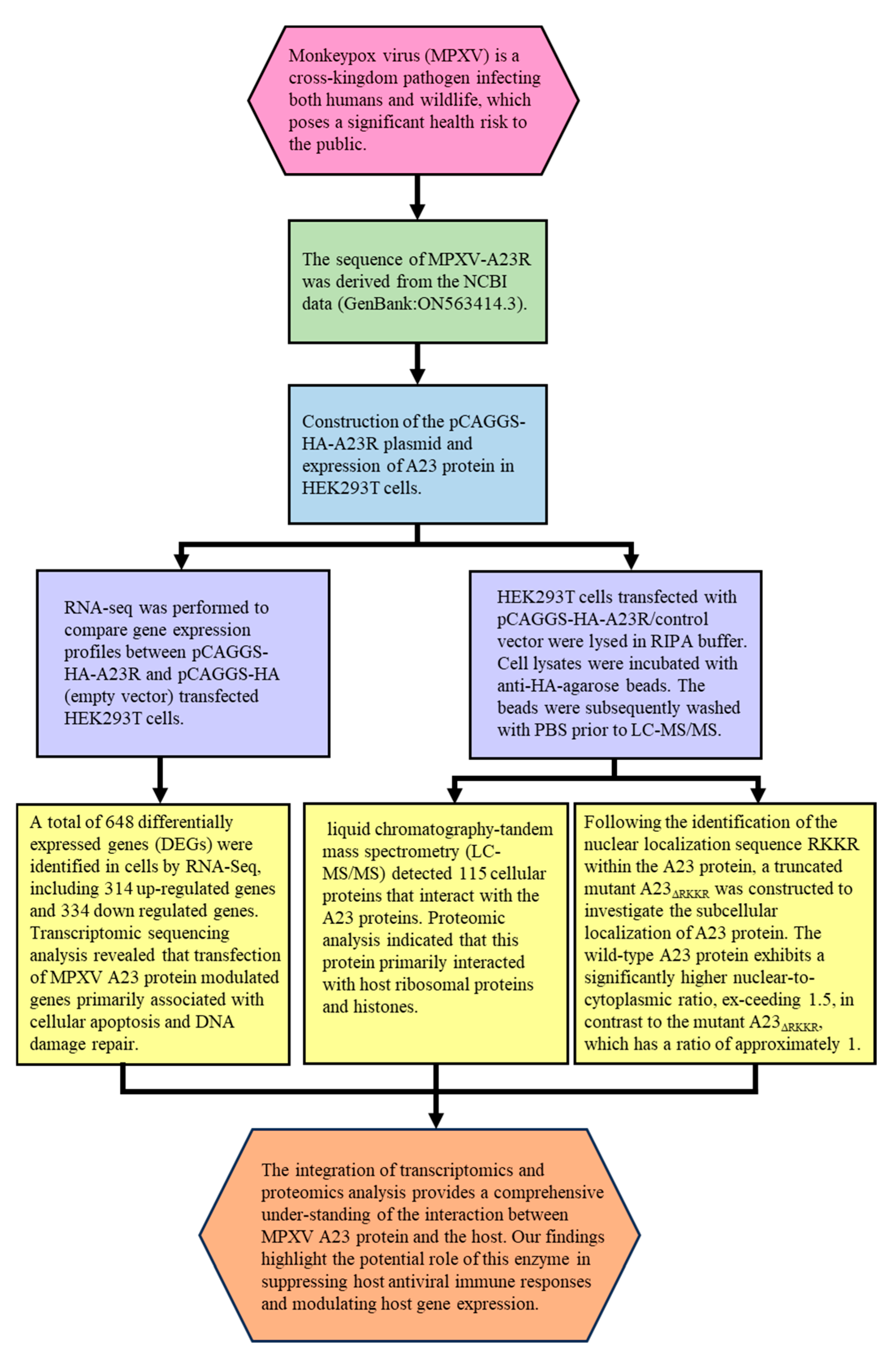
A Combined Transcriptomic and Proteomic Analysis of Monkeypox Virus A23 Protein on HEK293T Cells
Abstract
1. Introduction
2. Results
2.1. Plasmids Construction and Western Blot Analyses of A23 Protein
2.2. Transcriptome Sequencing Evaluation
2.3. Identification of DEGs
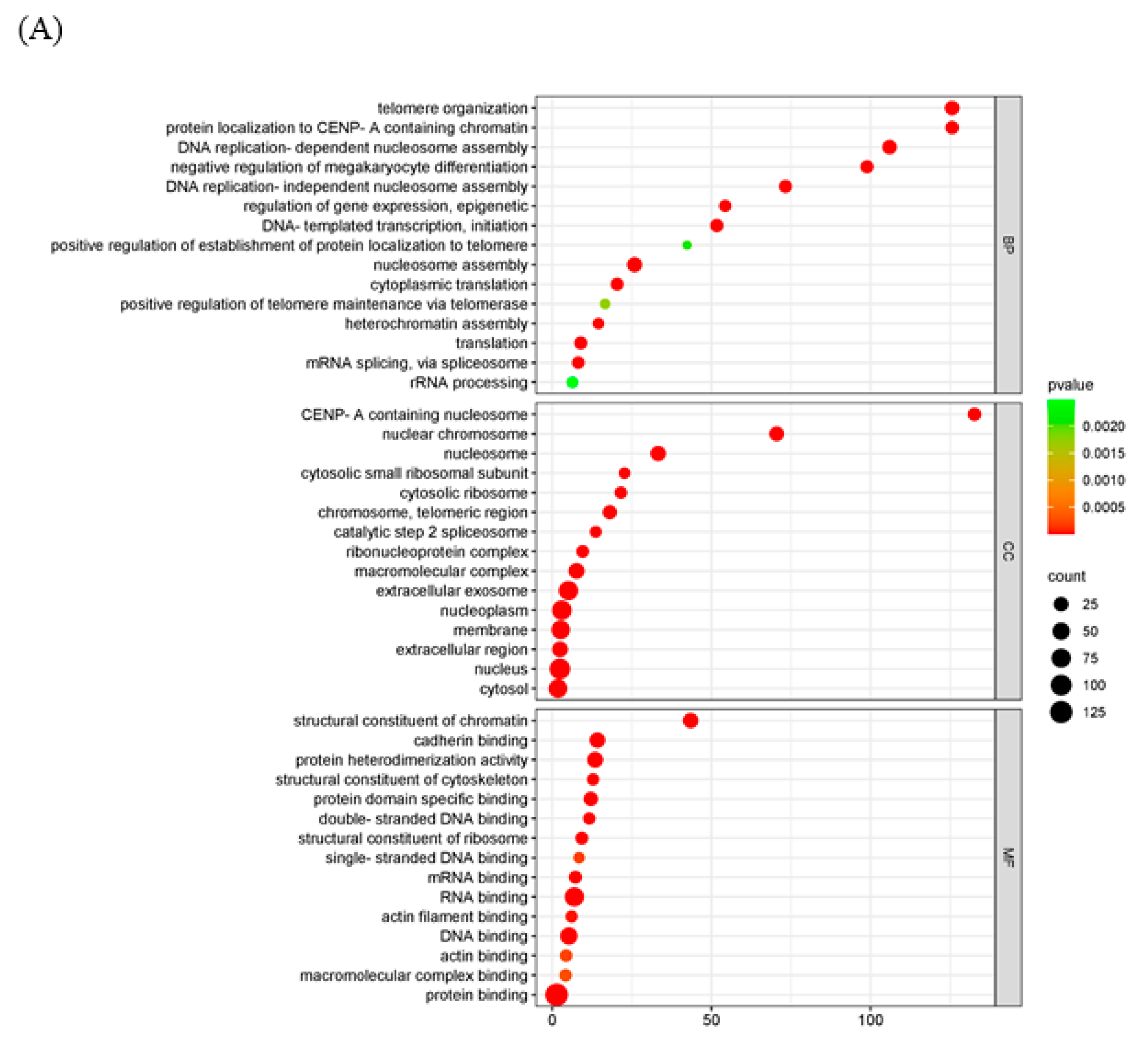
2.4. Functional and Pathway Enrichment Analysis of DEGs
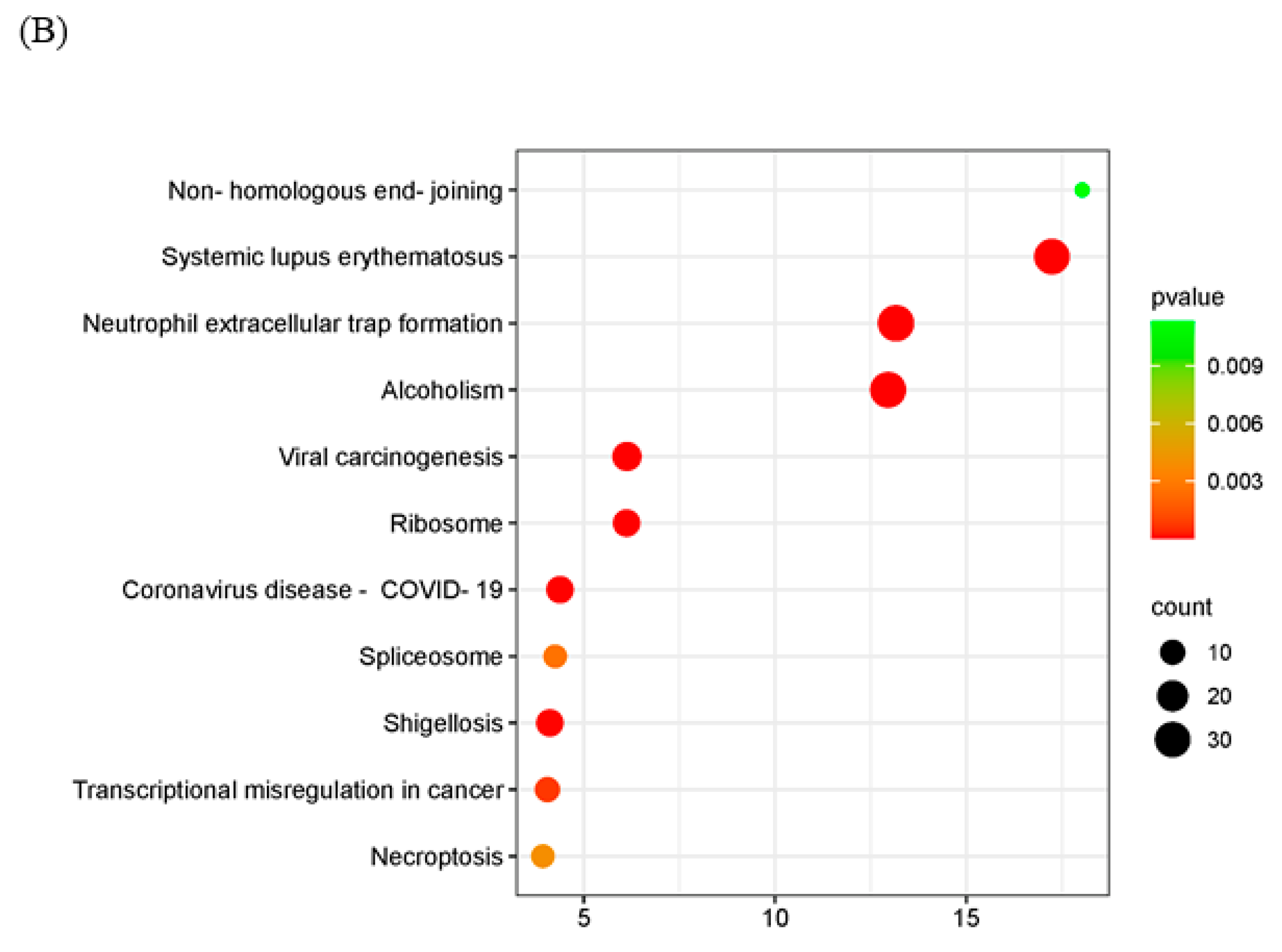
2.5. Interaction of MPXV A23R
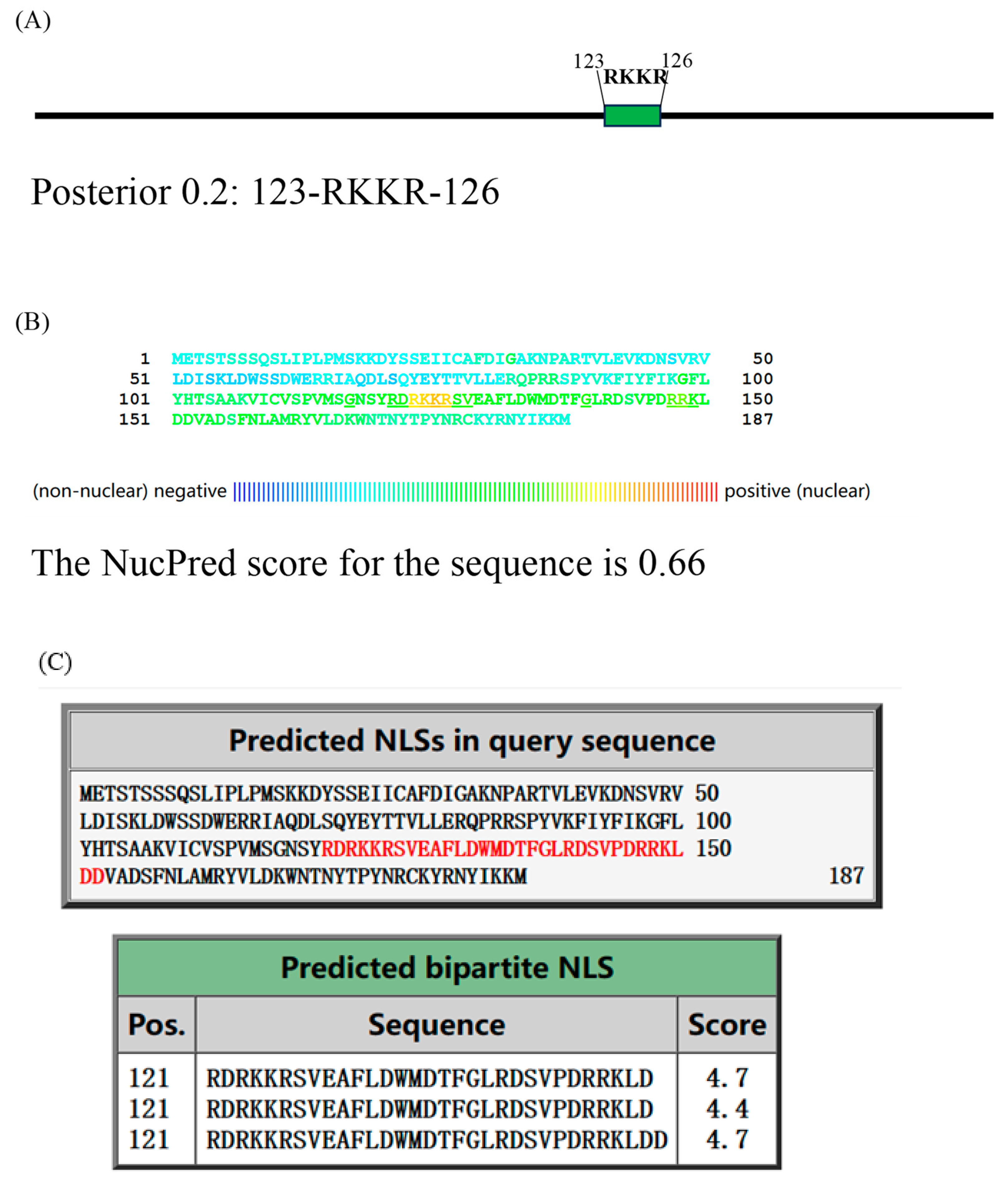
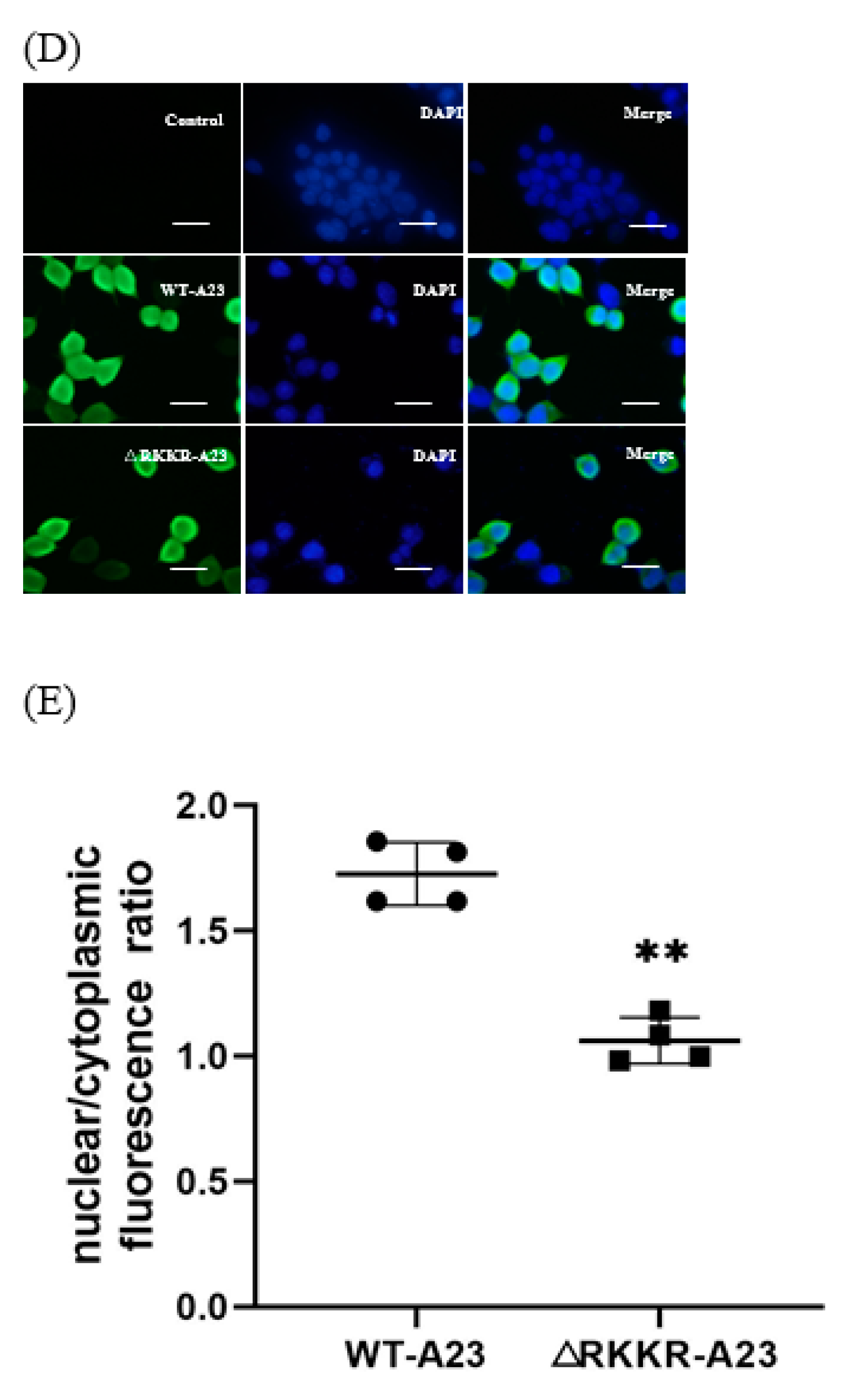
2.6. Mutation of the RKKR Impairs Nuclear Import of A23 Protein
3. Materials and Methods
3.1. Cell Culture and Transfections
3.2. Plasmid Construction and Transfection
3.3. RNA-Seq and Data Processing
3.4. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
3.5. LC-MS/MS and Data Processing
3.6. Western Blot Analyses and Immunofluorescence
3.7. Prediction of the Nuclear Location Signal
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Obeid, M.A.; Amawi, H.; Alshehri, A.; Adesokan, A. Monkeypox: Emerging virus of concern; antivirals and vaccines therapeutic options. Microb. Pathog. 2022, 173 Pt A, 105799. [Google Scholar] [CrossRef]
- Magnus, P.V.; Andersen, E.K.; Petersen, K.B.; Birch Andersen, A. A Pox-like Disease in Cynomolgus Monkeys. Acta Pathol. Microbiol. Scand. 1959, 46, 156–176. [Google Scholar] [CrossRef]
- Jezek, Z.; Marennikova, S.S.; Mutumbo, M.; Nakano, J.H.; Paluku, K.M.; Szczeniowski, M. Human monkeypox: A study of 2,510 contacts of 214 patients. J. Infect. Dis. 1986, 154, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Simpson, K.; Heymann, D.; Brown, C.S.; Edmunds, W.J.; Elsgaard, J.; Fine, P.; Hochrein, H.; Hoff, N.A.; Green, A.; Ihekweazu, C.; et al. Human monkeypox—After 40 years, an unintended consequence of smallpox eradication. Vaccine 2020, 38, 5077–5081. [Google Scholar] [CrossRef] [PubMed]
- Zardi, E.M.; Chello, C. Human Monkeypox—A Global Public Health Emergency. Int. J. Environ. Res. Public Health 2022, 19, 16781. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Das, P.; Sobti, R.C.; Kaur, T. Human monkeypox: Epidemiology, transmission, pathogenesis, immunology, diagnosis and therapeutics. Mol. Cell. Biochem. 2023, 478, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.; Kantele, A.; Koopmans, M.; Asogun, D.; Yinka-Ogunleye, A.; Ihekweazu, C.; Zumla, A. Human Monkeypox: Epidemiologic and Clinical Characteristics, Diagnosis, and Prevention. Infect. Dis. Clin. N. Am. 2019, 33, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Luna, N.; Ramirez, A.L.; Munoz, M.; Ballesteros, N.; Patino, L.H.; Castaneda, S.A.; Bonilla-Aldana, D.K.; Paniz-Mondolfi, A.; Ramirez, J.D. Phylogenomic analysis of the monkeypox virus (MPXV) 2022 outbreak: Emergence of a novel viral lineage? Travel Med. Infect. Dis. 2022, 49, 102402. [Google Scholar] [CrossRef] [PubMed]
- Lum, F.M.; Torres-Ruesta, A.; Tay, M.Z.; Lin, R.T.P.; Lye, D.C.; Renia, L.; Ng, L.F.P. Monkeypox: Disease epidemiology, host immunity and clinical interventions. Nat. Rev. Immunol. 2022, 22, 597–613. [Google Scholar] [CrossRef]
- Liem, J.; Liu, J. Stress Beyond Translation: Poxviruses and More. Viruses 2016, 8, 169. [Google Scholar] [CrossRef]
- Li, N.; Shi, K.; Rao, T.; Banerjee, S.; Aihara, H. Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase. Sci. Rep. 2020, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hwang, Y.; Perry, K.; Bushman, F.; Van Duyne, G.D. Structure and Metal Binding Properties of a Poxvirus Resolvase. J. Biol. Chem. 2016, 291, 11094–11104. [Google Scholar] [CrossRef] [PubMed]
- Sharples, G.J. The X philes: Structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 2001, 39, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Culyba, M.J.; Hwang, Y.; Hu, J.Y.; Minkah, N.; Ocwieja, K.E.; Bushman, F.D. Metal cofactors in the structure and activity of the fowlpox resolvase. J. Mol. Biol. 2010, 399, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Culyba, M.J.; Hwang, Y.; Minkah, N.; Bushman, F.D. DNA binding and cleavage by the fowlpox virus resolvase. J. Biol. Chem. 2009, 284, 1190–1201. [Google Scholar] [CrossRef] [PubMed]
- Culyba, M.J.; Minkah, N.; Hwang, Y.; Benhamou, O.M.; Bushman, F.D. DNA branch nuclease activity of vaccinia A22 resolvase. J. Biol. Chem. 2007, 282, 34644–34652. [Google Scholar] [CrossRef] [PubMed]
- Green, V.; Curtis, F.A.; Sedelnikova, S.; Rafferty, J.B.; Sharples, G.J. Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry. Mol. Microbiol. 2013, 89, 1240–1258. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garcia, A.D.; Otero, J.; Lebowitz, J.; Schuck, P.; Moss, B. Quaternary structure and cleavage specificity of a poxvirus holliday junction resolvase. J. Biol. Chem. 2006, 281, 11618–11626. [Google Scholar] [CrossRef] [PubMed]
- Grinsztejn, B.; Nguyen, B.Y.; Katlama, C.; Gatell, J.M.; Lazzarin, A.; Vittecoq, D.; Gonzalez, C.J.; Chen, J.; Harvey, C.M.; Isaacs, R.D.; et al. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: A phase II randomised controlled trial. Lancet 2007, 369, 1261–1269. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Brameier, M.; Krings, A.; MacCallum, R.M. NucPred—Predicting nuclear localization of proteins. Bioinformatics 2007, 23, 1159–1160. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Entani, T.; Takayama, S.; Tomita, M.; Yanagawa, H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 2008, 15, 940–949. [Google Scholar] [CrossRef]
- Brussow, H. Pandemic potential of poxviruses: From an ancient killer causing smallpox to the surge of monkeypox. Microb. Biotechnol. 2023, 16, 1723–1735. [Google Scholar] [CrossRef]
- Davison, A.J.; Moss, B. Structure of vaccinia virus late promoters. J. Mol. Biol. 1989, 210, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Broyles, S.S. Vaccinia virus transcription. J. Gen. Virol. 2003, 84 Pt 9, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, H.D.; West, S.C. Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 2014, 6, a023192. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.D.; Moss, B. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J. Virol. 2001, 75, 6460–6471. [Google Scholar] [CrossRef] [PubMed]
- Culyba, M.J.; Harrison, J.E.; Hwang, Y.; Bushman, F.D. DNA cleavage by the A22R resolvase of vaccinia virus. Virology 2006, 352, 466–476. [Google Scholar] [CrossRef]
- Rubins, K.H.; Hensley, L.E.; Relman, D.A.; Brown, P.O. Stunned silence: Gene expression programs in human cells infected with monkeypox or vaccinia virus. PLoS ONE 2011, 6, e15615. [Google Scholar] [CrossRef] [PubMed]
- Katsafanas, G.C.; Moss, B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2007, 2, 221–228. [Google Scholar] [CrossRef]
- Cao, S.; Dhungel, P.; Yang, Z. Going against the Tide: Selective Cellular Protein Synthesis during Virally Induced Host Shutoff. J. Virol. 2017, 91, e00071-17. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Chung, C.S.; Heine, H.G.; Chang, W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 2000, 74, 3353–3365. [Google Scholar] [CrossRef]
- Guerra, S.; Lopez-Fernandez, L.A.; Pascual-Montano, A.; Munoz, M.; Harshman, K.; Esteban, M. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 2003, 77, 6493–6506. [Google Scholar] [CrossRef]
- Pedley, S.; Cooper, R.J. The inhibition of HeLa cell RNA synthesis following infection with vaccinia virus. J. Gen. Virol. 1984, 65 Pt 10, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.P.; Roberts, B.E. Vaccinia virus induces cellular mRNA degradation. J. Virol. 1983, 47, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Parrish, S.; Moss, B. Characterization of a vaccinia virus mutant with a deletion of the D10R gene encoding a putative negative regulator of gene expression. J. Virol. 2006, 80, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Parrish, S.; Resch, W.; Moss, B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc. Natl. Acad. Sci. USA 2007, 104, 2139–2144. [Google Scholar] [CrossRef] [PubMed]
- Burgess, H.M.; Mohr, I. Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 2015, 17, 332–344. [Google Scholar] [CrossRef]
- Sivan, G.; Martin, S.E.; Myers, T.G.; Buehler, E.; Szymczyk, K.H.; Ormanoglu, P.; Moss, B. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3519–3524. [Google Scholar] [CrossRef]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Everett, H.; McFadden, G. Poxviruses and apoptosis: A time to die. Curr. Opin. Microbiol. 2002, 5, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, T.; Ray, C.A.; Pickup, D.J.; Howard, A.D.; Thornberry, N.A.; Peterson, E.P.; Salvesen, G. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J. Biol. Chem. 1994, 269, 19331–19337. [Google Scholar] [CrossRef]
- Quan, L.T.; Caputo, A.; Bleackley, R.C.; Pickup, D.J.; Salvesen, G.S. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J. Biol. Chem. 1995, 270, 10377–10379. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Ryerson, M.R.; Richards, M.M.; Kvansakul, M.; Hawkins, C.J.; Shisler, J.L. Vaccinia Virus Encodes a Novel Inhibitor of Apoptosis That Associates with the Apoptosome. J. Virol. 2017, 91, e01385-17. [Google Scholar] [CrossRef] [PubMed]
- Amara, A.; Mercer, J. Viral apoptotic mimicry. Nat. Rev. Microbiol. 2015, 13, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Alkhalil, A.; Hammamieh, R.; Hardick, J.; Ichou, M.A.; Jett, M.; Ibrahim, S. Gene expression profiling of monkeypox virus-infected cells reveals novel interfaces for host-virus interactions. Virol. J. 2010, 7, 173. [Google Scholar] [CrossRef] [PubMed]


| Sample | Clean Reads | GC pct | Q20% | Q30% | Error Rate | Total Map | Unique Map | Multi Map |
|---|---|---|---|---|---|---|---|---|
| A23R | 6.67G | 48.55% | 95.93% | 89.62% | 0.03% | 94.55% | 92.07% | 2.48% |
| Control | 6.47G | 49.7% | 96.28% | 90.41% | 0.03% | 94.97% | 92.49% | 2.48% |
| Gene_id | A23R | Control | log2FoldChange | p Value | Gene_Name |
|---|---|---|---|---|---|
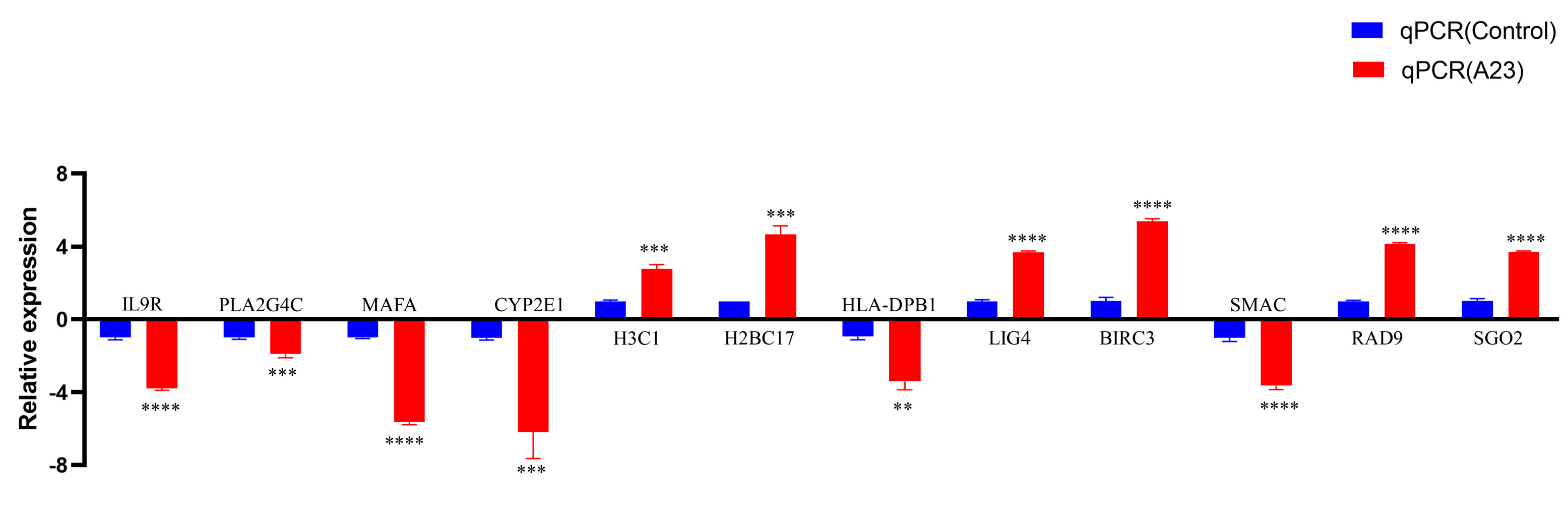
| ENSG00000124334 | 1.022537168 | 8.815369231 | −2.964356163 | 0.034866602 | IL9R |
| ENSG00000105499 | 3.06400515 | 13.713761 | −2.118526375 | 0.030348148 | PLA2G4C |
| ENSG00000182759 | 41.8522638 | 98.94567829 | −1.238935084 | 0.013213222 | MAFA |
| ENSG00000130649 | 6.126221523 | 21.5511823 | −1.794423926 | 0.019137366 | CYP2E1 |
| ENSG00000275714 | 9.188448457 | 1.39 × 10−17 | 6.219344696 | 0.007243414 | H3C1 |
| ENSG00000274641 | 11.22993507 | 1.957591471 | 2.445440274 | 0.041032031 | H2BC17 |
| ENSG00000223865 | 34.7070525 | 81.31149061 | −1.22536275 | 0.018875718 | HLA-DPB1 |
| ENSG00000174405 | 206.192132 | 94.04729284 | 1.131451016 | 0.012902837 | LIG4 |
| ENSG00000023445 | 20.4166311 | 5.876330133 | 1.774662417 | 0.033910222 | BIRC3 |
| ENSG00000284934 | 2.043272661 | 10.77472654 | −2.331102932 | 0.041032031 | SMAC |
| ENSG00000151164 | 24.49960836 | 2.937280525 | 3.00650683 | 0.000829981 | RAD9B |
| ENSG00000163535 | 702.2739732 | 337.9868831 | 1.054779239 | 0.013382118 | SGO2 |
| Accession | Protein_Name | Score Sequest HT | Abundance: A23R | Abundance: Control | Ratio of Abundance (A23R/Control) |
|---|---|---|---|---|---|
| P04908 | H2AC4 | 212.66 | 1.3 × 108 | ||
| P0C0S5 | H2AZ1 | 114.72 | 26,609,078 | ||
| P50914 | RPL14 | 9.07 | 1,190,588 | ||
| P62244 | RPS15A | 8.62 | 12,072,808 | ||
| P47914 | RPL29 | 6.38 | 3,892,084 | ||
| P62854 | RPS26 | 4.09 | 4,662,978 | ||
| P18621 | RPL17 | 1.65 | 5,741,302 | ||
| P62249 | RPS16 | 7.63 | 5,723,261 | 30,704 | 186.4011 |
| P42766 | RPL35 | 10.51 | 13,601,984 | 87,385.85 | 155.6543 |
| P15880 | RPS2 | 8.85 | 3,162,923 | 51,026.81 | 61.98551 |
| P83731 | RPL24 | 3.3 | 3,176,778 | 51,312.03 | 61.91098 |
| P62906 | RPL10A | 7.78 | 6,333,896 | 596,794.5 | 10.6132 |
| P68431 | H3C1 | 84.69 | 5.46 × 108 | 51,672,486 | 10.56361 |
| Q6FI13 | H2AC18 | 190.38 | 1.42 × 109 | 1.53 × 108 | 9.29268 |
| P16104 | H2AX | 197.19 | 16,754,252 | 2,945,901 | 5.687309 |
| P62805 | H4C1 | 141.17 | 1.83 × 109 | 3.45 × 108 | 5.314483 |
| P62280 | RPS11 | 21.92 | 22,282,617 | 5,319,779 | 4.188636 |
| P62857 | RPS28 | 17.31 | 11,419,641 | 2,801,809 | 4.07581 |
| Primer | Sequence (5′→3′) | Usage |
|---|---|---|
| A23R-F | 5′-CCGGAATTCATGGAACCAGCCACCAGC-3′ | pCAGGS-HA-A23R |
| A23R-R | 5′-CCGCTCGAGCATTTTGATATACGATATTACAAC-3′ | |
| A23R△RKKR-F | 5′-GCTATCGTGATAGCGTTGAAGCATTTCTGGATT-3′ | A23R△RKKR |
| A23R△RKKR-R | 5′-CAACGCTATCACGATAGCTATTACCGCTCATCA-3′ | |
| IL9R-F | 5′-CGTGCCCTCTCCAGCGATGTTCT-3′ | qRT-PCR |
| IL9R-R | 5′-GACGCGCTGGGCCACAAGTG-3′ | |
| PLA2G4C-F | 5′-CCTTGAGTTCACCTTGGCTGTCCTAA-3′ | qRT-PCR |
| PLA2G4C-R | 5′-AAGGAGCAGTGGAAGGCATTGGTC-3′ | |
| MAFA-F | 5′-TCCTTCGTTCTCTTCTCAGCC-3′ | qRT-PCR |
| MAFA-R | 5′-AAAGAAGGGGCTTCCTCCAAG-3′ | |
| CYP2E1-F | 5′-CACAGTCGTAGTGCCAACTCT-3′ | qRT-PCR |
| CYP2E1-R | 5′-CACACACTCGTTTTCCTGTGGA-3′ | |
| H3C1-F | 5′-GTGTTCCGCTGTGCTGTTTT-3′ | qRT-PCR |
| H3C1-R | 5′-TAGCGGTGGGGCTTTTTCAC-3′ | |
| H2BC17-F | 5′-TACAACAAGCGCTCGACCAT-3′ | qRT-PCR |
| H2BC17-R | 5′-AGCTGCGAGAGCTCACTTG-3′ | |
| HLA DPB1-F | 5′-CAGCTCTTTTCATTTTGCCATCC-3′ | qRT-PCR |
| HLA DPB1-R | 5′-CTGGAAAAGGTAATTCTCTGGAGTG-3′ |
| Assays | Methods | Results |
|---|---|---|
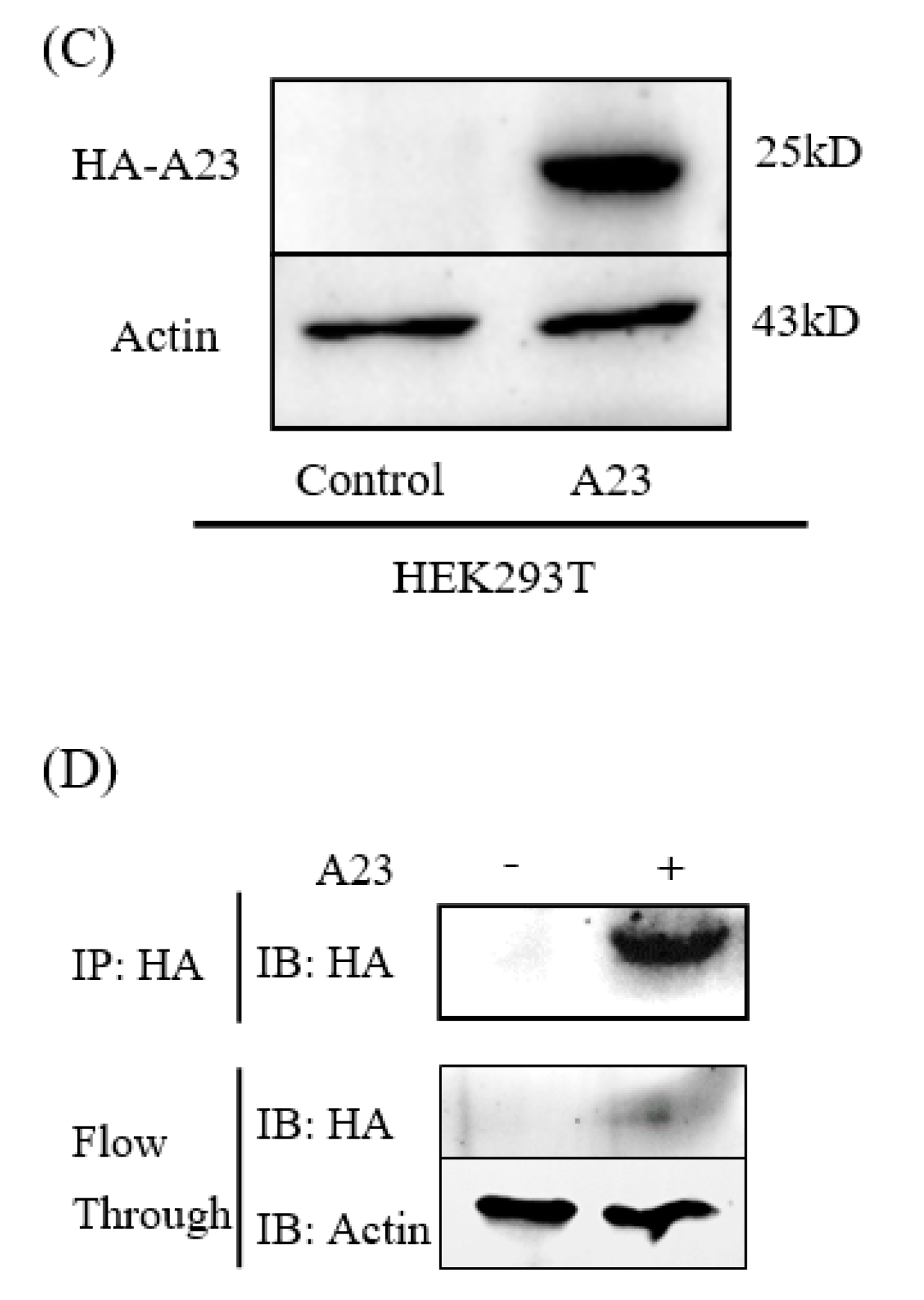
| Plasmid construction and Western blot analyses of A23 protein | A 573 bp gene encoding MPXV A23R resolvase was codon-optimized and synthesized by TSINGKE (TsingKe Biotechnology, China) based on the MPXV gene data (MPXV_USA_2022_MA001) published by NCBI (GenBank: ON563414.3). Primers were designed based on the MPXV-A23R gene sequence. The synthetic gene was inserted into the EcoR I and Xho I sites of pCAGGS-HA plasmid to obtain the pCAGGS-HA-A23R construct. The MPXV A23R coding sequence was amplified with PCR, then transformed into E. coli for homologous recombination. The resulting clones were verified by DNA sequencing (capillary sequencing). | The pCAGGS-HA-A23R plasmids were successfully constructed and expressed in HEK293T (Figure 1B,C). |
| Transcriptome sequencing evaluation | RNA-seq was performed to compare gene expression profiles between pCAGGS-HA-A23R- and pCAGGS-HA (empty vector)-transfected HEK293T cells. The raw sequences were quality controlled and filtered using fastp software (v0.23.1). The high-quality clean reads were aligned to the reference genome of Homo sapiens GRCh38 using Hisat2 (v2.0.5) with default parameters. | The transcriptome sequencing data were appropriate and reliable for further analysis. |
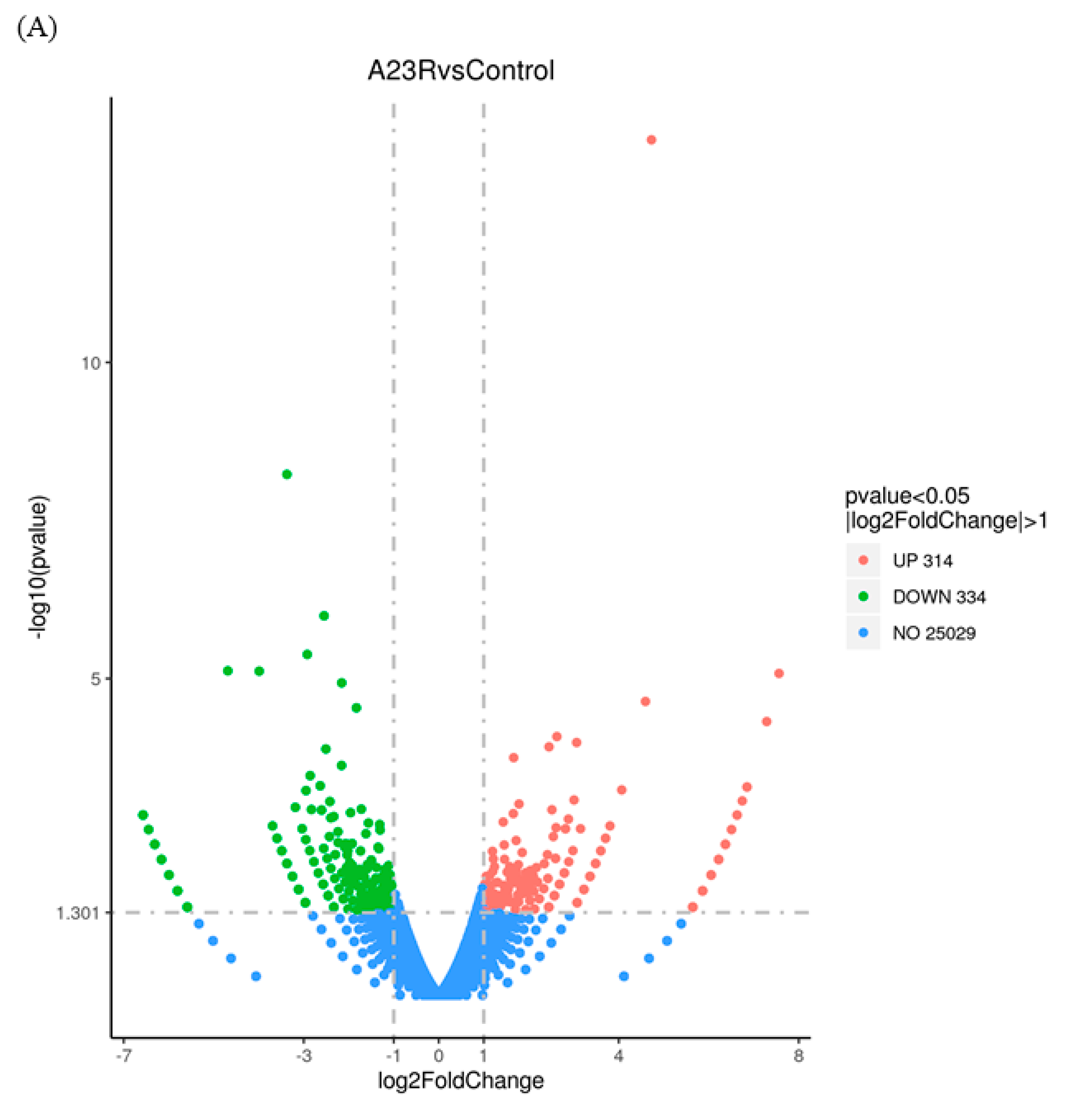
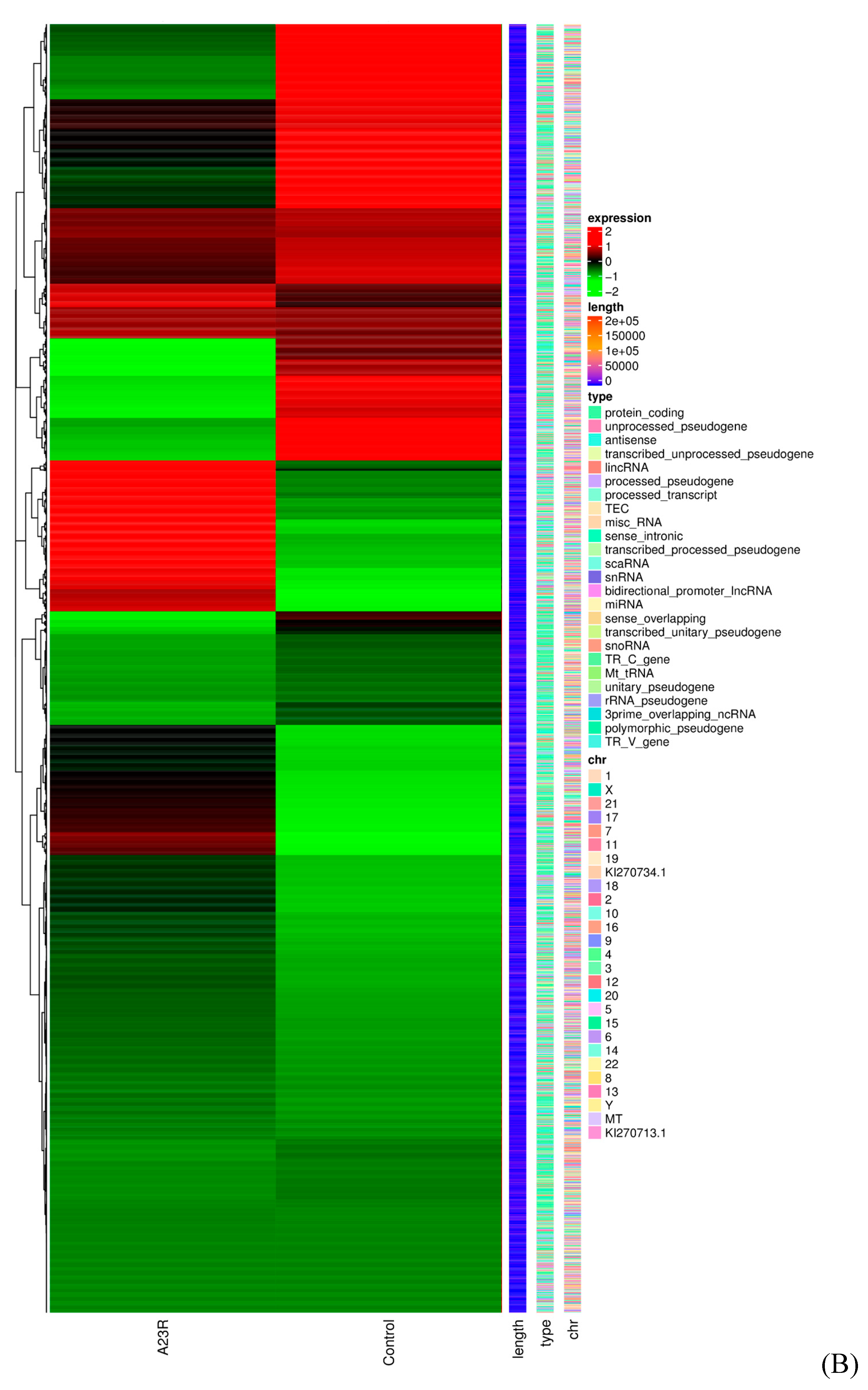
| Identification of DEGs | The gene expression quantification was calculated by FPKM (fragments per kilobase of transcript per million fragments mapped) values using featureCounts (v1.5.0-p3). The differential expression analysis was performed using the edgeR R package (v3.22.5) [23] with Padj ≤ 0.05 and |log2 (FC)| ≥ 1 as the difference significance criterion. | When compared with cells transfected with empty vectors, a total of 648 DEGs were screened in HEK293T cells transfected with pCAGGS-HA-A23R, of which 314 genes were upregulated, and 334 genes were downregulated (Figure 2 and Figure 3). |
| Functional and pathway enrichment analysis of DEGs | GO and KEGG enrichment analyses were implemented by the clusterProfiler R package (v3.8.1) with screening criteria of p < 0.05. A heatmap was plotted using https://www.bioinformatics.com.cn, an online platform for data analysis and visualization. | In the GO enrichment analysis and KEGG pathway enrichment analyses, transcriptional misregulation in cancer, arachidonic acid metabolism, and linoleic acid metabolism were statistically significantly enriched (Figure 4). |
| Interaction of MPXV A23R | HEK293T cells transfected with pCAGGS-HA-A23R/control vector were lysed in RIPA buffer. Cell lysates were incubated with anti-HA-agarose beads. The beads were subsequently washed with PBS prior to LC-MS/MS. According to the manufacturer’s instructions (Oebiotech, Shanghai, China). The samples were enzymatically digested into peptides, which were desalted and evaporated to dryness. Separation was performed using a Nano-HPLC liquid phase system (EASY-nLC1200). The enzymatic products were separated by capillary high-performance liquid chromatography. The mass spectrometry analysis was carried out with a Q-Exactive HF mass spectrometer (Thermo Scientific). | In the GO enrichment analysis and KEGG pathway enrichment analyses, viral carcinogenesis, ribosome and transcriptional misregulation in cancer were significantly enriched (Figure 5). |
| Mutation of the RKKR impairs nuclear import of A23 protein | Glass slides were mounted on the microscope stage and images were recorded through a 63×objective using a Nikon TI-S fluorescent inverted microscope. DAPI was acquired through a 385–470 nm band pass filter using 5% of the UV la-ser intensity; HA was acquired through a 505–530 nm band pass filter using 5% of the 488 nm laser intensity. For each single cell analyzed, the nuclear to cytoplasmic fluorescence ratio was calculated by dividing the nuclear HA fluorescence by the cytoplasmic HA fluorescence. | The A23 protein shows a high degree of co-localization with the nuclear staining DAPI, and a higher nuclear vs. cytoplasmic ratio than the mutant A23△RKKR (Figure 6). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, Y.; Li, M.; Wang, K.; Xiong, J.; Wang, T.; Wang, Y.; Guo, Y.; Kong, L.; Li, M. A Combined Transcriptomic and Proteomic Analysis of Monkeypox Virus A23 Protein on HEK293T Cells. Int. J. Mol. Sci. 2024, 25, 8678. https://doi.org/10.3390/ijms25168678
Wang Y, Li Y, Li M, Wang K, Xiong J, Wang T, Wang Y, Guo Y, Kong L, Li M. A Combined Transcriptomic and Proteomic Analysis of Monkeypox Virus A23 Protein on HEK293T Cells. International Journal of Molecular Sciences. 2024; 25(16):8678. https://doi.org/10.3390/ijms25168678
Chicago/Turabian StyleWang, Yihao, Yihan Li, Mingzhi Li, Keyi Wang, Jiaqi Xiong, Ting Wang, Yu Wang, Yunli Guo, Lingbao Kong, and Meifeng Li. 2024. "A Combined Transcriptomic and Proteomic Analysis of Monkeypox Virus A23 Protein on HEK293T Cells" International Journal of Molecular Sciences 25, no. 16: 8678. https://doi.org/10.3390/ijms25168678
APA StyleWang, Y., Li, Y., Li, M., Wang, K., Xiong, J., Wang, T., Wang, Y., Guo, Y., Kong, L., & Li, M. (2024). A Combined Transcriptomic and Proteomic Analysis of Monkeypox Virus A23 Protein on HEK293T Cells. International Journal of Molecular Sciences, 25(16), 8678. https://doi.org/10.3390/ijms25168678





