ABCC1 Is a ΔNp63 Target Gene Overexpressed in Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
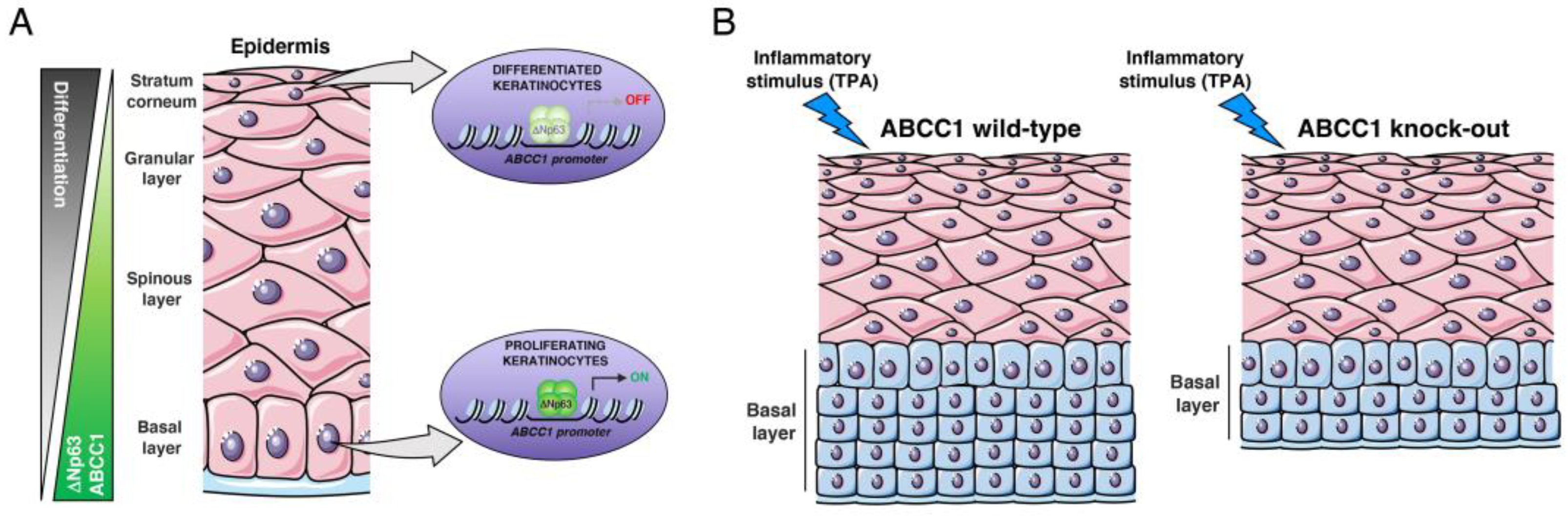
2.1. ΔNp63 Regulates the Expression of ABCC1
2.2. ABCC1 Expression Is Modulated during Keratinocyte Differentiation
2.3. ABCC1 Deletion Does Not Impair Skin Development
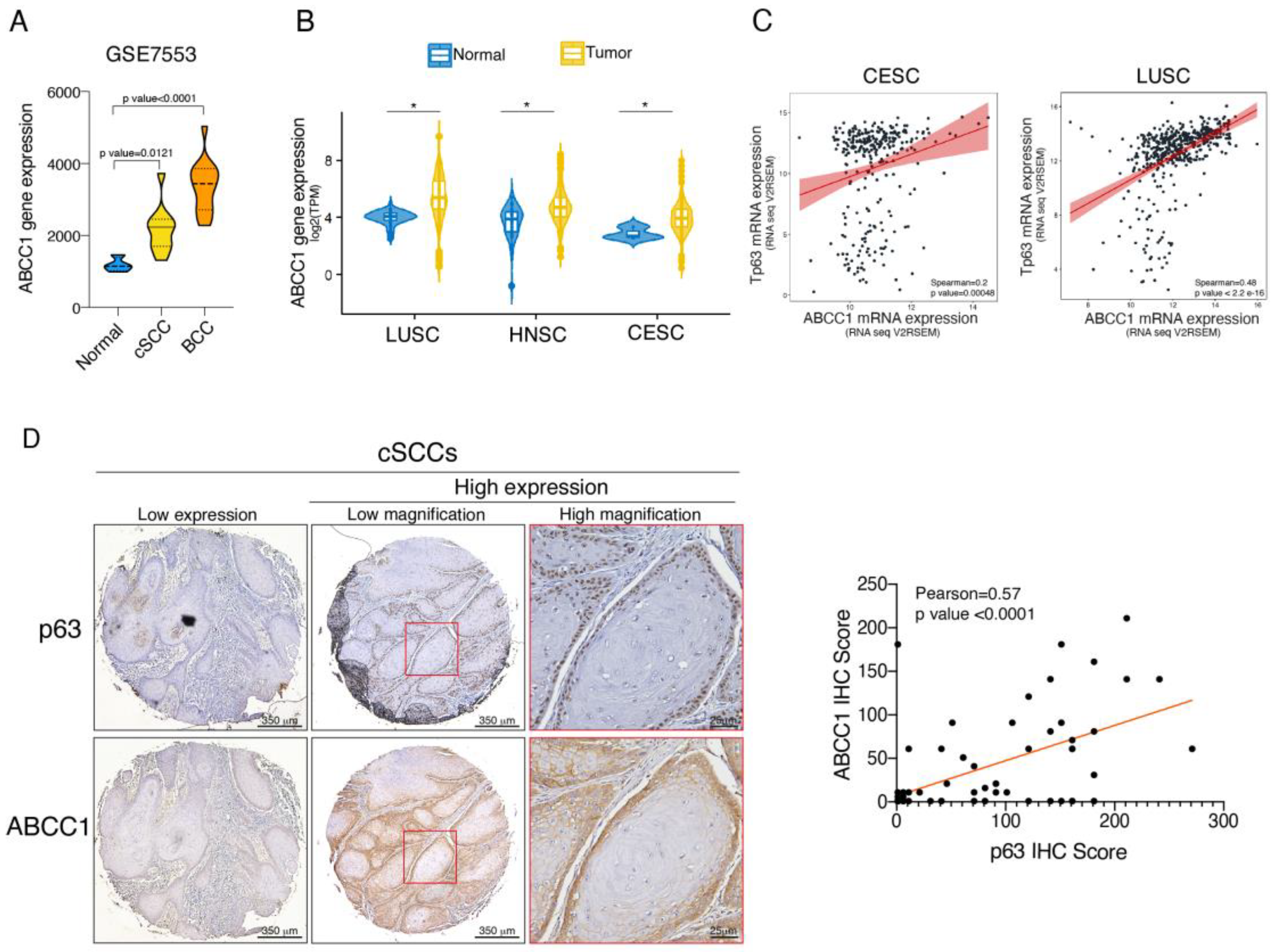
2.4. ABCC1 and p63 Protein Levels Are Increased in SCC
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Growth Curve Analysis
4.3. Animal Studies
4.4. Keratinocyte Isolation Assay
4.5. 12-O-tetradecanoylphorbol 13-acetate (TPA) Treatment
4.6. Protein Extraction and Immunoblotting Analysis
4.7. Chromatin Immunoprecipitation (ChIP) Assay
4.8. RNA Extraction and Real-Time PCR
4.9. Histological and Immunostaining Analysis
4.10. Bioinformatic Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Melino, G.; Memmi, E.M.; Pelicci, P.G.; Bernassola, F. Maintaining epithelial stemness with p63. Sci. Signal 2015, 8, re9. [Google Scholar] [CrossRef]
- Soares, E.; Zhou, H. Master regulatory role of p63 in epidermal development and disease. Cell Mol. Life Sci. 2018, 75, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- Osterburg, C.; Dotsch, V. Structural diversity of p63 and p73 isoforms. Cell Death Differ. 2022, 29, 921–937. [Google Scholar] [CrossRef] [PubMed]
- Trink, B.; Osada, M.; Ratovitski, E.; Sidransky, D. p63 transcriptional regulation of epithelial integrity and cancer. Cell Cycle 2007, 6, 240–245. [Google Scholar] [CrossRef]
- Candi, E.; Cipollone, R.; Rivetti di Val Cervo, P.; Gonfloni, S.; Melino, G.; Knight, R. p63 in epithelial development. Cell Mol. Life Sci. 2008, 65, 3126–3133. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Giovannini, S.; Wang, T.; Fang, J.; Li, P.; Shao, C.; Wang, Y.; TOR centre; Shi, Y.; Candi, E.; et al. p63: A crucial player in epithelial stemness regulation. Oncogene 2023, 42, 3371–3384. [Google Scholar] [CrossRef]
- Yang, A.; Schweitzer, R.; Sun, D.; Kaghad, M.; Walker, N.; Bronson, R.T.; Tabin, C.; Sharpe, A.; Caput, D.; Crum, C.; et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999, 398, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Mills, A.A.; Zheng, B.; Wang, X.J.; Vogel, H.; Roop, D.R.; Bradley, A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398, 708–713. [Google Scholar] [CrossRef]
- Romano, R.A.; Smalley, K.; Magraw, C.; Serna, V.A.; Kurita, T.; Raghavan, S.; Sinha, S. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development 2012, 139, 772–782. [Google Scholar] [CrossRef]
- Kouwenhoven, E.N.; Oti, M.; Niehues, H.; van Heeringen, S.J.; Schalkwijk, J.; Stunnenberg, H.G.; van Bokhoven, H.; Zhou, H. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 2015, 16, 863–878. [Google Scholar] [CrossRef]
- Kouwenhoven, E.N.; van Bokhoven, H.; Zhou, H. Gene regulatory mechanisms orchestrated by p63 in epithelial development and related disorders. Biochim. Biophys. Acta 2015, 1849, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.K.; Carroll, J.S.; Leong, C.O.; Cheng, F.; Brown, M.; Mills, A.A.; Brugge, J.S.; Ellisen, L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006, 8, 551–561. [Google Scholar] [CrossRef]
- Fisher, M.L.; Balinth, S.; Mills, A.A. p63-related signaling at a glance. J. Cell Sci. 2020, 133, jcs228015. [Google Scholar] [CrossRef] [PubMed]
- Westfall, M.D.; Mays, D.J.; Sniezek, J.C.; Pietenpol, J.A. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell Biol. 2003, 23, 2264–2276. [Google Scholar] [CrossRef] [PubMed]
- Sen, G.L.; Boxer, L.D.; Webster, D.E.; Bussat, R.T.; Qu, K.; Zarnegar, B.J.; Johnston, D.; Siprashvili, Z.; Khavari, P.A. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell 2012, 22, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Candi, E.; Rufini, A.; Terrinoni, A.; Giamboi-Miraglia, A.; Lena, A.M.; Mantovani, R.; Knight, R.; Melino, G. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc. Natl. Acad. Sci. USA 2007, 104, 11999–12004. [Google Scholar] [CrossRef] [PubMed]
- Fessing, M.Y.; Mardaryev, A.N.; Gdula, M.R.; Sharov, A.A.; Sharova, T.Y.; Rapisarda, V.; Gordon, K.B.; Smorodchenko, A.D.; Poterlowicz, K.; Ferone, G.; et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 2011, 194, 825–839. [Google Scholar] [CrossRef] [PubMed]
- LeBoeuf, M.; Terrell, A.; Trivedi, S.; Sinha, S.; Epstein, J.A.; Olson, E.N.; Morrisey, E.E.; Millar, S.E. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell 2010, 19, 807–818. [Google Scholar] [CrossRef]
- Mardaryev, A.N.; Gdula, M.R.; Yarker, J.L.; Emelianov, V.U.; Poterlowicz, K.; Sharov, A.A.; Sharova, T.Y.; Scarpa, J.A.; Joffe, B.; Solovei, I.; et al. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development 2014, 141, 101–111. [Google Scholar] [CrossRef]
- Yi, M.; Tan, Y.; Wang, L.; Cai, J.; Li, X.; Zeng, Z.; Xiong, W.; Li, G.; Li, X.; Tan, P.; et al. TP63 links chromatin remodeling and enhancer reprogramming to epidermal differentiation and squamous cell carcinoma development. Cell Mol. Life Sci. 2020, 77, 4325–4346. [Google Scholar] [CrossRef]
- Fierro, C.; Gatti, V.; La Banca, V.; De Domenico, S.; Scalera, S.; Corleone, G.; Fanciulli, M.; De Nicola, F.; Mauriello, A.; Montanaro, M.; et al. The long non-coding RNA NEAT1 is a DeltaNp63 target gene modulating epidermal differentiation. Nat. Commun. 2023, 14, 3795. [Google Scholar] [CrossRef]
- Gatti, V.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. DeltaNp63 in squamous cell carcinoma: Defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol. Oncol. 2019, 13, 981–1001. [Google Scholar] [CrossRef]
- Gatti, V.; Bongiorno-Borbone, L.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. p63 at the Crossroads between Stemness and Metastasis in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 2683. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.L.; Balinth, S.; Mills, A.A. DeltaNp63alpha in cancer: Importance and therapeutic opportunities. Trends Cell Biol. 2023, 33, 280–292. [Google Scholar] [CrossRef]
- Rothenberg, S.M.; Ellisen, L.W. The molecular pathogenesis of head and neck squamous cell carcinoma. J. Clin. Investig. 2012, 122, 1951–1957. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef]
- Saladi, S.V.; Ross, K.; Karaayvaz, M.; Tata, P.R.; Mou, H.; Rajagopal, J.; Ramaswamy, S.; Ellisen, L.W. ACTL6A Is Co-Amplified with p63 in Squamous Cell Carcinoma to Drive YAP Activation, Regenerative Proliferation, and Poor Prognosis. Cancer Cell 2017, 31, 35–49. [Google Scholar] [CrossRef]
- Maier, S.; Wilbertz, T.; Braun, M.; Scheble, V.; Reischl, M.; Mikut, R.; Menon, R.; Nikolov, P.; Petersen, K.; Beschorner, C.; et al. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum. Pathol. 2011, 42, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Botti, E.; Spallone, G.; Moretti, F.; Marinari, B.; Pinetti, V.; Galanti, S.; De Meo, P.D.; De Nicola, F.; Ganci, F.; Castrignano, T.; et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl. Acad. Sci. USA 2011, 108, 13710–13715. [Google Scholar] [CrossRef]
- Yang, X.; Lu, H.; Yan, B.; Romano, R.A.; Bian, Y.; Friedman, J.; Duggal, P.; Allen, C.; Chuang, R.; Ehsanian, R.; et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011, 71, 3688–3700. [Google Scholar] [CrossRef]
- Compagnone, M.; Gatti, V.; Presutti, D.; Ruberti, G.; Fierro, C.; Markert, E.K.; Vousden, K.H.; Zhou, H.; Mauriello, A.; Anemone, L.; et al. DeltaNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc. Natl. Acad. Sci. USA 2017, 114, 13254–13259. [Google Scholar] [CrossRef] [PubMed]
- Gatti, V.; Fierro, C.; Compagnone, M.; Giangrazi, F.; Markert, E.K.; Bongiorno-Borbone, L.; Melino, G.; Peschiaroli, A. DeltaNp63 regulates the expression of hyaluronic acid-related genes in breast cancer cells. Oncogenesis 2018, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.A.; George, A.L.; Sakakibara, N.; Mahmood, K.; Ponnamperuma, R.M.; King, K.E.; Weinberg, W.C. Molecular Mechanisms of p63-Mediated Squamous Cancer Pathogenesis. Int. J. Mol. Sci. 2019, 20, 3590. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.M.; Deeley, R.G.; Cole, S.P. Toxicological relevance of the multidrug resistance protein 1, MRP1 (ABCC1) and related transporters. Toxicology 2001, 167, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Mohle, L.; Stefan, K.; Bascunana, P.; Brackhan, M.; Bruning, T.; Eiriz, I.; El Menuawy, A.; van Genderen, S.; Santos-Garcia, I.; Gorska, A.M.; et al. ABC Transporter C1 Prevents Dimethyl Fumarate from Targeting Alzheimer’s Disease. Biology 2023, 12, 932. [Google Scholar] [CrossRef] [PubMed]
- Krohn, M.; Lange, C.; Hofrichter, J.; Scheffler, K.; Stenzel, J.; Steffen, J.; Schumacher, T.; Bruning, T.; Plath, A.S.; Alfen, F.; et al. Cerebral amyloid-beta proteostasis is regulated by the membrane transport protein ABCC1 in mice. J. Clin. Investig. 2011, 121, 3924–3931. [Google Scholar] [CrossRef] [PubMed]
- Gatti, V.; Fierro, C.; Compagnone, M.; La Banca, V.; Mauriello, A.; Montanaro, M.; Scalera, S.; De Nicola, F.; Candi, E.; Ricci, F.; et al. DeltaNp63-Senataxin circuit controls keratinocyte differentiation by promoting the transcriptional termination of epidermal genes. Proc. Natl. Acad. Sci. USA 2022, 119, e2104718119. [Google Scholar] [CrossRef] [PubMed]
- Gatti, V.; Bernassola, F.; Talora, C.; Melino, G.; Peschiaroli, A. The Impact of the Ubiquitin System in the Pathogenesis of Squamous Cell Carcinomas. Cancers 2020, 12, 1595. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Xie, F.; Pan, Y.; Chen, Z.; Chen, H.; Liu, X.; Tian, K.; Xu, D. A comprehensive prognostic score for head and neck squamous cancer driver genes and phenotype traits. Discov. Oncol. 2023, 14, 193. [Google Scholar] [CrossRef]
- Corchado-Cobos, R.; Garcia-Sancha, N.; Gonzalez-Sarmiento, R.; Perez-Losada, J.; Canueto, J. Cutaneous Squamous Cell Carcinoma: From Biology to Therapy. Int. J. Mol. Sci. 2020, 21, 2956. [Google Scholar] [CrossRef]
- Winge, M.C.G.; Kellman, L.N.; Guo, K.; Tang, J.Y.; Swetter, S.M.; Aasi, S.Z.; Sarin, K.Y.; Chang, A.L.S.; Khavari, P.A. Advances in cutaneous squamous cell carcinoma. Nat. Rev. Cancer 2023, 23, 430–449. [Google Scholar] [CrossRef]
- Deeley, R.G.; Westlake, C.; Cole, S.P. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol. Rev. 2006, 86, 849–899. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, K.M.; Haber, M.; Fletcher, J.I. Targeting multidrug resistance-associated protein 1 (MRP1)-expressing cancers: Beyond pharmacological inhibition. Drug Resist. Updat. 2021, 59, 100795. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.P.; Sparks, K.E.; Fraser, K.; Loe, D.W.; Grant, C.E.; Wilson, G.M.; Deeley, R.G. Pharmacological characterization of multidrug resistant MRP-transfected human tumor cells. Cancer Res. 1994, 54, 5902–5910. [Google Scholar] [PubMed]
- Allen, J.D.; Brinkhuis, R.F.; van Deemter, L.; Wijnholds, J.; Schinkel, A.H. Extensive contribution of the multidrug transporters P-glycoprotein and Mrp1 to basal drug resistance. Cancer Res. 2000, 60, 5761–5766. [Google Scholar] [PubMed]
- Nedeljkovic, M.; Tanic, N.; Prvanovic, M.; Milovanovic, Z.; Tanic, N. Friend or foe: ABCG2, ABCC1 and ABCB1 expression in triple-negative breast cancer. Breast Cancer 2021, 28, 727–736. [Google Scholar] [CrossRef]
- Takebayashi, Y.; Akiyama, S.; Natsugoe, S.; Hokita, S.; Niwa, K.; Kitazono, M.; Sumizawa, T.; Tani, A.; Furukawa, T.; Aikou, T. The expression of multidrug resistance protein in human gastrointestinal tract carcinomas. Cancer 1998, 82, 661–666. [Google Scholar] [CrossRef]
- Tsuzuki, H.; Fujieda, S.; Sunaga, H.; Sugimoto, C.; Tanaka, N.; Saito, H. Expression of multidrug resistance-associated protein (MRP) in head and neck squamous cell carcinoma. Cancer Lett. 1998, 126, 89–95. [Google Scholar] [CrossRef]
- Larbcharoensub, N.; Leopairat, J.; Sirachainan, E.; Narkwong, L.; Bhongmakapat, T.; Rasmeepaisarn, K.; Janvilisri, T. Association between multidrug resistance-associated protein 1 and poor prognosis in patients with nasopharyngeal carcinoma treated with radiotherapy and concurrent chemotherapy. Hum. Pathol. 2008, 39, 837–845. [Google Scholar] [CrossRef]
- Eijdems, E.W.; De Haas, M.; Coco-Martin, J.M.; Ottenheim, C.P.; Zaman, G.J.; Dauwerse, H.G.; Breuning, M.H.; Twentyman, P.R.; Borst, P.; Baas, F. Mechanisms of MRP over-expression in four human lung-cancer cell lines and analysis of the MRP amplicon. Int. J. Cancer 1995, 60, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Li, H.; Gao, P.; Shang, G.; Zhang, D.D.; Zhang, N.; Jiang, T. Nrf2 pathway regulates multidrug-resistance-associated protein 1 in small cell lung cancer. PLoS ONE 2013, 8, e63404. [Google Scholar] [CrossRef]
- Kurz, E.U.; Cole, S.P.; Deeley, R.G. Identification of DNA-protein interactions in the 5′ flanking and 5′ untranslated regions of the human multidrug resistance protein (MRP1) gene: Evaluation of a putative antioxidant response element/AP-1 binding site. Biochem. Biophys. Res. Commun. 2001, 285, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Manohar, C.F.; Bray, J.A.; Salwen, H.R.; Madafiglio, J.; Cheng, A.; Flemming, C.; Marshall, G.M.; Norris, M.D.; Haber, M.; Cohn, S.L. MYCN-mediated regulation of the MRP1 promoter in human neuroblastoma. Oncogene 2004, 23, 753–762. [Google Scholar] [CrossRef]
- Si, X.; Gao, Z.; Xu, F.; Zheng, Y. SOX2 upregulates side population cells and enhances their chemoresistant ability by transactivating ABCC1 expression contributing to intrinsic resistance to paclitaxel in melanoma. Mol. Carcinog. 2020, 59, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jiang, Y.Y.; Xie, J.J.; Mayakonda, A.; Hazawa, M.; Chen, L.; Xiao, J.F.; Li, C.Q.; Huang, M.L.; Ding, L.W.; et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat. Commun. 2018, 9, 3619. [Google Scholar] [CrossRef] [PubMed]
- Minich, T.; Riemer, J.; Schulz, J.B.; Wielinga, P.; Wijnholds, J.; Dringen, R. The multidrug resistance protein 1 (Mrp1), but not Mrp5, mediates export of glutathione and glutathione disulfide from brain astrocytes. J. Neurochem. 2006, 97, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Koontongkaew, S. The tumor microenvironment contribution to development, growth, invasion and metastasis of head and neck squamous cell carcinomas. J. Cancer 2013, 4, 66–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, R.; Fang, J. Exploring the frontiers: Tumor immune microenvironment and immunotherapy in head and neck squamous cell carcinoma. Discov. Oncol. 2024, 15, 22. [Google Scholar] [CrossRef]
- Krohn, M.; Zoufal, V.; Mairinger, S.; Wanek, T.; Paarmann, K.; Brüning, T.; Eiriz, I.; Brackhan, M.; Langer, O.; Pahnke, J. Generation and Characterization of an Abcc1 Humanized Mouse Model (hABCC1(flx/flx)) with Knockout Capability. Mol. Pharmacol. 2019, 96, 138–147. [Google Scholar] [CrossRef]
- Malatesta, M.; Peschiaroli, A.; Memmi, E.M.; Zhang, J.; Antonov, A.; Green, D.R.; Barlev, N.A.; Garabadgiu, A.V.; Zhou, P.; Melino, G.; et al. The Cul4A-DDB1 E3 ubiquitin ligase complex represses p73 transcriptional activity. Oncogene 2013, 32, 4721–4726. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Banca, V.; De Domenico, S.; Nicolai, S.; Gatti, V.; Scalera, S.; Maugeri, M.; Mauriello, A.; Montanaro, M.; Pahnke, J.; Candi, E.; et al. ABCC1 Is a ΔNp63 Target Gene Overexpressed in Squamous Cell Carcinoma. Int. J. Mol. Sci. 2024, 25, 8741. https://doi.org/10.3390/ijms25168741
La Banca V, De Domenico S, Nicolai S, Gatti V, Scalera S, Maugeri M, Mauriello A, Montanaro M, Pahnke J, Candi E, et al. ABCC1 Is a ΔNp63 Target Gene Overexpressed in Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2024; 25(16):8741. https://doi.org/10.3390/ijms25168741
Chicago/Turabian StyleLa Banca, Veronica, Sara De Domenico, Sara Nicolai, Veronica Gatti, Stefano Scalera, Marcello Maugeri, Alessandro Mauriello, Manuela Montanaro, Jens Pahnke, Eleonora Candi, and et al. 2024. "ABCC1 Is a ΔNp63 Target Gene Overexpressed in Squamous Cell Carcinoma" International Journal of Molecular Sciences 25, no. 16: 8741. https://doi.org/10.3390/ijms25168741




