M., Kan, L. L.-Y., Lai, A. Y.-T., Chan, B. C.-L., Leung, P.-C., & Wong, C.-K. (2024). Integrating 16S rRNA Sequencing, Microflora Metabolism, and Network Pharmacology to Investigate the Mechanism of SBL in Alleviating HDM-Induced Allergic Rhinitis. International Journal of Molecular Sciences, 25(16), 8655. https://doi.org/10.3390/ijms25168655





