Anti-Neuroinflammatory Effect of Ombuin from Rhamnus erythroxylon Pall. Leaves in LPS-Induced BV-2 Microglia by Targeting Src and Suppressing the PI3K-AKT/NF-κB Signaling Pathway
Abstract
1. Introduction
2. Results
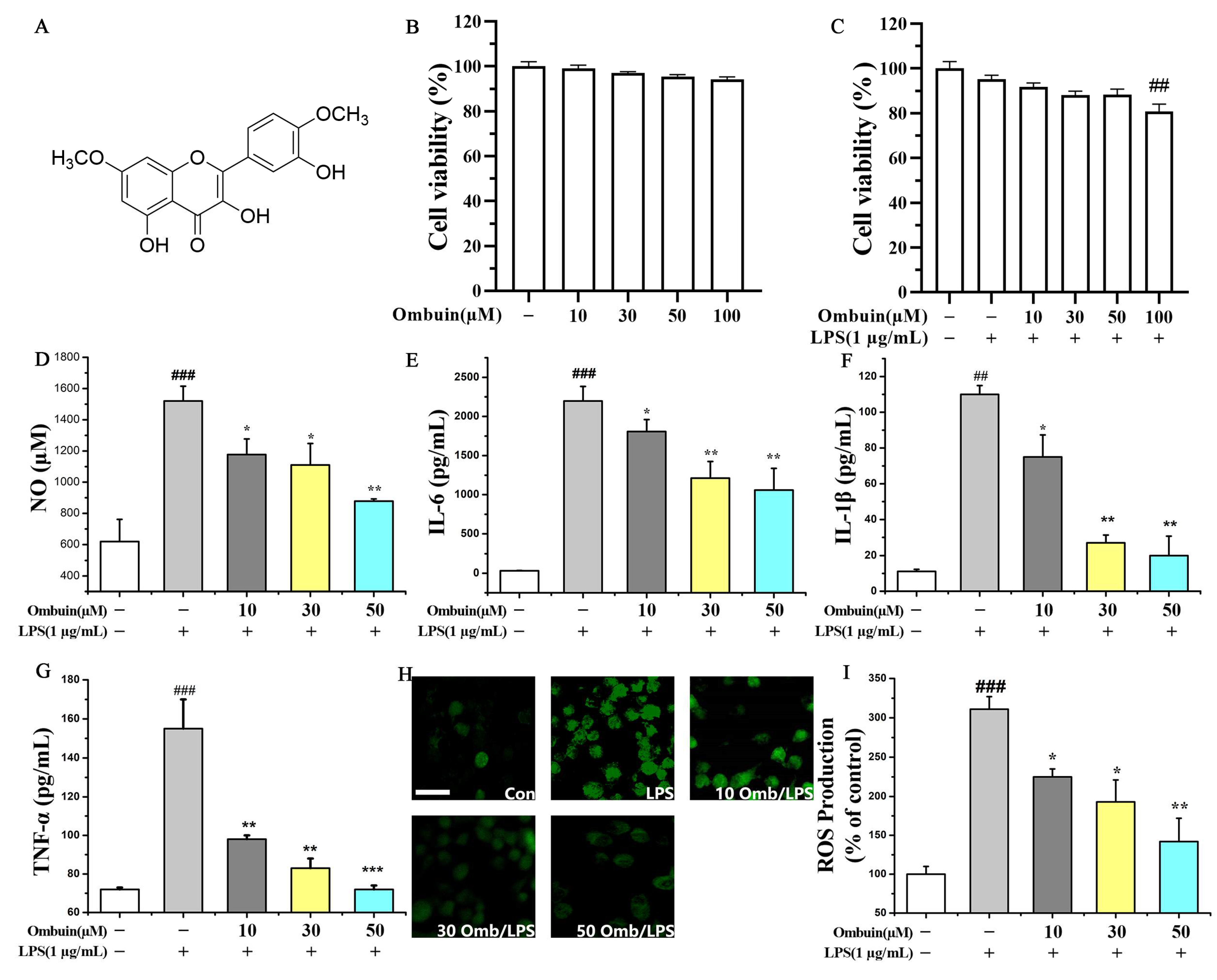
2.1. Ombuin Inhibited LPS-Induced Neuroinflammation in BV-2 Cells
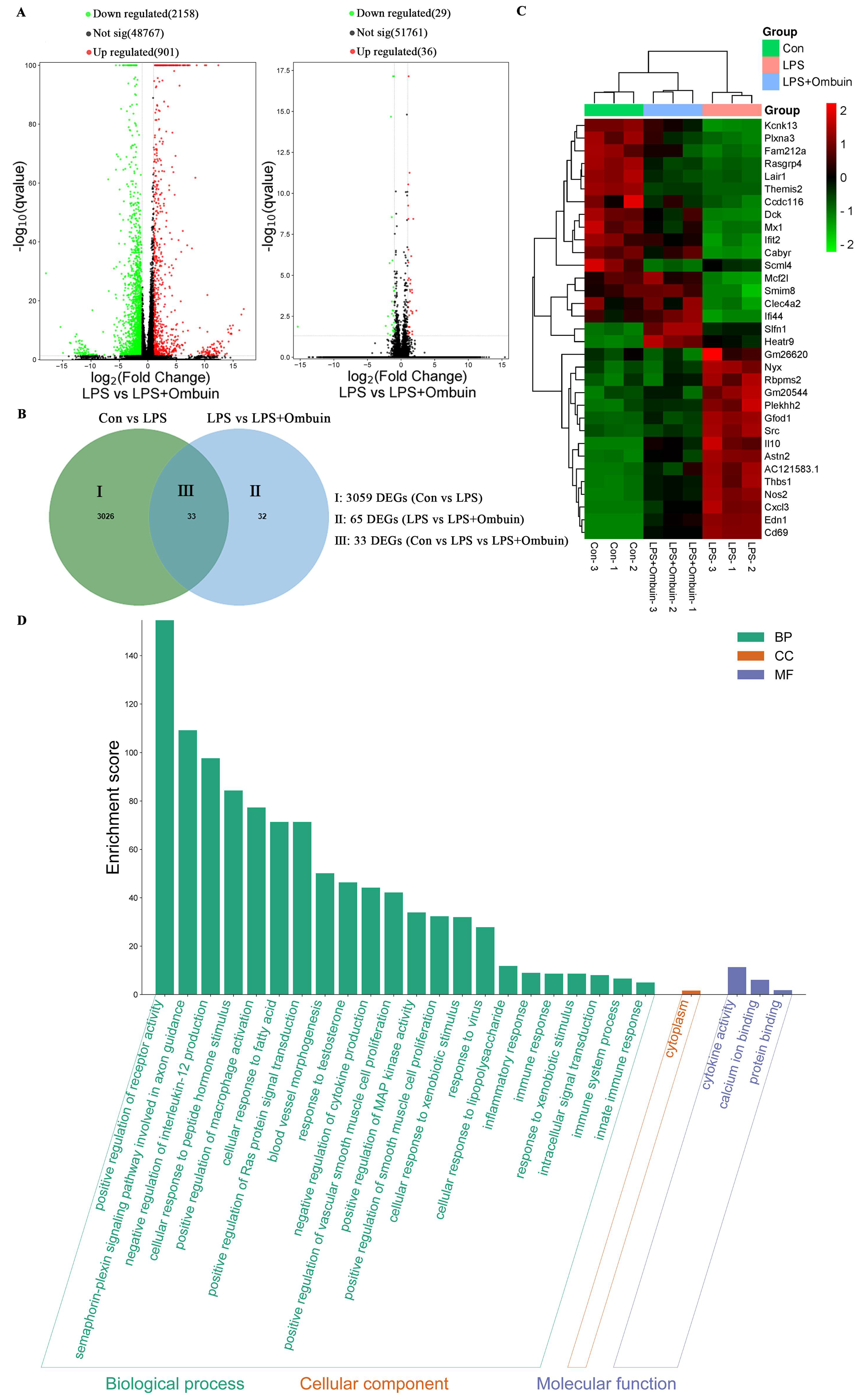
2.2. Network Pharmacology Predicted the Targets for Neuroinflammation of Ombuin
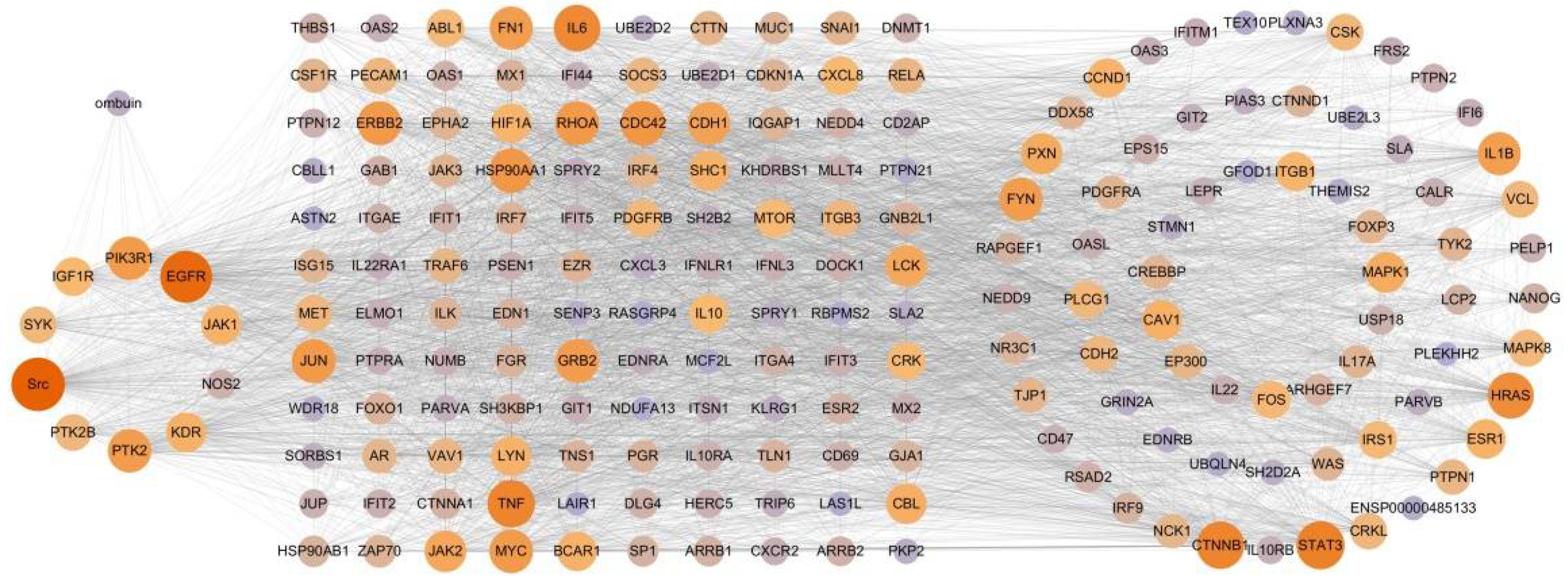
2.3. Therapeutic Network of Ombuin in Neuroinflammatory Diseases
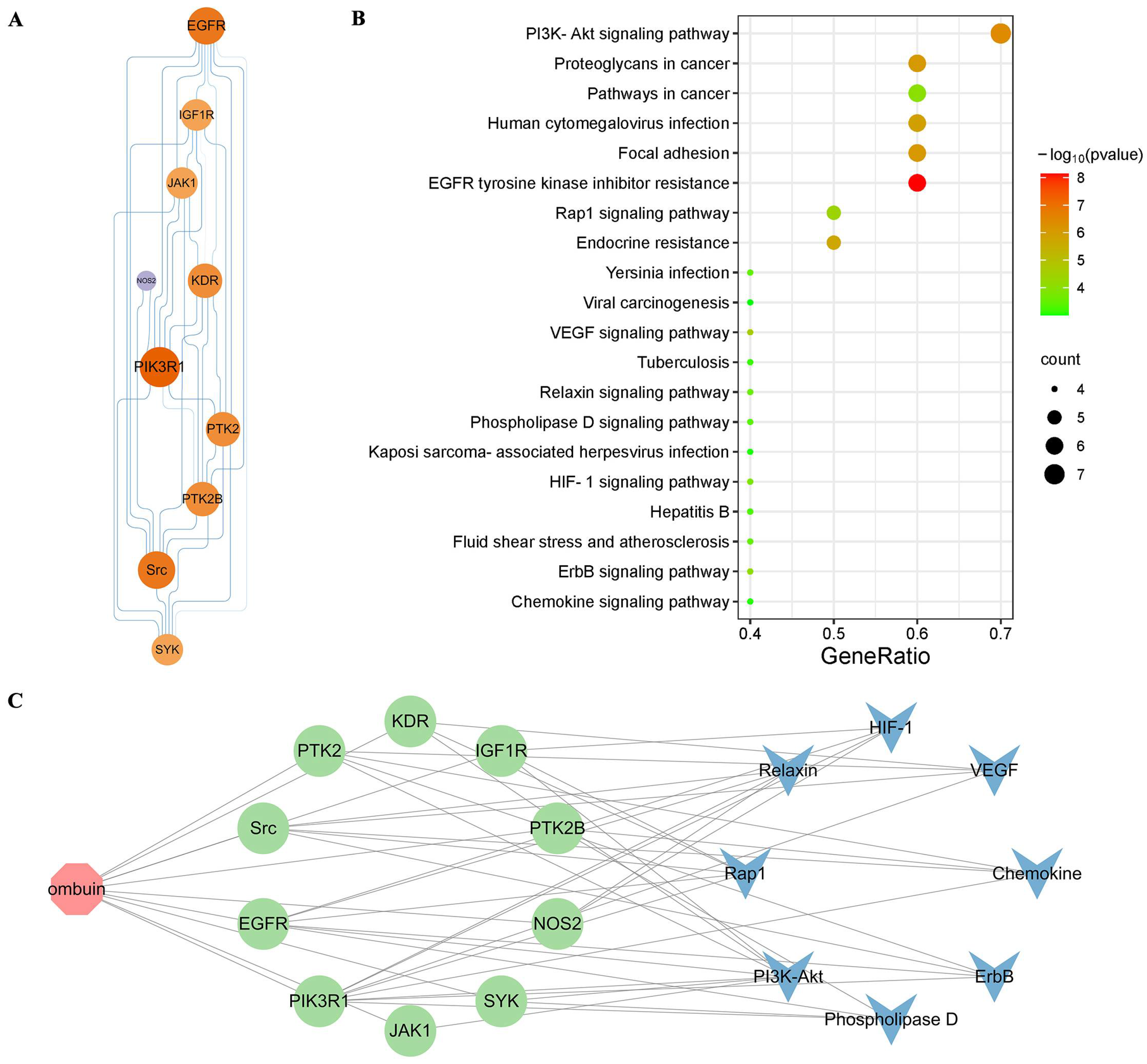
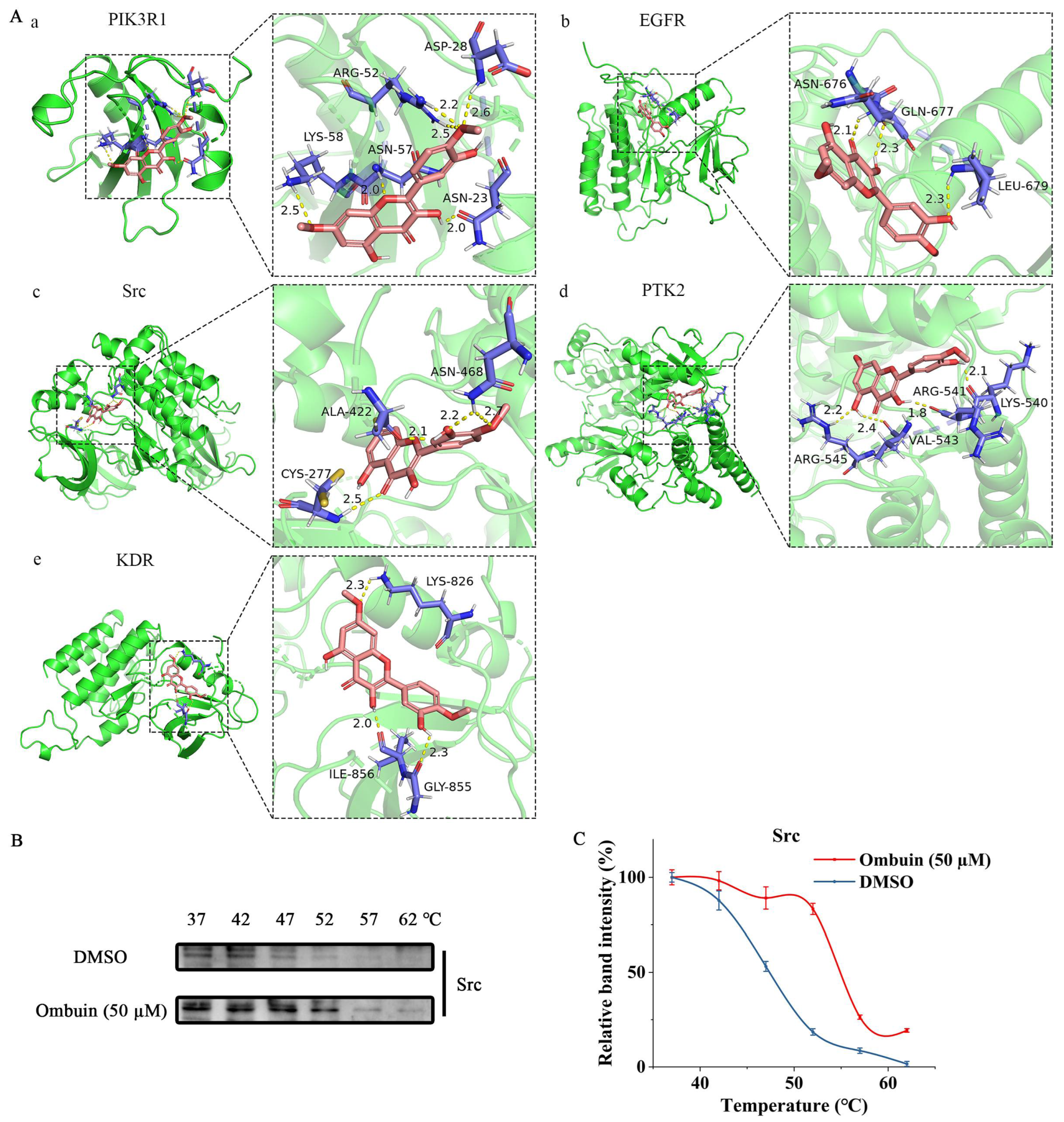
2.4. Potential Mechanism of Anti-Neuroinflammation of Ombuin
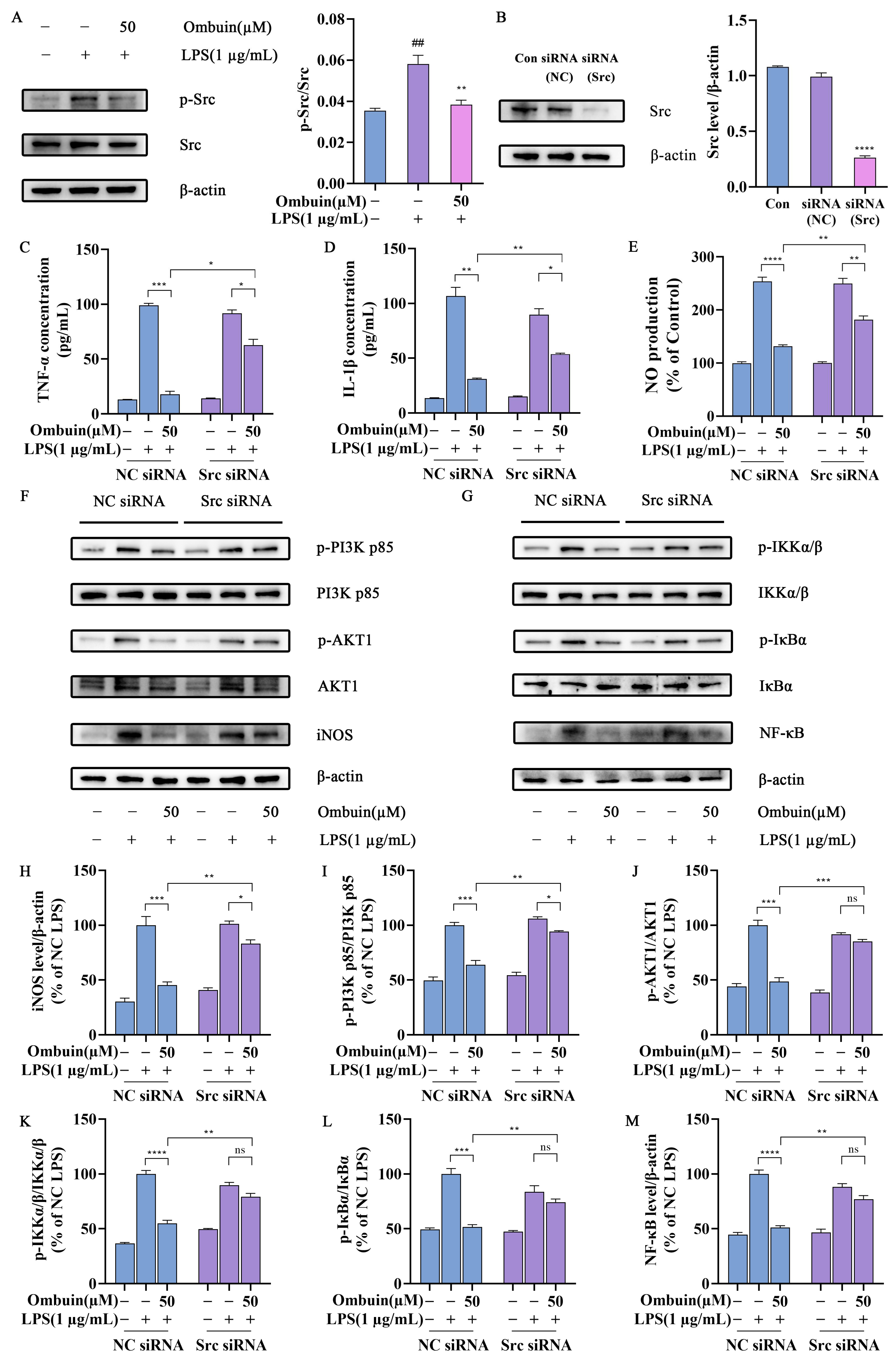
2.5. Src Was Necessary for Ombuin-Mediated Anti-Inflammatory Activity
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. MTT Test
4.4. Griess Test
4.5. Measurement of Pro-Inflammatory Factors
4.6. ROS Detection Assay
4.7. Network Pharmacology Analysis
4.8. Transcriptome Analysis
4.9. Building a “Component–Target–Pathway” Network
4.10. Molecular Docking
4.11. Cell Heat Transfer Analysis (CETSA)
4.12. Western Blot
4.13. Src siRNA Transfection
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woodburn, S.C.; Bollinger, J.L.; Wohleb, E.S. The semantics of microglia activation: Neuroinflammation, homeostasis, and stress. J. Neuroinflamm. 2021, 18, 258. [Google Scholar] [CrossRef]
- Maher, P. The Potential of Flavonoids for the Treatment of Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3056. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Zhou, Y.L.; He, D.H.; Liu, W.; Fan, X.Z.; Wang, Q.; Pan, H.F.; Cheng, Y.X.; Liu, Y.Q. Centipeda minima extract exerts antineuroinflammatory effects via the inhibition of NF-κB signaling pathway. Phytomedicine 2020, 67, 153164. [Google Scholar] [CrossRef]
- Singh, D. Astrocytic and microglial cells as the modulators of neuroinflammation in Alzheimer’s disease. J. Neuroinflamm. 2022, 19, 206. [Google Scholar] [CrossRef]
- Decandia, D.; Gelfo, F.; Landolfo, E.; Balsamo, F.; Petrosini, L.; Cutuli, D. Dietary Protection against Cognitive Impairment, Neuroinflammation and Oxidative Stress in Alzheimer’s Disease Animal Models of Lipopolysaccharide-Induced Inflammation. Int. J. Mol. Sci. 2023, 24, 5921. [Google Scholar] [CrossRef]
- Stöberl, N.; Maguire, E.; Salis, E.; Shaw, B.; Hall-Roberts, H. Human iPSC-derived glia models for the study of neuroinflammation. J. Neuroinflamm. 2023, 20, 231. [Google Scholar] [CrossRef]
- Kwon, H.S.; Koh, S.H. Neuroinflammation in neurodegenerative disorders: The roles of microglia and astrocytes. Transl. Neurodegener. 2020, 9, 42. [Google Scholar] [CrossRef]
- Muzio, L.; Viotti, A.; Martino, G. Microglia in neuroinflammation and neurodegeneration: From understanding to therapy. Front. Neurosci. 2021, 15, 742065. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.I.; Cheng, C.I.; Kang, Y.F.; Chang, P.C.; Lin, I.P.; Kuo, Y.H.; Jhou, A.J.; Lin, M.Y.; Chen, C.Y.; Lee, C.H. Hispidulin Inhibits Neuroinflammation in Lipopolysaccharide-Activated BV2 Microglia and Attenuates the Activation of Akt, NF-κB, and STAT3 Pathway. Neurotox. Res. 2020, 38, 163–174. [Google Scholar] [CrossRef]
- Ismail, E.N.; Jantan, I.; Vidyadaran, S.; Jamal, J.A.; Azmi, N. Phyllanthus amarus prevents LPS-mediated BV2 microglial activation via MyD88 and NF-κB signaling pathways. BMC Complement. Med. Ther. 2020, 20, 202. [Google Scholar] [CrossRef]
- Jana, M.; Jana, A.; Liu, X.; Ghosh, S.; Pahan, K. Involvement of phosphatidylinositol 3-kinase-mediated up-regulation of I kappa B alpha in anti-inflammatory effect of gemfibrozil in microglia. J. Immunol. 2007, 179, 4142–4152. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, G.R.; Mohana, T.; Athesh, K.; Hillary, V.E.; Vasconcelos, A.B.S.; de Franca, M.N.F.; Montalvão, M.M.; Ceasar, S.A.; Jothi, G.; Sridharan, G.; et al. Anti-inflammatory natural products modulate interleukins and their related signaling markers in inflammatory bowel disease: A systematic review. J. Pharm. Anal. 2023, 13, 1408–1428. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Dong, F.; Su, L.; Tan, Z.X.; Lei, M.; Li, L.; Wen, D.; Zhang, F. The role and therapeutic potential of nuclear factor κB (NF-κB) in ischemic stroke. Biomed. Pharmacother. 2024, 171, 116140. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.J.; Hwang, Y.G.; Sharma, N.; Tran, H.Q.; Dang, D.K.; Jang, C.G.; Jeong, J.H.; Nah, S.Y.; Nabeshima, T.; Kim, H.C. Role of protein kinase Cδ in dopaminergic neurotoxic events. Food Chem. Toxicol. 2018, 121, 254–261. [Google Scholar] [CrossRef]
- Xu, P.; Huang, M.W.; Xiao, C.X.; Long, F.; Wang, Y.; Liu, S.Y.; Jia, W.W.; Wu, W.J.; Yang, D.; Hu, J.F.; et al. Matairesinol Suppresses Neuroinflammation and Migration Associated with Src and ERK1/2-NF-κB Pathway in Activating BV2 Microglia. Neurochem. Res. 2017, 42, 2850–2860. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C.; Lin, C.C.; Hsiao, L.D.; Kuo, J.M.; Tseng, H.C.; Yang, C.M. Lipopolysaccharide-Induced Matrix Metalloproteinase-9 Expression Associated with Cell Migration in Rat Brain Astrocytes. Int. J. Mol. Sci. 2020, 21, 259. [Google Scholar] [CrossRef] [PubMed]
- Lowell, C.A. Src-family kinases: Rheostats of immune cell signaling. Mol. Immunol. 2004, 41, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Gage, M.; Putra, M.; Wachter, L.; Dishman, K.; Gard, M.; Gomez-Estrada, C.; Thippeswamy, T. Saracatinib, a Src Tyrosine Kinase Inhibitor, as a Disease Modifier in the Rat DFP Model: Sex Differences, Neurobehavior, Gliosis, Neurodegeneration, and Nitro-Oxidative Stress. Antioxidants 2022, 11, 61. [Google Scholar] [CrossRef]
- Tai, W.; Ye, X.; Bao, X.; Zhao, B.; Wang, X.; Zhang, D. Inhibition of Src tyrosine kinase activity by squamosamide derivative FLZ attenuates neuroinflammation in both in vivo and in vitro Parkinson’s disease models. Neuropharmacology 2013, 75, 201–212. [Google Scholar] [CrossRef]
- Bosco, F.; Ruga, S.; Citraro, R.; Leo, A.; Guarnieri, L.; Maiuolo, J.; Oppedisano, F.; Macrì, R.; Scarano, F.; Nucera, S.; et al. The Effects of Andrographis paniculata (Burm.F.) Wall. Ex Nees and Andrographolide on Neuroinflammation in the Treatment of Neurodegenerative Diseases. Nutrients 2023, 15, 3428. [Google Scholar] [CrossRef]
- Barber, K.; Mendonca, P.; Evans, J.A.; Soliman, K.F.A. Antioxidant and Anti-Inflammatory Mechanisms of Cardamonin through Nrf2 Activation and NF-kB Suppression in LPS-Activated BV-2 Microglial Cells. Int. J. Mol. Sci. 2023, 24, 10872. [Google Scholar] [CrossRef] [PubMed]
- Elbandy, M. Anti-Inflammatory Effects of Marine Bioactive Compounds and Their Potential as Functional Food Ingredients in the Prevention and Treatment of Neuroinflammatory Disorders. Molecules 2023, 28, 2. [Google Scholar] [CrossRef] [PubMed]
- Al-Azzawie, H.F.; Alhamdani, M.S.S. Hypoglycemic and antioxidant effect of oleuropein in alloxan-diabetic rabbits. Life Sci. 2006, 78, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Ketha, A.; Hieu, H.V.; Tatipamula, V.B. In vitro antimycobacterial studies of flavonols from Bauhinia vahlii Wight and Arn. 3 Biotech 2021, 11, 128. [Google Scholar] [CrossRef]
- Rao, Y.K.; Fang, S.H.; Tzeng, Y.M. Antiinflammatory activities of flavonoids and a triterpene caffeate isolated from Bauhinia variegata. Phytother. Res. 2008, 22, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Ranjana; Nooreen, Z.; Bushra, U.; Jyotshna; Bawankule, D.U.; Shanker, K.; Ahmad, A.; Tandon, S. Standardization and xanthine oxidase inhibitory potential of Zanthoxylum armatum fruits. J. Ethnopharmacol. 2019, 230, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Chen, Y. Transcriptomics: Advances and approaches. Sci. China Life Sci. 2013, 56, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Streit, W.J.; Mrak, R.E.; Griffin, W.S.T. Microglia and neuroinflammation: A pathological perspective. J. Neuroinflamm. 2004, 1, 14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- El-Bakoush, A.; Olajide, O.A. Formononetin inhibits neuroinflammation and increases estrogen receptor beta (ERβ) protein expression in BV2 microglia. Int. Immunopharmacol. 2018, 61, 325–337. [Google Scholar] [CrossRef]
- Zheng, Y.; Fang, W.; Fan, S.; Liao, W.; Xiong, Y.; Liao, S.; Li, Y.; Xiao, S.; Liu, J. Neurotropin inhibits neuroinflammation via suppressing NF-κB and MAPKs signaling pathways in lipopolysaccharide-stimulated BV2 cells. J. Pharmacol. Sci. 2018, 136, 242–248. [Google Scholar] [CrossRef]
- Badoer, E. Microglia: Activation in acute and chronic inflammatory states and in response to cardiovascular dysfunction. Int. J. Biochem. Cell Biol. 2010, 42, 1580–1585. [Google Scholar] [CrossRef] [PubMed]
- Karimi, A.; Majlesi, M.; Rafieian-Kopaei, M. Herbal versus synthetic drugs; beliefs and facts. J. Nephropharmacol. 2015, 4, 27–30. [Google Scholar]
- So, Y.J.; Lee, J.U.; Yang, G.S.; Yang, G.; Kim, S.W.; Lee, J.H.; Kim, J.U. The Potentiality of Natural Products and Herbal Medicine as Novel Medications for Parkinson’s Disease: A Promising Therapeutic Approach. Int. J. Mol. Sci. 2024, 25, 1071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, H.; Li, C.; Han, B.; Zhang, Y. Exploring Chinese herbal medicine for ischemic stroke: Insights into microglia and signaling pathways. Front. Pharmacol. 2024, 15, 1333006. [Google Scholar]
- Ross, F.C.; Mayer, D.E.; Horn, J.; Cryan, J.F.; Del Rio, D.; Randolph, E.; Gill, C.I.R.; Gupta, A.; Ross, R.P.; Stanton, C.; et al. Potential of dietary polyphenols for protection from age-related decline and neurodegeneration: A role for gut microbiota? Nutr. Neurosci. 2024, 1–19. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Q.; Zhou, C.C.; Jiang, S.G.; Lan, W.Q.; Zhang, F.; Tao, X.; Chen, W.S. Natural products for the treatment of chemotherapy-related cognitive impairment and prospects of nose-to-brain drug delivery. Front. Pharmacol. 2024, 15, 1292807. [Google Scholar]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Cianciulli, A.; Porro, C.; Calvello, R.; Trotta, T.; Lofrumento, D.D.; Panaro, M.A. Microglia Mediated Neuroinflammation: Focus on PI3K Modulation. Biomolecules 2020, 10, 137. [Google Scholar] [CrossRef]
- Panneerselvan, P.; Vasanthakumar, K.; Muthuswamy, K.; Krishnan, V.; Subramaniam, S. Insights on the functional dualism of nitric oxide in the hallmarks of cancer. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 189001. [Google Scholar]
- Russell, T.M.; Richardson, D.R. The good Samaritan glutathione-S-transferase P1: An evolving relationship in nitric oxide metabolism mediated by the direct interactions between multiple effector molecules. Redox Biol. 2023, 59, 102568. [Google Scholar]
- Dilshara, M.G.; Lee, K.T.; Jayasooriya, R.G.P.T.; Kang, C.H.; Park, S.R.; Choi, Y.H.; Choi, I.W.; Hyun, J.W.; Chang, W.Y.; Kim, Y.S.; et al. Downregulation of NO and PGE2 in LPS-stimulated BV2 microglial cells by trans-isoferulic acid via suppression of PI3K/Akt-dependent NF-κB and activation of Nrf2-mediated HO-1. Int. Immunopharmacol. 2014, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Du, R.W.; Du, R.H.; Bu, W.G. β-Arrestin 2 mediates the anti-inflammatory effects of fluoxetine in lipopolysaccharide-stimulated microglial cells. J. Neuroimmune Pharmacol. 2014, 9, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, C.P.; Tansey, M.G. A survey from 2012 of evidence for the role of neuroinflammation in neurotoxin animal models of Parkinson’s disease and potential molecular targets. Exp. Neurol. 2014, 256, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, C.; Wang, J.; Zhao, S.; Zhang, K.; Wang, J.; Zhang, W.; Wu, C.; Yang, J. Pseudoginsenoside-F11 (PF11) exerts anti-neuroinflammatory effects on LPS-activated microglial cells by inhibiting TLR4-mediated TAK1/IKK/NF-κB, MAPKs and Akt signaling pathways. Neuropharmacology 2014, 79, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Akanchise, T.; Angelova, A. Potential of Nano-Antioxidants and Nanomedicine for Recovery from Neurological Disorders Linked to Long COVID Syndrome. Antioxidants 2023, 12, 393. [Google Scholar] [CrossRef] [PubMed]
- Patten, D.A.; Germain, M.; Kelly, M.A.; Slack, R.S. Reactive oxygen species: Stuck in the middle of neurodegeneration. J. Alzheimers Dis. 2010, 20 (Suppl. 2), S357–S367. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Sun, L.; Jiang, Z.; Zeng, T.; Wu, P.; Huang, J.; Liu, H.; Xiao, P. Neuropharmacological insights into Gardenia jasminoides Ellis: Harnessing therapeutic potential for central nervous system disorders. Phytomedicine 2024, 125, 155374. [Google Scholar] [CrossRef] [PubMed]
- Li, A.P.; He, S.S.; Zhang, W.N.; Zhang, L.C.; Liu, Y.T.; Li, K.; Qin, X.M. Exploration the active compounds of Astragali Radix in treatment of adriamycin nephropathy by network pharmacology combined with transcriptomic approach. J. Ethnopharmacol. 2020, 258, 112537. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Wang, H.; Yang, M.; Xia, H.; Wu, Y.; Liu, Q.; Tang, H. miR-153-3p via PIK3R1 Is Involved in Cigarette Smoke-Induced Neurotoxicity in the Brain. Toxics 2023, 11, 969. [Google Scholar] [CrossRef]
- Qu, W.S.; Tian, D.S.; Guo, Z.B.; Fang, J.; Zhang, Q.; Yu, Z.Y.; Xie, M.J.; Zhang, H.Q.; Lü, J.G.; Wang, W. Inhibition of EGFR/MAPK signaling reduces microglial inflammatory response and the associated secondary damage in rats after spinal cord injury. J. Neuroinflamm. 2012, 9, 178. [Google Scholar] [CrossRef]
- Lee, S.; Jo, M.; Kwon, Y.; Jeon, Y.M.; Kim, S.; Lee, K.J.; Kim, H.J. PTK2 regulates tau-induced neurotoxicity via phosphorylation of p62 at Ser403. J. Neurogenet. 2023, 37, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Mathew, A.; Dourvetakis, K.D.; Sanchez-Guerrero, E.; Pangeni, R.P.; Gurusamy, N.; Aenlle, K.K.; Ravindran, G.; Twahir, A.; Isler, D.; et al. Recent Research Trends in Neuroinflammatory and Neurodegenerative Disorders. Cells 2024, 13, 511. [Google Scholar] [CrossRef] [PubMed]
- Check, J.; Byrd, C.L.; Menio, J.; Rippe, R.A.; Hines, I.N.; Wheeler, M.D. Src kinase participates in LPS-induced activation of NADPH oxidase. Mol. Immunol. 2010, 47, 756–762. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, X.; Dai, X.; Lu, S.; Gui, B.; Jin, W.; Zhang, S.; Zhang, S.; Qian, Y. Lithium ameliorates lipopolysaccharide-induced microglial activation via inhibition of toll-like receptor 4 expression by activating the PI3K/Akt/FoxO1 pathway. J. Neuroinflamm. 2014, 11, 140. [Google Scholar] [CrossRef]
- Khadaroo, R.G.; Kapus, A.; Powers, K.A.; Cybulsky, M.I.; Marshall, J.C.; Rotstein, O.D. Oxidative stress reprograms lipopolysaccharide signaling via Src kinase-dependent pathway in RAW 264.7 macrophage cell line. J. Biol. Chem. 2003, 278, 47834–47841. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, G.; Combs, C.K. Inhibition of Src kinase activity attenuates amyloid associated microgliosis in a murine model of Alzheimer’s disease. J. Neuroinflamm. 2012, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Moghbeli, M. PI3K/AKT pathway as a pivotal regulator of epithelial-mesenchymal transition in lung tumor cells. Cancer Cell Int. 2024, 24, 165. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-κB in biology and targeted therapy: New insights and translational implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar]
- Li, C.; Liu, M.; Deng, L.; Luo, D.; Ma, R.; Lu, Q. Oxyberberine ameliorates TNBS-induced colitis in rats through suppressing inflammation and oxidative stress via Keap1/Nrf2/NF-κB signaling pathways. Phytomedicine 2023, 116, 154899. [Google Scholar] [CrossRef]
- Palmieri, E.M.; Gonzalez-Cotto, M.; Baseler, W.A.; Davies, L.C.; Ghesquière, B.; Maio, N.; Rice, C.M.; Rouault, T.A.; Cassel, T.; Higashi, R.M.; et al. Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat. Commun. 2020, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.Y.; Joo, S.H.; Lee, S.-O.; Yoon, G.; Cho, S.-S.; Choi, Y.H.; Park, J.W.; Shim, J.-H. Activation of p38 and JNK by ROS Contributes to Deoxybouvardin-Mediated Intrinsic Apoptosis in Oxaliplatin-Sensitive and -Resistant Colorectal Cancer Cells. Antioxidants 2024, 13, 866. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Yang, L.; Wan, C.X.; Xia, Y.Z.; Zhang, C.; Chen, M.H.; Wang, Z.D.; Li, Z.R.; Li, X.M.; Geng, Y.D.; et al. Anti-neuroinflammatory effect of Sophoraflavanone G from Sophora alopecuroides in LPS-activated BV2 microglia by MAPK, JAK/STAT and Nrf2/HO-1 signaling pathways. Phytomedicine 2016, 23, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Deng, Y.; Song, H.X.; Fang, Y.Y.; Gan, C.L.; Lin, J.J.; Luo, J.J.; Zheng, X.W.; Aschner, M.; Jiang, Y.M. Protective Effects of Sodium Para-Aminosalicylic Acid on Lead and Cadmium Co-Exposure in SH-SY5Y Cells. Brain Sci. 2023, 13, 382. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, Y.; Qiao, N.; Han, S.; Gao, J.; Lv, X.; Yuan, L.; Zhang, L.; Wei, Z. Anti-Neuroinflammatory Effect of Ombuin from Rhamnus erythroxylon Pall. Leaves in LPS-Induced BV-2 Microglia by Targeting Src and Suppressing the PI3K-AKT/NF-κB Signaling Pathway. Int. J. Mol. Sci. 2024, 25, 8789. https://doi.org/10.3390/ijms25168789
Bian Y, Qiao N, Han S, Gao J, Lv X, Yuan L, Zhang L, Wei Z. Anti-Neuroinflammatory Effect of Ombuin from Rhamnus erythroxylon Pall. Leaves in LPS-Induced BV-2 Microglia by Targeting Src and Suppressing the PI3K-AKT/NF-κB Signaling Pathway. International Journal of Molecular Sciences. 2024; 25(16):8789. https://doi.org/10.3390/ijms25168789
Chicago/Turabian StyleBian, Yanjie, Nan Qiao, Suyun Han, Jixiang Gao, Xiaofang Lv, Lihuan Yuan, Linjing Zhang, and Zuofu Wei. 2024. "Anti-Neuroinflammatory Effect of Ombuin from Rhamnus erythroxylon Pall. Leaves in LPS-Induced BV-2 Microglia by Targeting Src and Suppressing the PI3K-AKT/NF-κB Signaling Pathway" International Journal of Molecular Sciences 25, no. 16: 8789. https://doi.org/10.3390/ijms25168789
APA StyleBian, Y., Qiao, N., Han, S., Gao, J., Lv, X., Yuan, L., Zhang, L., & Wei, Z. (2024). Anti-Neuroinflammatory Effect of Ombuin from Rhamnus erythroxylon Pall. Leaves in LPS-Induced BV-2 Microglia by Targeting Src and Suppressing the PI3K-AKT/NF-κB Signaling Pathway. International Journal of Molecular Sciences, 25(16), 8789. https://doi.org/10.3390/ijms25168789



