Protein Fold Usages in Ribosomes: Another Glance to the Past
Abstract
1. Introduction
- Ce texte cite une «certaine encyclopédie chinoise » où il est écrit que «Les animaux se divisent en: (a) appartenant à l’Empereur, (b) embaumés, (c) apprivoisés, (d) cochons de lait, (e) sirènes, (f) fabuleux, (g) chiens en libertés, (h) inclus dans la présente classification, (i) qui s’agitent comme des fous, (j) innombrables, (k) dessinés avec un pinceau très fin en poil de chameau, (l) et caetera, (m) qui viennent de casser la cruche, (n) qui de loin semblent des mouches» (Borges). Préface de «les mots et les choses» Michel Foucault.
1.1. About Usages in Complex Systems
1.2. “To Be or Not to Be” a Protein Fold
1.3. Folding or Misfolding: That’s a Question of Ribosome?
2. Results
2.1. Ribosomal Protein Fold Distribution
2.1.1. Overview
2.1.2. OB and SH3 Domains—The Small Barrel SRFs
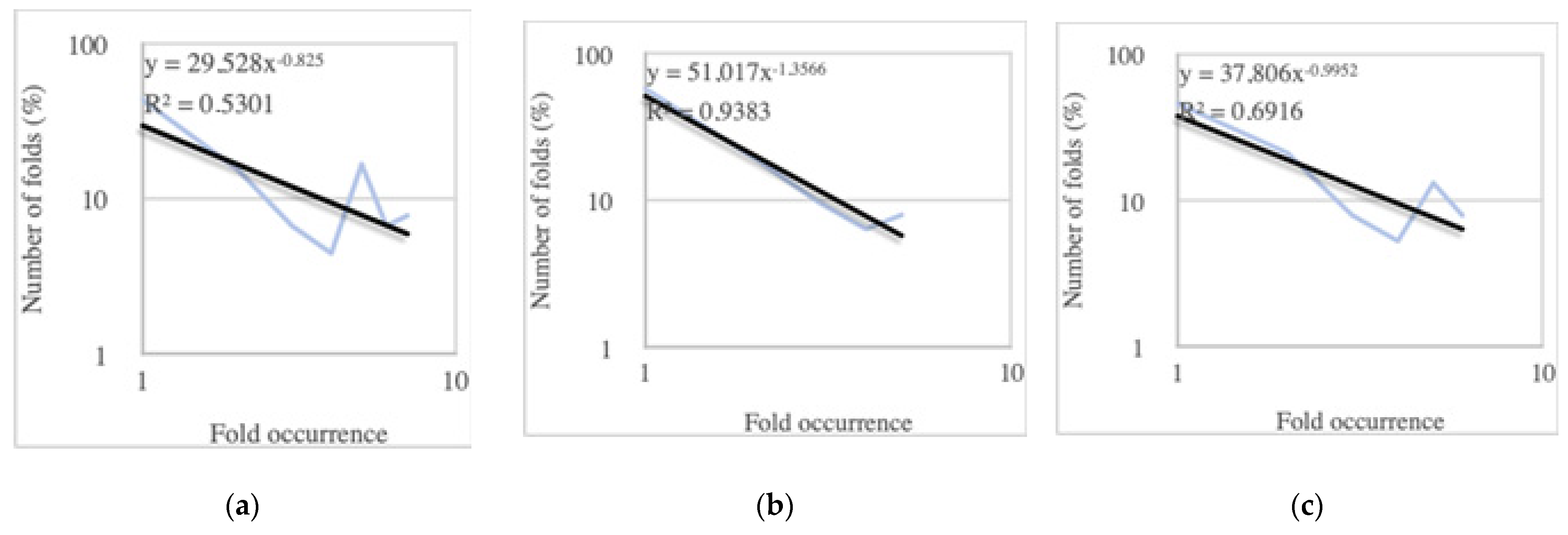
2.1.3. The Premise of the Power-Law Behavior
2.1.4. Different Fold Usages in the LSU and SSU
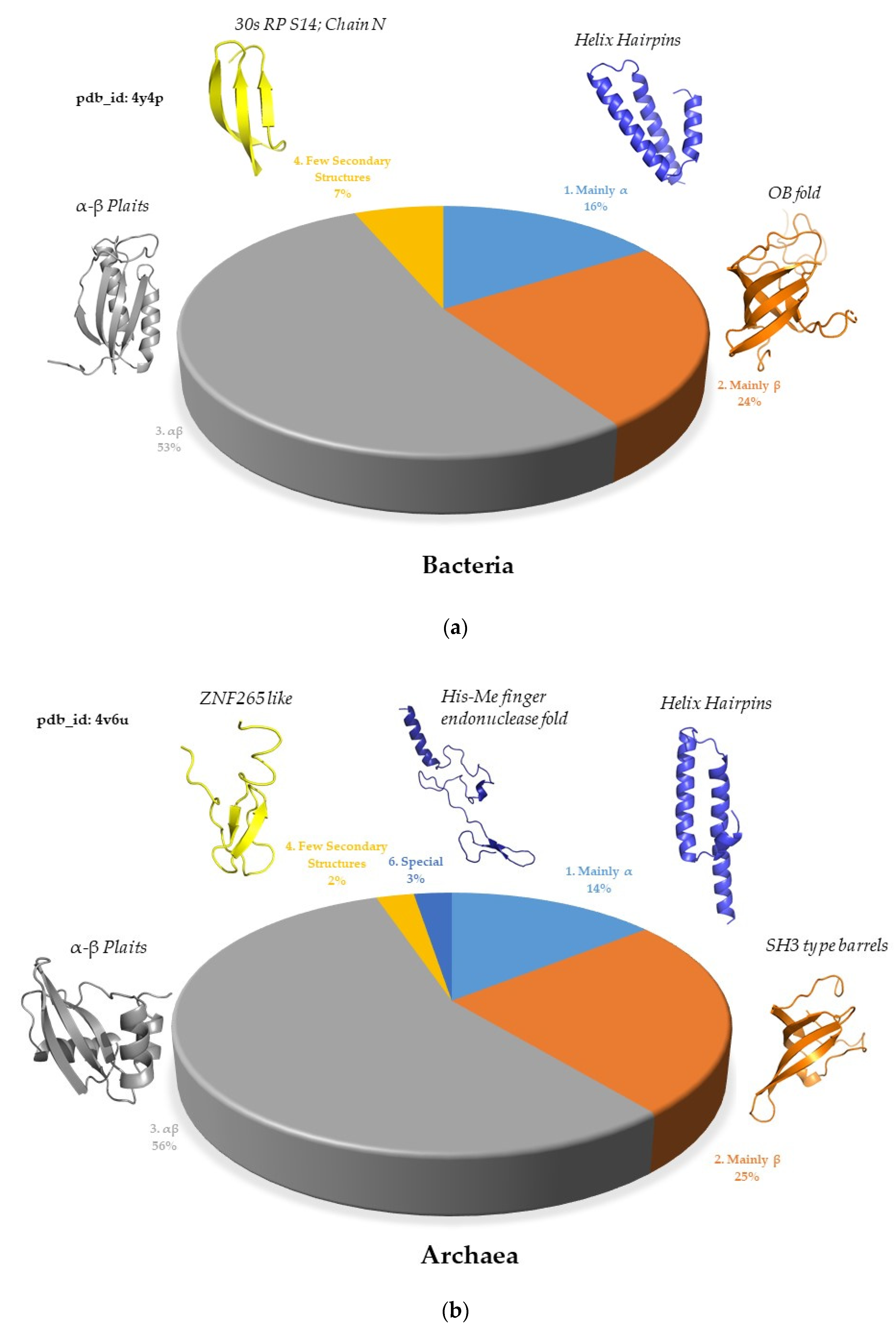
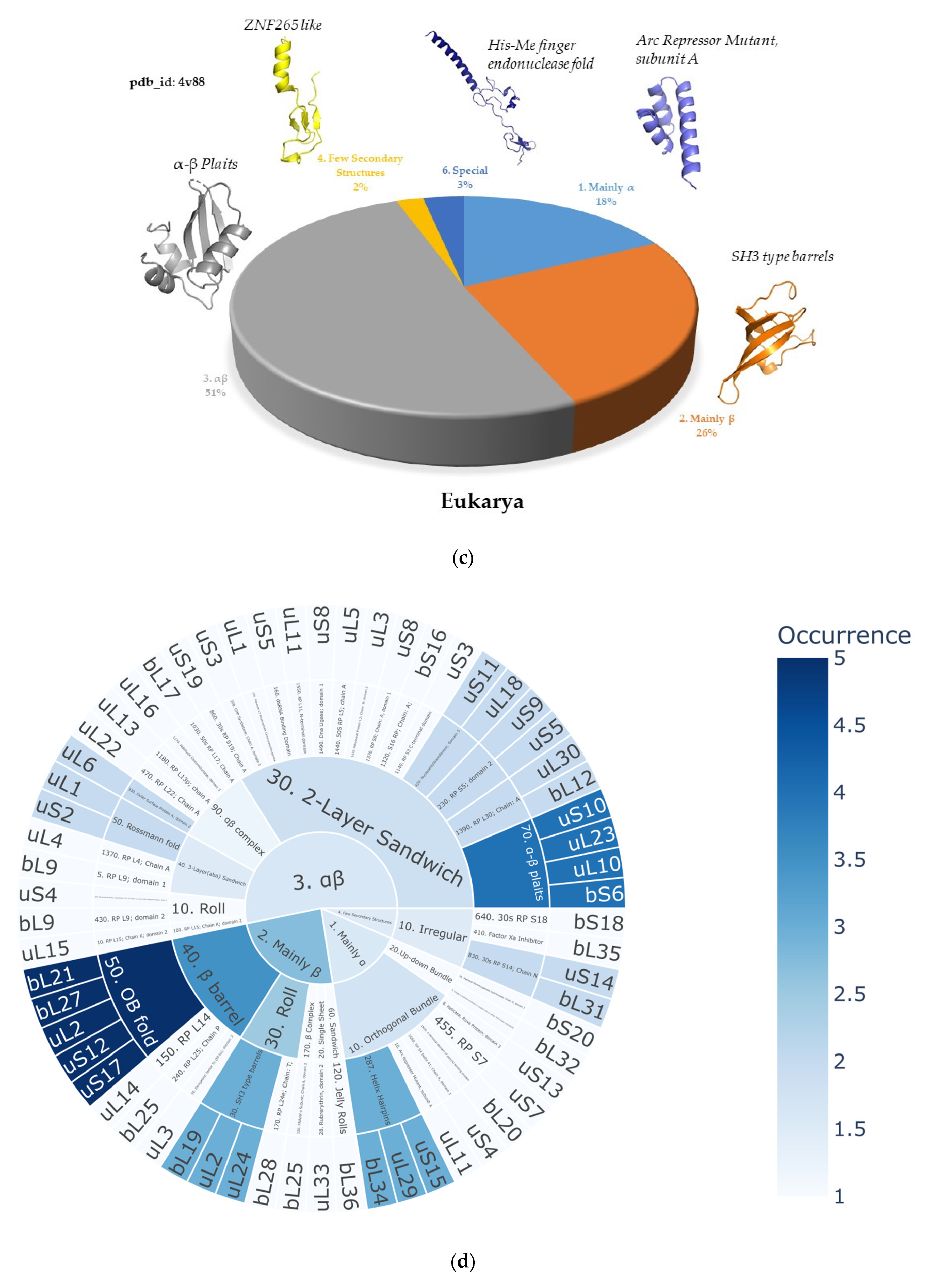
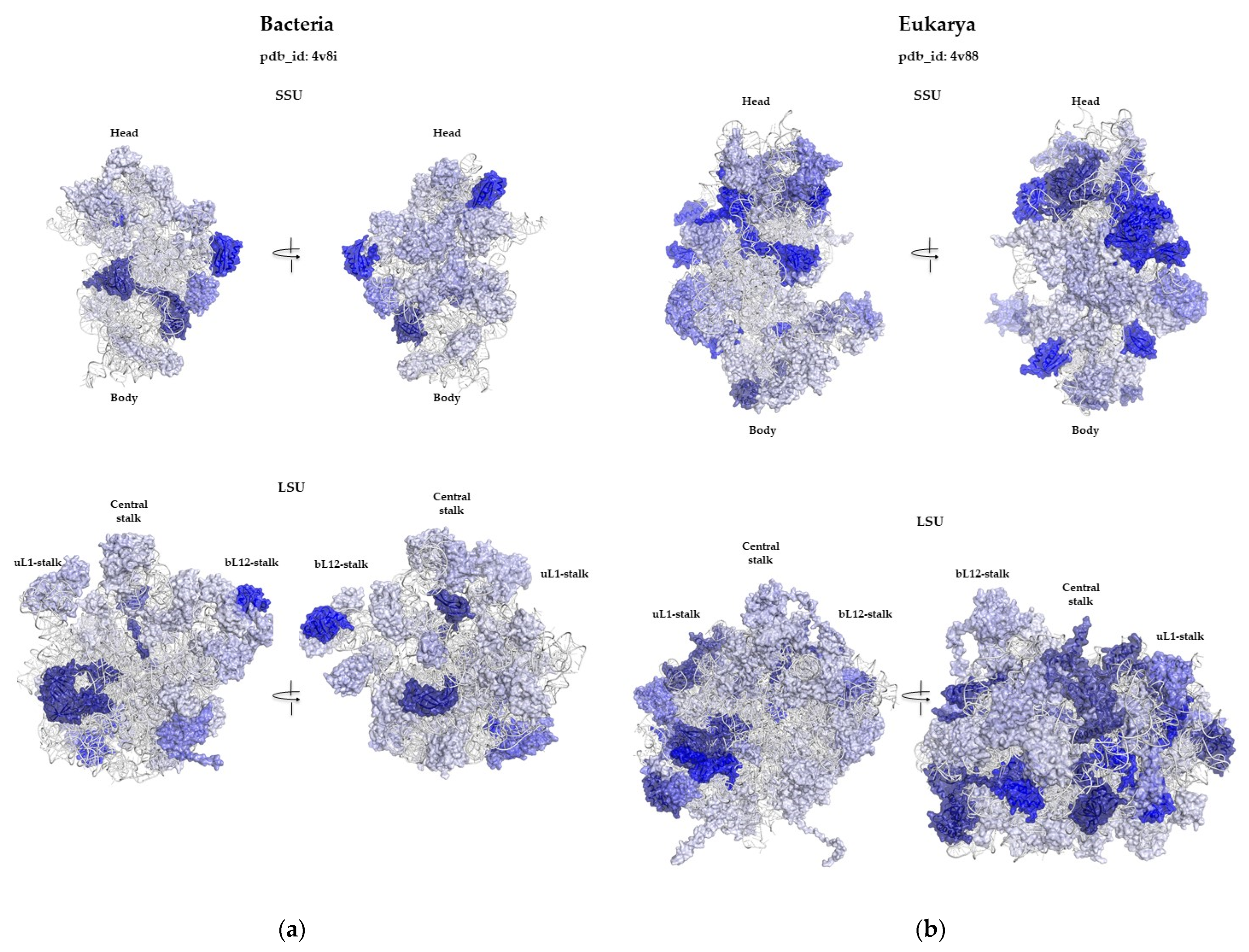
2.2. Ribosomal Protein Folds Specific to Each Kingdom
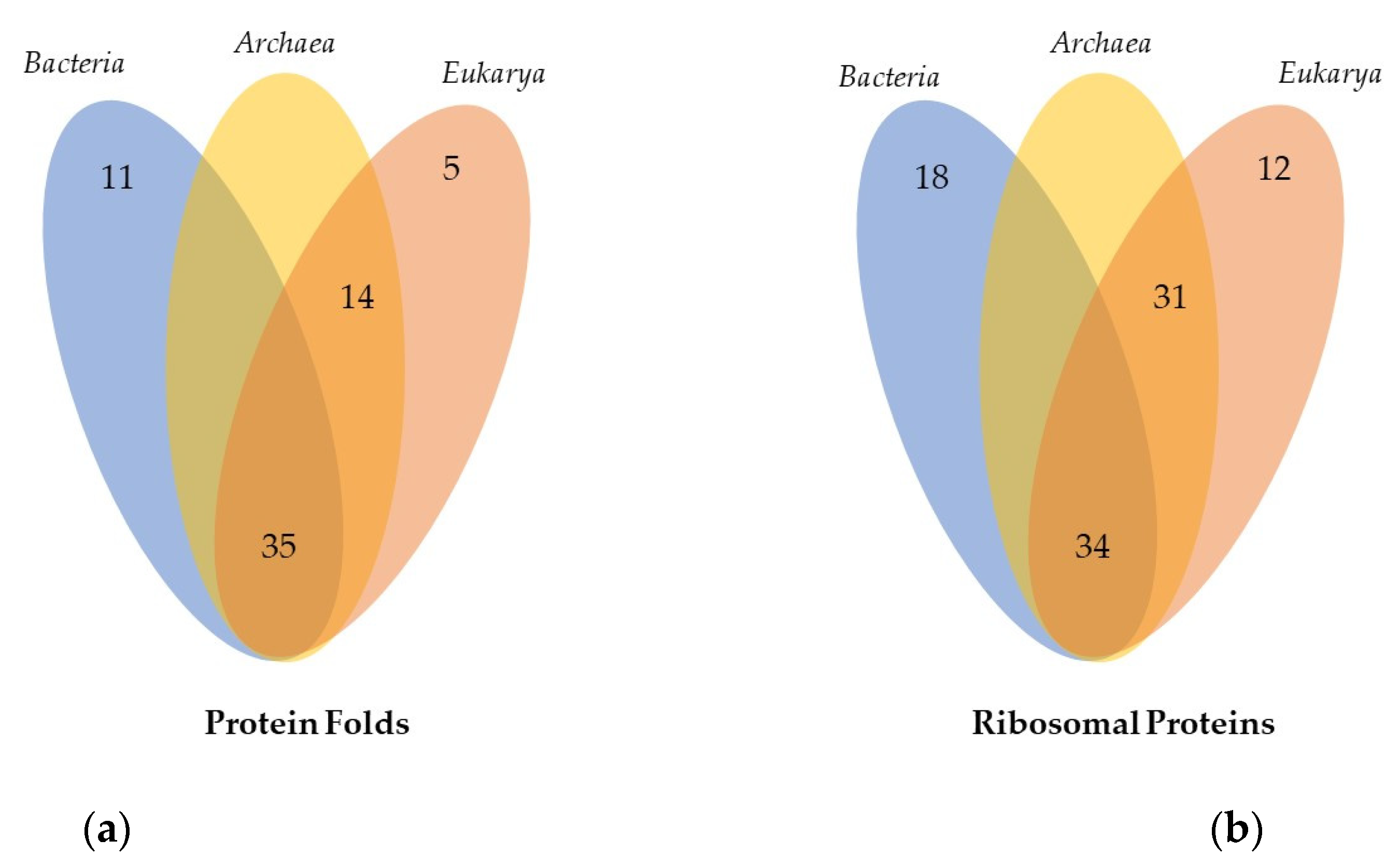
2.2.1. Minimal Fold Content
2.2.2. Different Behavior of Kingdom Specific Folds in Bacteria and Archaea/Eukarya
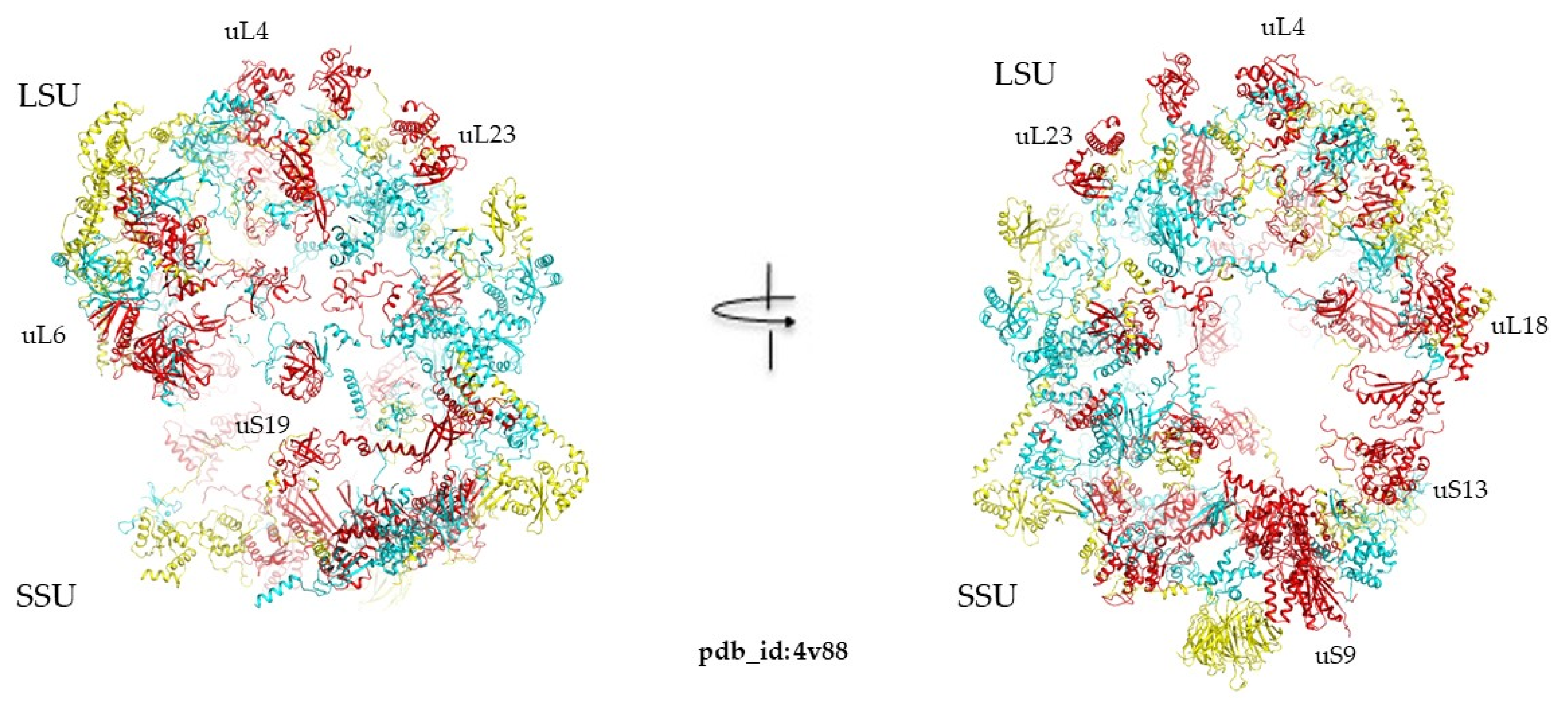
2.3. Imitation Game: Fold Mimicry across Kingdoms
2.4. Fold Evolution/Convergence
2.5. The Inexorable Expansion of Fold Extensions
3. Discussion
3.1. What’s New from the Past?
3.2. Ancestral Evolutionary Mechanisms with Different Selective Pressure
3.3. Imitation Games at the Ribosome Age
3.4. Order First, Disorder Next?
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newman, M. Power Laws, Pareto Distributions and Zipf’s Law. Contemp. Phys. 2005, 46, 323–351. [Google Scholar] [CrossRef]
- Caetano-Anollés, G. The Compressed Vocabulary of Microbial Life. Front. Microbiol. 2021, 12, 655990. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, M. Patterns of Protein-Fold Usage in Eight Microbial Genomes: A Comprehensive Structural Census. Proteins 1998, 33, 518–534. [Google Scholar] [CrossRef]
- Abeln, S.; Deane, C.M. Fold Usage on Genomes and Protein Fold Evolution. Proteins Struct. Funct. Bioinform. 2005, 60, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.; Ferrer-i-Cancho, R.; Gustison, M.L. Linguistic Laws in Biology. Trends Ecol. Evol. 2022, 37, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Are There Laws of Genome Evolution? PLoS Comput. Biol. 2011, 7, e1002173. [Google Scholar] [CrossRef] [PubMed]
- Sikorav, J. The Utility of Scientific Papers. Scientometrics 2005, 21, 49–68. [Google Scholar] [CrossRef]
- Ohta, M.; Yamamoto, K. Power-Law Relation and Complexity in the Shape of Chinese Character (Kanji). J. Phys. Soc. Jpn. 2019, 88, 064803. [Google Scholar] [CrossRef]
- Chierichetti, F.; Kumar, R.; Pang, B. On the Power Laws of Language: Word Frequency Distributions. In Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval, Tokyo, Japan, 7–11 August 2017; Association for Computing Machinery: New York, NY, USA, 2017; pp. 385–394. [Google Scholar]
- Seržant, I.A.; Moroz, G. Universal Attractors in Language Evolution Provide Evidence for the Kinds of Efficiency Pressures Involved. Humanit. Soc. Sci. Commun. 2022, 9, 58. [Google Scholar] [CrossRef]
- Piantadosi, S.T. Zipf’s Word Frequency Law in Natural Language: A Critical Review and Future Directions. Psychon. Bull. Rev. 2014, 21, 1112–1130. [Google Scholar] [CrossRef]
- Breaker, R.R. The Biochemical Landscape of Riboswitch Ligands. Biochemistry 2022, 61, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, T. Correlation between the Abundance of Escherichia Coli Transfer RNAs and the Occurrence of the Respective Codons in Its Protein Genes: A Proposal for a Synonymous Codon Choice That Is Optimal for the E. coli Translational System. J. Mol. Biol. 1981, 151, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but Not the Same: The Causes and Consequences of Codon Bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef]
- Komar, A.A.; Samatova, E.; Rodnina, M.V. Translation Rates and Protein Folding. J. Mol. Biol. 2023, 436, 168384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. A Code within the Genetic Code: Codon Usage Regulates Co-Translational Protein Folding. Cell Commun. Signal. 2020, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Orengo, C.A.; Jones, D.T.; Thornton, J.M. Protein Superfamilies and Domain Superfolds. Nature 1994, 372, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Luscombe, N.M.; Gerstein, M. Protein Family and Fold Occurrence in Genomes: Power-Law Behaviour and Evolutionary Model. J. Mol. Biol. 2001, 313, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Buchan, D.W.A.; Shepherd, A.J.; Lee, D.; Pearl, F.M.G.; Rison, S.C.G.; Thornton, J.M.; Orengo, C.A. Gene3D: Structural Assignment for Whole Genes and Genomes Using the CATH Domain Structure Database. Genome Res. 2002, 12, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.I.; Brenner, S.E.; Bash, P.A.; Koonin, E.V. Distribution of Protein Folds in the Three Superkingdoms of Life. Genome Res. 1999, 9, 17–26. [Google Scholar] [CrossRef]
- Hegyi, H.; Lin, J.; Greenbaum, D.; Gerstein, M. Structural Genomics Analysis: Characteristics of Atypical, Common, and Horizontally Transferred Folds. Proteins 2002, 47, 126–141. [Google Scholar] [CrossRef]
- Romei, M.; Carpentier, M.; Chomilier, J.; Lecointre, G. Origins and Functional Significance of Eukaryotic Protein Folds. J. Mol. Evol. 2023, 91, 854–864. [Google Scholar] [CrossRef]
- Romei, M.; Sapriel, G.; Imbert, P.; Jamay, T.; Chomilier, J.; Lecointre, G.; Carpentier, M. Protein Folds as Synapomorphies of the Tree of Life. Evol. Int. J. Org. Evol. 2022, 76, 1706–1719. [Google Scholar] [CrossRef] [PubMed]
- Kauko, A.; Lehto, K. Eukaryote Specific Folds: Part of the Whole. Proteins 2018, 86, 868–881. [Google Scholar] [CrossRef]
- Ponting, C.P.; Russell, R.R. The Natural History of Protein Domains. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 45–71. [Google Scholar] [CrossRef]
- Maggi, L.; Carloni, P.; Rossetti, G. Vibrational Energy in Proteins Correlates with Topology. J. Phys. Chem. Lett. 2018, 9, 6393–6398. [Google Scholar] [CrossRef]
- Xie, J.; Lai, L. Protein Topology and Allostery. Curr. Opin. Struct. Biol. 2020, 62, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, R.D.; Daggett, V. Protein Folds and Protein Folding. Protein Eng. Des. Sel. 2011, 24, 11–19. [Google Scholar] [CrossRef]
- Henzler-Wildman, K.; Kern, D. Dynamic Personalities of Proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Abrusán, G.; Marsh, J.A. Alpha Helices Are More Robust to Mutations than Beta Strands. PLOS Comput. Biol. 2016, 12, e1005242. [Google Scholar] [CrossRef]
- Tóth-Petróczy, Á.; Tawfik, D.S. The Robustness and Innovability of Protein Folds. Curr. Opin. Struct. Biol. 2014, 26, 131–138. [Google Scholar] [CrossRef]
- Bukhari, S.A.; Caetano-Anollés, G. Origin and Evolution of Protein Fold Designs Inferred from Phylogenomic Analysis of CATH Domain Structures in Proteomes. PLoS Comput. Biol. 2013, 9, e1003009. [Google Scholar] [CrossRef] [PubMed]
- Laurino, P.; Tóth-Petróczy, Á.; Meana-Pañeda, R.; Lin, W.; Truhlar, D.G.; Tawfik, D.S. An Ancient Fingerprint Indicates the Common Ancestry of Rossmann-Fold Enzymes Utilizing Different Ribose-Based Cofactors. PLOS Biol. 2016, 14, e1002396. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Caetano-Anollés, G. The Evolutionary History of Protein Fold Families and Proteomes Confirms That the Archaeal Ancestor Is More Ancient than the Ancestors of Other Superkingdoms. BMC Evol. Biol. 2012, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Ranea, J.A.G.; Buchan, D.W.A.; Thornton, J.M.; Orengo, C.A. Evolution of Protein Superfamilies and Bacterial Genome Size. J. Mol. Biol. 2004, 336, 871–887. [Google Scholar] [CrossRef] [PubMed]
- Winstanley, H.F.; Abeln, S.; Deane, C.M. How Old Is Your Fold? Bioinformatics 2005, 21, i449–i458. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V. Ribosome Structure and the Mechanism of Translation. Cell 2002, 108, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, S.; Ben-Shem, A.; Garreau De Loubresse, N.; Jenner, L.; Yusupova, G.; Yusupov, M. One Core, Two Shells: Bacterial and Eukaryotic Ribosomes. Nat. Struct. Mol. Biol. 2012, 19, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Bashan, A.; Agmon, I.; Zarivach, R.; Schluenzen, F.; Harms, J.; Berisio, R.; Bartels, H.; Franceschi, F.; Auerbach, T.; Hansen, H.A.S.; et al. Structural Basis of the Ribosomal Machinery for Peptide Bond Formation, Translocation, and Nascent Chain Progression. Mol. Cell 2003, 11, 91–102. [Google Scholar] [CrossRef]
- Klinge, S.; Voigts-Hoffmann, F.; Leibundgut, M.; Ban, N. Atomic Structures of the Eukaryotic Ribosome. Trends Biochem. Sci. 2012, 37, 189–198. [Google Scholar] [CrossRef]
- Wilson, D.N.; Doudna Cate, J.H. The Structure and Function of the Eukaryotic Ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4, a011536. [Google Scholar] [CrossRef]
- Armache, J.-P.; Anger, A.M.; Márquez, V.; Franckenberg, S.; Fröhlich, T.; Villa, E.; Berninghausen, O.; Thomm, M.; Arnold, G.J.; Beckmann, R.; et al. Promiscuous Behaviour of Archaeal Ribosomal Proteins: Implications for Eukaryotic Ribosome Evolution. Nucleic Acids Res. 2013, 41, 1284–1293. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Tirumalai, M.R.; Rivas, M.; Tran, Q.; Fox, G.E. The Peptidyl Transferase Center: A Window to the Past. Microbiol. Mol. Biol. Rev. 2021, 85, e00104-21. [Google Scholar] [CrossRef]
- Agmon, I. The Dimeric Proto-Ribosome: Structural Details and Possible Implications on the Origin of Life. Int. J. Mol. Sci. 2009, 10, 2921–2934. [Google Scholar] [CrossRef]
- Agmon, I. Three Biopolymers and Origin of Life Scenarios. Life 2024, 14, 277. [Google Scholar] [CrossRef]
- Belousoff, M.J.; Davidovich, C.; Zimmerman, E.; Caspi, Y.; Wekselman, I.; Rozenszajn, L.; Shapira, T.; Sade-Falk, O.; Taha, L.; Bashan, A.; et al. Ancient Machinery Embedded in the Contemporary Ribosome. Biochem. Soc. Trans. 2010, 38, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Opron, K.; Burton, Z.F. Ribosome Structure, Function, and Early Evolution. Int. J. Mol. Sci. 2018, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, M.; Root-Bernstein, R. The Ribosome as a Missing Link in the Evolution of Life. J. Theor. Biol. 2015, 367, 130–158. [Google Scholar] [CrossRef]
- Root-Bernstein, R.; Root-Bernstein, M. The Ribosome as a Missing Link in Prebiotic Evolution II: Ribosomes Encode Ribosomal Proteins That Bind to Common Regions of Their Own mRNAs and rRNAs. J. Theor. Biol. 2016, 397, 115–127. [Google Scholar] [CrossRef]
- Root-Bernstein, R.; Root-Bernstein, M. The Ribosome as a Missing Link in Prebiotic Evolution III: Over-Representation of tRNA- and rRNA-like Sequences and Plieofunctionality of Ribosome-Related Molecules Argues for the Evolution of Primitive Genomes from Ribosomal RNA Modules. Int. J. Mol. Sci. 2019, 20, 140. [Google Scholar] [CrossRef]
- Fox, G.E. Origin and Evolution of the Ribosome. Cold Spring Harb. Perspect. Biol. 2010, 2, a003483. [Google Scholar] [CrossRef]
- Rodgers, M.L.; Woodson, S.A. A Roadmap for rRNA Folding and Assembly during Transcription. Trends Biochem. Sci. 2021, 46, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.H.; Williamson, J.R. Structure and Dynamics of Bacterial Ribosome Biogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160181. [Google Scholar] [CrossRef]
- Steitz, T.A. A Structural Understanding of the Dynamic Ribosome Machine. Nat. Rev. Mol. Cell Biol. 2008, 9, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Noller, H.F.; Lancaster, L.; Mohan, S.; Zhou, J. Ribosome Structural Dynamics in Translocation: Yet Another Functional Role for Ribosomal RNA. Q. Rev. Biophys. 2017, 50, e12. [Google Scholar] [CrossRef]
- Noller, H.F. Evolution of Protein Synthesis from an RNA World. Cold Spring Harb. Perspect. Biol. 2012, 4, a003681. [Google Scholar] [CrossRef]
- Bose, T.; Fridkin, G.; Davidovich, C.; Krupkin, M.; Dinger, N.; Falkovich, A.H.; Peleg, Y.; Agmon, I.; Bashan, A.; Yonath, A. Origin of Life: Protoribosome Forms Peptide Bonds and Links RNA and Protein Dominated Worlds. Nucleic Acids Res. 2022, 50, 1815–1828. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.I.; Koonin, E.V. On the Origin of the Translation System and the Genetic Code in the RNA World by Means of Natural Selection, Exaptation, and Subfunctionalization. Biol. Direct 2007, 2, 14. [Google Scholar] [CrossRef]
- Grosjean, H.; Westhof, E. An Integrated, Structure- and Energy-Based View of the Genetic Code. Nucleic Acids Res. 2016, 44, 8020–8040. [Google Scholar] [CrossRef]
- Hartman, H.; Smith, T. The Evolution of the Ribosome and the Genetic Code. Life 2014, 4, 227–249. [Google Scholar] [CrossRef]
- Koonin, E.V.; Novozhilov, A.S. Origin and Evolution of the Universal Genetic Code. Annu. Rev. Genet. 2017, 51, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Lescoute, A.; Westhof, E. Topology of Three-Way Junctions in Folded RNAs. RNA 2006, 12, 83–93. [Google Scholar] [CrossRef]
- Cruz, J.A.; Westhof, E. Sequence-Based Identification of 3D Structural Modules in RNA with RMDetect. Nat. Methods 2011, 8, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Vitas, M.; Dobovišek, A. In the Beginning Was a Mutualism—On the Origin of Translation. Orig. Life Evol. Biosph. 2018, 48, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P. The Universal Tree of Life: An Update. Front. Microbiol. 2015, 6, 717. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.S.; Gulen, B.; Norris, A.M.; Kovacs, N.A.; Bernier, C.R.; Lanier, K.A.; Fox, G.E.; Harvey, S.C.; Wartell, R.M.; Hud, N.V.; et al. History of the Ribosome and the Origin of Translation. Proc. Natl. Acad. Sci. USA 2015, 112, 15396–15401. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Caetano-Anollés, D. Computing the Origin and Evolution of the Ribosome from Its Structure—Uncovering Processes of Macromolecular Accretion Benefiting Synthetic Biology. Comput. Struct. Biotechnol. J. 2015, 13, 427–447. [Google Scholar] [CrossRef] [PubMed]
- Harish, A.; Caetano-Anollés, G. Ribosomal History Reveals Origins of Modern Protein Synthesis. PLoS ONE 2012, 7, e32776. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N.; Nierhaus, K.H. Ribosomal Proteins in the Spotlight. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 243–267. [Google Scholar] [CrossRef]
- Graifer, D.; Karpova, G. Roles of Ribosomal Proteins in the Functioning of Translational Machinery of Eukaryotes. Biochimie 2015, 109, 1–17. [Google Scholar] [CrossRef]
- Brodersen, D.E.; Clemons, W.M.; Carter, A.P.; Wimberly, B.T.; Ramakrishnan, V. Crystal Structure of the 30 s Ribosomal Subunit from Thermus thermophilus: Structure of the Proteins and Their Interactions with 16 s RNA1. J. Mol. Biol. 2002, 316, 725–768. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.J.; Moore, P.B.; Steitz, T.A. The Roles of Ribosomal Proteins in the Structure Assembly, and Evolution of the Large Ribosomal Subunit. J. Mol. Biol. 2004, 340, 141–177. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Oldfield, C.J.; Xue, B.; Mizianty, M.J.; Dunker, A.K.; Kurgan, L.; Uversky, V.N. A Creature with a Hundred Waggly Tails: Intrinsically Disordered Proteins in the Ribosome. Cell. Mol. Life Sci. 2014, 71, 1477–1504. [Google Scholar] [CrossRef]
- Timsit, Y.; Acosta, Z.; Allemand, F.; Chiaruttini, C.; Springer, M. The Role of Disordered Ribosomal Protein Extensions in the Early Steps of Eubacterial 50 S Ribosomal Subunit Assembly. Int. J. Mol. Sci. 2009, 10, 817–834. [Google Scholar] [CrossRef]
- Calidas, D.; Lyon, H.; Culver, G.M. The N-Terminal Extension of S12 Influences Small Ribosomal Subunit Assembly in Escherichia coli. RNA 2014, 20, 321–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tutuncuoglu, B.; Jakovljevic, J.; Wu, S.; Gao, N.; Woolford, J.L. The N-Terminal Extension of Yeast Ribosomal Protein L8 Is Involved in Two Major Remodeling Events during Late Nuclear Stages of 60S Ribosomal Subunit Assembly. RNA 2016, 22, 1386–1399. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.G.; Shamsuzzaman, M.; Kondopaka, M.; Pascual, C.; Zengel, J.M.; Lindahl, L. The Extended Loops of Ribosomal Proteins uL4 and uL22 of Escherichia coli Contribute to Ribosome Assembly and Protein Translation. Nucleic Acids Res. 2016, 44, 5798–5810. [Google Scholar] [CrossRef]
- Rhodin, M.H.J.; Dinman, J.D. A Flexible Loop in Yeast Ribosomal Protein L11 Coordinates P-Site tRNA Binding. Nucleic Acids Res. 2010, 38, 8377–8389. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yutin, N.; Puigbò, P.; Koonin, E.V.; Wolf, Y.I. Phylogenomics of Prokaryotic Ribosomal Proteins. PLoS ONE 2012, 7, e36972. [Google Scholar] [CrossRef]
- Melnikov, S.; Manakongtreecheep, K.; Söll, D. Revising the Structural Diversity of Ribosomal Proteins across the Three Domains of Life. Mol. Biol. Evol. 2018, 35, 1588–1598. [Google Scholar] [CrossRef]
- Alva, V.; Söding, J.; Lupas, A.N. A Vocabulary of Ancient Peptides at the Origin of Folded Proteins. eLife 2015, 4, e09410. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.N.; Alva, V. Ribosomal Proteins as Documents of the Transition from Unstructured (Poly)Peptides to Folded Proteins. J. Struct. Biol. 2017, 198, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, N.A.; Petrov, A.S.; Lanier, K.A.; Williams, L.D. Frozen in Time: The History of Proteins. Mol. Biol. Evol. 2017, 34, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Poirot, O.; Timsit, Y. Neuron-like Networks between Ribosomal Proteins within the Ribosome. Sci. Rep. 2016, 6, 26485. [Google Scholar] [CrossRef] [PubMed]
- Timsit, Y.; Bennequin, D. Nervous-like Circuits in the Ribosome Facts, Hypotheses and Perspectives. Int. J. Mol. Sci. 2019, 20, 2911. [Google Scholar] [CrossRef] [PubMed]
- Timsit, Y.; Sergeant-Perthuis, G.; Bennequin, D. Evolution of Ribosomal Protein Network Architectures. Sci. Rep. 2021, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Timsit, Y.; Grégoire, S.-P. Towards the Idea of Molecular Brains. Int. J. Mol. Sci. 2021, 22, 11868. [Google Scholar] [CrossRef] [PubMed]
- Youkharibache, P.; Veretnik, S.; Li, Q.; Stanek, K.A.; Mura, C.; Bourne, P.E. The Small β-Barrel Domain: A Survey-Based Structural Analysis. Structure 2019, 27, 6–26. [Google Scholar] [CrossRef] [PubMed]
- Ajawatanawong, P.; Baldauf, S.L. Evolution of Protein Indels in Plants, Animals and Fungi. BMC Evol. Biol. 2013, 13, 140. [Google Scholar] [CrossRef]
- Wolf, Y.; Madej, T.; Babenko, V.; Shoemaker, B.; Panchenko, A.R. Long-Term Trends in Evolution of Indels in Protein Sequences. BMC Evol. Biol. 2007, 7, 19. [Google Scholar] [CrossRef][Green Version]
- Deryusheva, E.I.; Selivanova, O.M.; Serdyuk, I.N. Loops and Repeats in Proteins as Footprints of Molecular Evolution. Biochemistry 2012, 77, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Tóth-Petróczy, Á.; Tawfik, D.S. Protein Insertions and Deletions Enabled by Neutral Roaming in Sequence Space. Mol. Biol. Evol. 2013, 30, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Light, S.; Sagit, R.; Sachenkova, O.; Ekman, D.; Elofsson, A. Protein Expansion Is Primarily Due to Indels in Intrinsically Disordered Regions. Mol. Biol. Evol. 2013, 30, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.R. “To Be, or Not to Be”: Shakespeare against Philosophy. Shakespeare 2018, 14, 341–359. [Google Scholar] [CrossRef]
- Sikosek, T.; Chan, H.S. Biophysics of Protein Evolution and Evolutionary Protein Biophysics. J. R. Soc. Interface 2014, 11, 20140419. [Google Scholar] [CrossRef] [PubMed]
- Valas, R.E.; Yang, S.; Bourne, P.E. Nothing about Protein Structure Classification Makes Sense except in the Light of Evolution. Curr. Opin. Struct. Biol. 2009, 19, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Nassar, R.; Dignon, G.L.; Razban, R.M.; Dill, K.A. The Protein Folding Problem: The Role of Theory. J. Mol. Biol. 2021, 433, 167126. [Google Scholar] [CrossRef] [PubMed]
- Onuchic, J.N.; Wolynes, P.G. Theory of Protein Folding. Curr. Opin. Struct. Biol. 2004, 14, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Mirny, L.; Shakhnovich, E. Protein Folding Theory: From Lattice to All-Atom Models. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 361–396. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Argos, P. Protein Folding. Annu. Rev. Biochem. 1981, 50, 497–532. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Røgen, P.; Paci, E.; Vendruscolo, M.; Dobson, C.M. Protein Folding and the Organization of the Protein Topology Universe. Trends Biochem. Sci. 2005, 30, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein Folding and Misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A.R.; Daggett, V. Protein Folding and Unfolding at Atomic Resolution. Cell 2002, 108, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Mirny, L.A.; Shakhnovich, E.I. Universally Conserved Positions in Protein Folds: Reading Evolutionary Signals about Stability, Folding Kinetics and Function. J. Mol. Biol. 1999, 291, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Gruebele, M. Protein Folding: The Free Energy Surface. Curr. Opin. Struct. Biol. 2002, 12, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, I.; Mushegian, A.R.; Koonin, E.V. Is Protein Folding a Thermodynamically Unfavorable, Active, Energy-Dependent Process? Int. J. Mol. Sci. 2022, 23, 521. [Google Scholar] [CrossRef]
- Agard, D.A. To Fold or Not to Fold. Science 1993, 260, 1903–1904. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Rothman, J.E.; Schekman, R. Molecular Mechanism of Protein Folding in the Cell. Cell 2011, 146, 851–854. [Google Scholar] [CrossRef]
- Levitt, M.; Chothia, C. Structural Patterns in Globular Proteins. Nature 1976, 261, 552–558. [Google Scholar] [CrossRef]
- Chothia, C. Principles That Determine the Structure of Proteins. Annu. Rev. Biochem. 1984, 53, 537–572. [Google Scholar] [CrossRef] [PubMed]
- Chothia, C.; Finkelstein, A.V. The Classification and Origins of Protein Folding Patterns. Annu. Rev. Biochem. 1990, 59, 1007–1035. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: A Structural Classification of Proteins Database for the Investigation of Sequences and Structures. J. Mol. Biol. 1995, 247, 536–540. [Google Scholar] [CrossRef]
- Orengo, C.; Michie, A.; Jones, S.; Jones, D.; Swindells, M.; Thornton, J. CATH—A Hierarchic Classification of Protein Domain Structures. Structure 1997, 5, 1093–1109. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Dancing Protein Clouds: The Strange Biology and Chaotic Physics of Intrinsically Disordered Proteins. J. Biol. Chem. 2016, 291, 6681–6688. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing Protein Intrinsic Disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef]
- Towse, C.-L.; Daggett, V. When a Domain Is Not a Domain, and Why It Is Important to Properly Filter Proteins in Databases: Conflicting Definitions and Fold Classification Systems for Structural Domains Make Filtering of Such Databases Imperative. BioEssays 2012, 34, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Abel, K.; Yoder, M.D.; Hilgenfeld, R.; Jurnak, F. An Alpha to Beta Conformational Switch in EF-Tu. Structure 1996, 4, 1153–1159. [Google Scholar] [CrossRef]
- Polekhina, G.; Thirup, S.; Kjeldgaard, M.; Nissen, P.; Lippmann, C.; Nyborg, J. Helix Unwinding in the Effector Region of Elongation Factor EF-Tu-GDP. Structure 1996, 4, 1141–1151. [Google Scholar] [CrossRef]
- Bhattacharjee, N.; Biswas, P. Statistical Analysis and Molecular Dynamics Simulations of Ambivalent α-Helices. BMC Bioinform. 2010, 11, 519. [Google Scholar] [CrossRef]
- Guo, J.-T.; Jaromczyk, J.W.; Xu, Y. Analysis of Chameleon Sequences and Their Implications in Biological Processes. Proteins 2007, 67, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, I.B.; Rackovsky, S. On the Properties and Sequence Context of Structurally Ambivalent Fragments in Proteins. Protein Sci. Publ. Protein Soc. 2003, 12, 2420–2433. [Google Scholar] [CrossRef] [PubMed]
- Young, M.; Kirshenbaum, K.; Dill, K.A.; Highsmith, S. Predicting Conformational Switches in Proteins. Protein Sci. Publ. Protein Soc. 1999, 8, 1752–1764. [Google Scholar] [CrossRef]
- Pauwels, K.; Lebrun, P.; Tompa, P. To Be Disordered or Not to Be Disordered: Is That Still a Question for Proteins in the Cell? Cell. Mol. Life Sci. 2017, 74, 3185–3204. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Basile, W.; Salvatore, M.; Bassot, C.; Elofsson, A. Why Do Eukaryotic Proteins Contain More Intrinsically Disordered Regions? PLOS Comput. Biol. 2019, 15, e1007186. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Galzitskaya, O.V. Occurrence of Disordered Patterns and Homorepeats in Eukaryotic and Bacterial Proteomes. Mol. BioSyst. 2012, 8, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, J.B.; Nunez-Castilla, J.; Siltberg-Liberles, J. Evolution of Intrinsic Disorder in Eukaryotic Proteins. Cell. Mol. Life Sci. 2017, 74, 3163–3174. [Google Scholar] [CrossRef] [PubMed]
- Lermyte, F. Roles, Characteristics, and Analysis of Intrinsically Disordered Proteins: A Minireview. Life 2020, 10, 320. [Google Scholar] [CrossRef]
- Fuxreiter, M. Electrostatics Tunes Protein Interactions to Context. Proc. Natl. Acad. Sci. USA 2022, 119, e2209201119. [Google Scholar] [CrossRef]
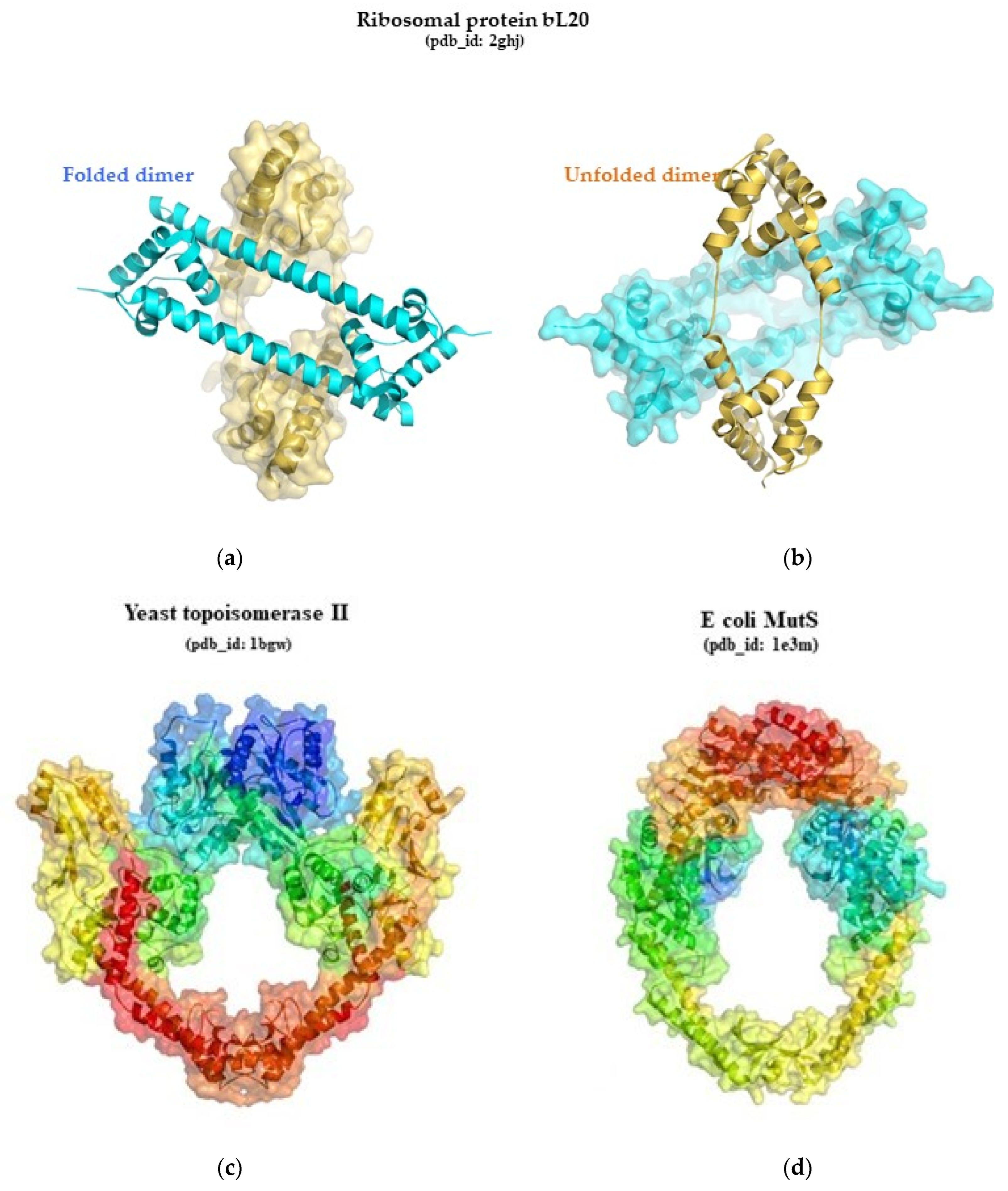
- Timsit, Y.; Allemand, F.; Chiaruttini, C.; Springer, M. Coexistence of Two Protein Folding States in the Crystal Structure of Ribosomal Protein L20. EMBO Rep. 2006, 7, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Dishman, A.F.; Tyler, R.C.; Fox, J.C.; Kleist, A.B.; Prehoda, K.E.; Babu, M.M.; Peterson, F.C.; Volkman, B.F. Evolution of Fold Switching in a Metamorphic Protein. Science 2021, 371, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Dishman, A.F.; Volkman, B.F. Metamorphic Protein Folding as Evolutionary Adaptation. Trends Biochem. Sci. 2023, 48, 665–672. [Google Scholar] [CrossRef]
- Porter, L.L.; Artsimovitch, I.; Ramírez-Sarmiento, C.A. Metamorphic Proteins and How to Find Them. Curr. Opin. Struct. Biol. 2024, 86, 102807. [Google Scholar] [CrossRef]
- Porter, L.L. Fluid Protein Fold Space and Its Implications. BioEssays 2023, 45, e2300057. [Google Scholar] [CrossRef]
- Schafer, J.W.; Porter, L.L. Evolutionary Selection of Proteins with Two Folds. Nat. Commun. 2023, 14, 5478. [Google Scholar] [CrossRef]
- Knauer, S.H.; Artsimovitch, I.; Rösch, P. Transformer Proteins. Cell Cycle 2012, 11, 4289–4290. [Google Scholar] [CrossRef][Green Version]
- Knauer, S.H.; Rösch, P.; Artsimovitch, I. Transformation: The next Level of Regulation. RNA Biol. 2012, 9, 1418–1423. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Das, M.; Chen, N.; LiWang, A.; Wang, L.-P. Identification and Characterization of Metamorphic Proteins: Current and Future Perspectives. Biopolymers 2021, 112, e23473. [Google Scholar] [CrossRef]
- Bahramali, G.; Goliaei, B.; Minuchehr, Z.; Salari, A. Chameleon Sequences in Neurodegenerative Diseases. Biochem. Biophys. Res. Commun. 2016, 472, 209–216. [Google Scholar] [CrossRef]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef] [PubMed]
- Collinge, J. Mammalian Prions and Their Wider Relevance in Neurodegenerative Diseases. Nature 2016, 539, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Hornemann, S.; Wider, G.; Billeter, M.; Glockshuber, R.; Wüthrich, K. NMR Structure of the Mouse Prion Protein Domain PrP(121–231). Nature 1996, 382, 180–182. [Google Scholar] [CrossRef] [PubMed]
- Zahn, R.; Liu, A.; Lührs, T.; Riek, R.; Von Schroetter, C.; López García, F.; Billeter, M.; Calzolai, L.; Wider, G.; Wüthrich, K. NMR Solution Structure of the Human Prion Protein. Proc. Natl. Acad. Sci. USA 2000, 97, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.M.; Baldwin, M.; Nguyen, J.; Gasset, M.; Serban, A.; Groth, D.; Mehlhorn, I.; Huang, Z.; Fletterick, R.J.; Cohen, F.E. Conversion of Alpha-Helices into Beta-Sheets Features in the Formation of the Scrapie Prion Proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 10962–10966. [Google Scholar] [CrossRef]
- Stubbs, G.; Stöhr, J. Structural Biology of PrP Prions. Cold Spring Harb. Perspect. Med. 2017, 7, a024455. [Google Scholar] [CrossRef] [PubMed]
- Nafe, R.; Hattingen, E. The Spectrum of Molecular Pathways in Gliomas—An Up-to-Date Review. Biomedicines 2023, 11, 2281. [Google Scholar] [CrossRef] [PubMed]
- Louros, N.; Schymkowitz, J.; Rousseau, F. Mechanisms and Pathology of Protein Misfolding and Aggregation. Nat. Rev. Mol. Cell Biol. 2023, 24, 912–933. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Wu, S.; Gregg, K. Prions Are the Greatest Protein Misfolding Problem, and Yeast Has Several Solutions. PLOS Pathog. 2023, 19, e1011333. [Google Scholar] [CrossRef]
- Spagnolli, G.; Rigoli, M.; Orioli, S.; Sevillano, A.M.; Faccioli, P.; Wille, H.; Biasini, E.; Requena, J.R. Full Atomistic Model of Prion Structure and Conversion. PLOS Pathog. 2019, 15, e1007864. [Google Scholar] [CrossRef]
- Kovač, V.; Čurin Šerbec, V. Prion Protein: The Molecule of Many Forms and Faces. Int. J. Mol. Sci. 2022, 23, 1232. [Google Scholar] [CrossRef] [PubMed]
- King, O.D.; Gitler, A.D.; Shorter, J. The Tip of the Iceberg: RNA-Binding Proteins with Prion-like Domains in Neurodegenerative Disease. Brain Res. 2012, 1462, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Scheckel, C.; Imeri, M.; Schwarz, P.; Aguzzi, A. Ribosomal Profiling during Prion Disease Uncovers Progressive Translational Derangement in Glia but Not in Neurons. eLife 2020, 9, e62911. [Google Scholar] [CrossRef]
- Louka, A.; Zacco, E.; Temussi, P.A.; Tartaglia, G.G.; Pastore, A. RNA as the Stone Guest of Protein Aggregation. Nucleic Acids Res. 2020, 48, 11880–11889. [Google Scholar] [CrossRef]
- Iben, S. To Aggregate or Not to Aggregate—Is It a Matter of the Ribosome? BioEssays 2023, 45, 2200230. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Shewmaker, F.P.; Kryndushkin, D.; Nemecek, J.; McGlinchey, R.; Bateman, D. The Relationship of Prions and Translation. WIREs RNA 2010, 1, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Mura, C.; Veretnik, S.; Bourne, P.E. The Urfold: Structural Similarity Just above the Superfold Level? Protein Sci. Publ. Protein Soc. 2019, 28, 2119–2126. [Google Scholar] [CrossRef] [PubMed]
- Alva, V.; Koretke, K.K.; Coles, M.; Lupas, A.N. Cradle-Loop Barrels and the Concept of Metafolds in Protein Classification by Natural Descent. Curr. Opin. Struct. Biol. 2008, 18, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Konagurthu, A.S.; Subramanian, R.; Allison, L.; Abramson, D.; Stuckey, P.J.; Garcia De La Banda, M.; Lesk, A.M. Universal Architectural Concepts Underlying Protein Folding Patterns. Front. Mol. Biosci. 2021, 7, 612920. [Google Scholar] [CrossRef]
- Alvarez-Carreño, C.; Penev, P.I.; Petrov, A.S.; Williams, L.D. Fold Evolution before LUCA: Common Ancestry of SH3 Domains and OB Domains. Mol. Biol. Evol. 2021, 38, 5134–5143. [Google Scholar] [CrossRef]
- Kopec, K.O.; Lupas, A.N. β-Propeller Blades as Ancestral Peptides in Protein Evolution. PLoS ONE 2013, 8, e77074. [Google Scholar] [CrossRef]
- Longo, L.M.; Jabłońska, J.; Vyas, P.; Kanade, M.; Kolodny, R.; Ben-Tal, N.; Tawfik, D.S. On the Emergence of P-Loop NTPase and Rossmann Enzymes from a Beta-Alpha-Beta Ancestral Fragment. eLife 2020, 9, e64415. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, K.E.; Kinch, L.N.; Dustin Schaeffer, R.; Pei, J.; Grishin, N.V. A Fifth of the Protein World: Rossmann-like Proteins as an Evolutionarily Successful Structural Unit. J. Mol. Biol. 2021, 433, 166788. [Google Scholar] [CrossRef] [PubMed]
- Alva, V.; Remmert, M.; Biegert, A.; Lupas, A.N.; Söding, J. A Galaxy of Folds. Protein Sci. Publ. Protein Soc. 2010, 19, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Khersonsky, O.; Tawfik, D.S. Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annu. Rev. Biochem. 2010, 79, 471–505. [Google Scholar] [CrossRef] [PubMed]
- Nagano, N.; Orengo, C.A.; Thornton, J.M. One Fold with Many Functions: The Evolutionary Relationships between TIM Barrel Families Based on Their Sequences, Structures and Functions. J. Mol. Biol. 2002, 321, 741–765. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Koonin, E.V. Divergence and Convergence in Enzyme Evolution. J. Biol. Chem. 2012, 287, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Beeby, M.; Ferreira, J.L.; Tripp, P.; Albers, S.-V.; Mitchell, D.R. Propulsive Nanomachines: The Convergent Evolution of Archaella, Flagella and Cilia. FEMS Microbiol. Rev. 2020, 44, 253–304. [Google Scholar] [CrossRef] [PubMed]
- Timsit, Y.; Lescot, M.; Valiadi, M.; Not, F. Bioluminescence and Photoreception in Unicellular Organisms: Light-Signalling in a Bio-Communication Perspective. Int. J. Mol. Sci. 2021, 22, 11311. [Google Scholar] [CrossRef]
- Hall, R. Molecular Mimicry. Adv. Parasitol. 1994, 34, 81–132. [Google Scholar] [CrossRef]
- Tsonis, P.A.; Dwivedi, B. Molecular Mimicry: Structural Camouflage of Proteins and Nucleic Acids. Biochim. Biophys. Acta 2008, 1783, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Nissen, P.; Kjeldgaard, M.; Nyborg, J. Macromolecular Mimicry. EMBO J. 2000, 19, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Chemes, L.B.; de Prat-Gay, G.; Sánchez, I.E. Convergent Evolution and Mimicry of Protein Linear Motifs in Host-Pathogen Interactions. Curr. Opin. Struct. Biol. 2015, 32, 91–101. [Google Scholar] [CrossRef]
- Beckett, D. Functional Switches in Transcription Regulation; Molecular Mimicry and Plasticity in Protein-Protein Interactions. Biochemistry 2004, 43, 7983–7991. [Google Scholar] [CrossRef]
- Putnam, C.D.; Tainer, J.A. Protein Mimicry of DNA and Pathway Regulation. DNA Repair. 2005, 4, 1410–1420. [Google Scholar] [CrossRef]
- Timsit, Y. Convergent Evolution of MutS and Topoisomerase II for Clamping DNA Crossovers and Stacked Holliday Junctions. J. Biomol. Struct. Dyn. 2001, 19, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Beckmann, R.; Cate, J.H.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A New System for Naming Ribosomal Proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Lecompte, O. Comparative Analysis of Ribosomal Proteins in Complete Genomes: An Example of Reductive Evolution at the Domain Scale. Nucleic Acids Res. 2002, 30, 5382–5390. [Google Scholar] [CrossRef]
- Sulima, S.O.; Dinman, J.D. The Expanding Riboverse. Cells 2019, 8, 1205. [Google Scholar] [CrossRef]
- Dinman, J.D. Pathways to Specialized Ribosomes: The Brussels Lecture. J. Mol. Biol. 2016, 428, 2186–2194. [Google Scholar] [CrossRef]
- Chen, Y.-X.; Xu, Z.; Ge, X.; Hong, J.-Y.; Sanyal, S.; Lu, Z.J.; Javid, B. Selective Translation by Alternative Bacterial Ribosomes. Proc. Natl. Acad. Sci. USA 2020, 117, 19487–19496. [Google Scholar] [CrossRef]
- Martinez-Seidel, F.; Beine-Golovchuk, O.; Hsieh, Y.-C.; Eshraky, K.E.; Gorka, M.; Cheong, B.-E.; Jimenez-Posada, E.V.; Walther, D.; Skirycz, A.; Roessner, U.; et al. Spatially Enriched Paralog Rearrangements Argue Functionally Diverse Ribosomes Arise during Cold Acclimation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6160. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.G.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Achsel, T.; Stark, H.; Lührmann, R. The Sm Domain Is an Ancient RNA-Binding Motif with Oligo(U) Specificity. Proc. Natl. Acad. Sci. USA 2001, 98, 3685–3689. [Google Scholar] [CrossRef]
- Theobald, D.L.; Mitton-Fry, R.M.; Wuttke, D.S. Nucleic Acid Recognition by OB-Fold Proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 115–133. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G. OB(Oligonucleotide/Oligosaccharide Binding)-Fold: Common Structural and Functional Solution for Non-Homologous Sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Dalgarno, D.C.; Botfield, M.C.; Rickles, R.J. SH3 Domains and Drug Design: Ligands, Structure, and Biological Function. Biopolymers 1997, 43, 383–400. [Google Scholar] [CrossRef]
- Nikonov, S.; Nevskaya, N.; Eliseikina, I.; Fomenkova, N.; Nikulin, A.; Ossina, N.; Garber, M.; Jonsson, B.H.; Briand, C.; Al-Karadaghi, S.; et al. Crystal Structure of the RNA Binding Ribosomal Protein L1 from Thermus Thermophilus. EMBO J. 1996, 15, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, K.E.; Kinch, L.N.; Schaeffer, R.D.; Grishin, N.V. Functional Analysis of Rossmann-like Domains Reveals Convergent Evolution of Topology and Reaction Pathways. PLOS Comput. Biol. 2019, 15, e1007569. [Google Scholar] [CrossRef]
- Pillai, A.S.; Hochberg, G.K.A.; Thornton, J.W. Simple Mechanisms for the Evolution of Protein Complexity. Protein Sci. 2022, 31, e4449. [Google Scholar] [CrossRef]
- Jiang, H.; Blouin, C. Insertions and the Emergence of Novel Protein Structure: A Structure-Based Phylogenetic Study of Insertions. BMC Bioinform. 2007, 8, 444. [Google Scholar] [CrossRef]
- Goldman, A.D.; Beatty, J.T.; Landweber, L.F. The TIM Barrel Architecture Facilitated the Early Evolution of Protein-Mediated Metabolism. J. Mol. Evol. 2016, 82, 17–26. [Google Scholar] [CrossRef] [PubMed]
- CATH: Protein Structure Classification Database; University College London: London, UK, 2024.
- Worbs, M.; Huber, R.; Wahl, M.C. Crystal Structure of Ribosomal Protein L4 Shows RNA-Binding Sites for Ribosome Incorporation and Feedback Control of the S10 Operon. EMBO J. 2000, 19, 807–818. [Google Scholar] [CrossRef]
- Nakashima, T.; Yao, M.; Kawamura, S.; Iwasaki, K.; Kimura, M.; Tanaka, I. Ribosomal Protein L5 Has a Highly Twisted Concave Surface and Flexible Arms Responsible for rRNA Binding. RNA 2001, 7, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.; White, S.W. Ribosomal Protein Structures: Insights into the Architecture, Machinery and Evolution of the Ribosome. Trends Biochem. Sci. 1998, 23, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Romero, P.; Obradovic, Z.; Garner, E.C.; Brown, C.J. Intrinsic Protein Disorder in Complete Genomes. Genome Inform. 2000, 11, 161–171. [Google Scholar]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and Combining Predictors of Mostly Disordered Proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.; Söding, J.; Steinegger, M. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2024, 42, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Laiho, A.; Toronen, P.; Salgado, M. DALI shines a light on remote homologs: One hundred discoveries. Protein Sci. 2023, 23, e4519. [Google Scholar] [CrossRef]
- PyMOL Molecular Graphics System, Version 2.2.2; Schrodinger, LLC: New York, NY, USA, 2020.





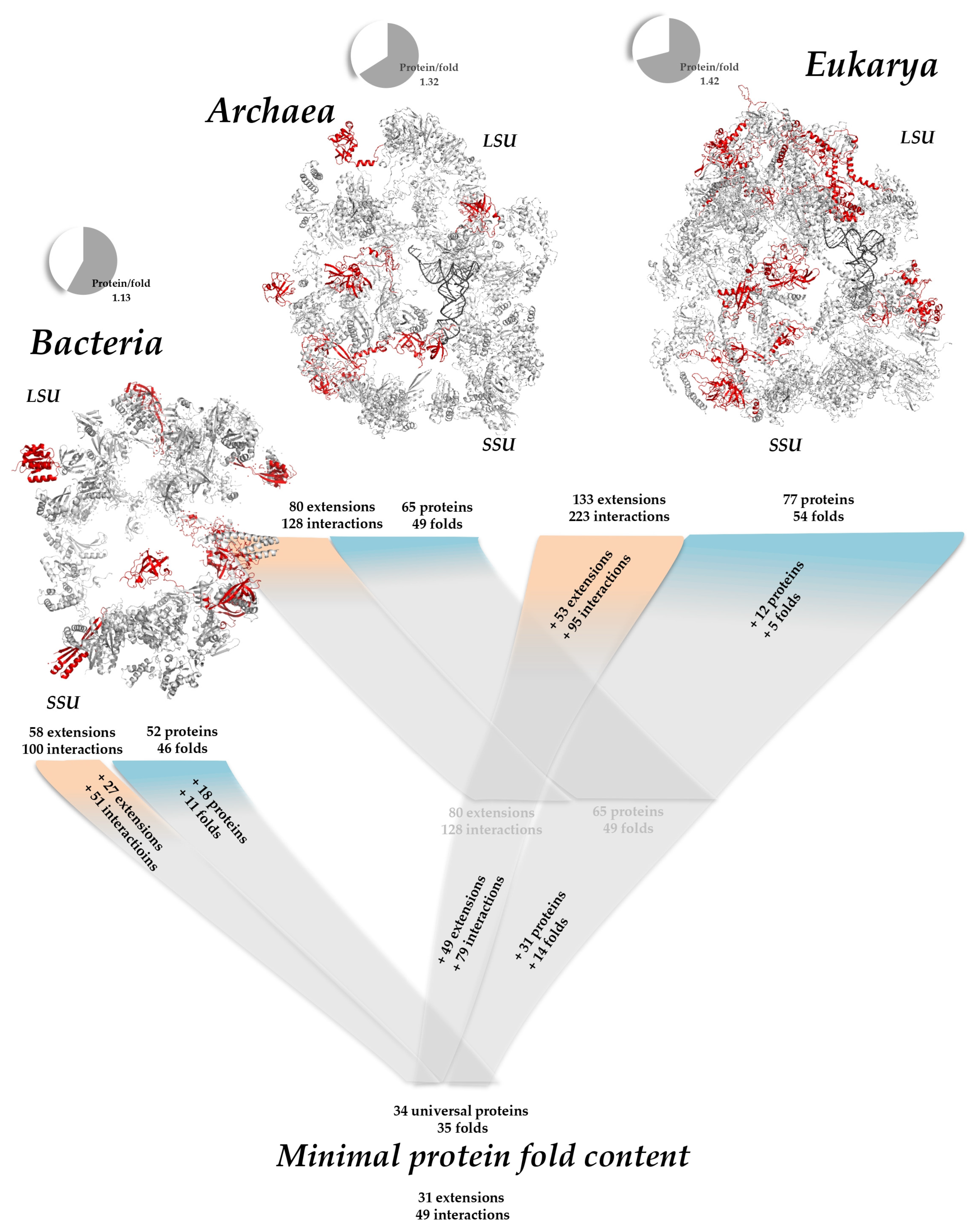
| Kingdom | Proteins | Folds | Protein/Fold Ratio | Nucleotides | |
|---|---|---|---|---|---|
| Bacteria | LSU | 33 | 32 | 1.03 | 3036 † |
| SSU | 19 | 20 | 0.95 | 1521 † | |
| Total | 52 | 46 * | 1.13 | 4557 † | |
| Archaea | LSU | 39 | 33 | 1.18 | 3175 † |
| SSU | 26 | 25 | 1.04 | 1495 † | |
| Total | 65 | 49 * | 1.32 | 4670 † | |
| Eukarya | LSU | 44 | 36 | 1.22 | 3675 † |
| SSU | 33 | 29 | 1.14 | 1800 † | |
| Total | 77 | 54 * | 1.42 | 5475 † |
| Specific Ribosomal Protein | Specific Ribosomal Protein Fold | Specific Ribosomal Protein | Universal Ribosomal Protein Fold | |
|---|---|---|---|---|
| Bacteria | bL9 | 3.40.5 RP L9; domain 1 & 3.10.430 RP L9; domain 2 | bL12 | 3.30.1390 RP L30; Chain: A |
| bL17 | 3.90.1030 50s RP L17; Chain A | bL19 | 2.30.30 SH3 type barrels | |
| bL20 | 1.10.1900 c-terminal domain of poly(a) binding domain | bL21 | 2.40.50 OB fold | |
| bL25 | 2.40.240 RP L25; Chain P & 2.170.120 RNApol α Subunit; Chain A, domain 2 | bL27 | 2.40.50 OB fold | |
| bL35 | 4.10.410 Factor Xa Inhibitor | bL28 | 2.30.170 RP L24e; Chain: T | |
| bL36 | 2.60.120 Jelly Rolls | bL31 | 4.10.830 30s RP S14; Chain N | |
| bS16 | 3.30.1320 S16 RP; Chain: A | bL32 | 1.20.5 Single α-helices involved in coiled-coils or other helix-helix interfaces | |
| bS18 | 4.10.640 30s RP S18 | bL34 | 1.10.287 Helix Hairpins | |
| bS20 | 1.20.58 Methane Monooxygenase Hydroxylase; Chain G, domain 1 | bS6 | 3.30.70 α-β Plaits | |
| Archaea and Eukarya | eL8 | 3.30.1330 60s RP L30; Chain: A | eL14 | 2.30.30 SH3 type barrels |
| eL15 | 3.40.1120 RP L15e | eL18 | 3.100.10 RP L15; Chain K; domain 2 | |
| eL19 | 1.10.1650 50s RP L19e, Chain O, domain 1 | eL21 | 2.30.30 SH3 type barrels | |
| eL20 | 3.10.20 UB roll | eL24 | 2.30.170 RP L24e; Chain: T | |
| eL30 | 3.30.1330 60s RP L30; Chain: A | eL27 | 2.30.30 SH3 type barrels | |
| eL31 | 3.10.440 RP L31e; Chain: W | eL32 | 3.30.70 α-β Plaits | |
| eL33 | 2.40.10 Thrombin, subunit H | eL41 | 1.20.5 Single α-helices involved in coiled-coils or other helix-helix interfaces | |
| eL34 | 6.20.340 His-Me finger endonuclease fold | eS4 (3 folds) | 2.30.30 SH3 type barrels & 2.40.50 OB fold & 3.10.290 Structural Genomics Hypothetical 15.5 Kd Protein In mrcA-pckA Intergenic Region; Chain A | |
| eL37 | 2.20.25 N-terminal domain TfIIb | eS8 | 3.10.290 Structural Genomics Hypothetical 15.5 Kd Protein In mrcA-pckA Intergenic Region; Chain A | |
| eL39 | 1.10.1620 Atp Synthase ε Chain; Chain: I | eS19 | 1.10.10 Arc Repressor Mutant, subunit A | |
| eL40 | 4.10.1060 ZNF265 like | eS24 | 3.30.70 α-β Plaits | |
| eL43 | 2.20.25 N-terminal domain TfIIb | eS28 | 2.40.50 OB fold | |
| eS1 | 3.10.20 UB roll & 3.30.479 Tetrahydropterin Synthase; Chain A | |||
| eS6 | 3.10.20 UB roll | |||
| eS21 | 3.30.1230 Hypothetical Cytosolic Protein; Chain A | |||
| eS27 | 2.20.25 N-terminal domain TfIIb | |||
| eS31 | 6.20.50 N-terminal domain TfIIb | |||
| Eukarya | eL22 | 3.30.1360 Gyrase A; domain 2 | eL6 | 2.30.30 SH3 type barrels |
| eL29 | 6.10.140 Helix Hairpins | eL13 | 1.10.10 Arc Repressor Mutant, subunit A | |
| eL38 | 3.30.720 Signal recognition particle alu RNA binding heterodimer, srp9/1 | eL36 | 1.10.10 Arc Repressor Mutant, subunit A | |
| RACK1 | 2.130.10 Methylamine Dehydrogenase; Chain H | eS10 | 1.10.10 Arc Repressor Mutant, subunit A | |
| eS26 | 3.30.1740 First Zn finger domain of poly(adp-ribose) polymerase-1 | eS25 | 1.10.10 Arc Repressor Mutant, subunit A | |
| eS30 | 1.20.5 Single α-helices involved in coiled-coils or other helix-helix interfaces | |||
| Eukarya specific ribosomal protein | Archaea and Eukarya specific ribosomal protein fold | |||
| eS7 | 2.20.25 N-terminal domain TfIIb | |||
| eS12 | 3.30.1330 60s RP L30; Chain: A |
| Eukarya | Bacteria | |||
|---|---|---|---|---|
| Protein | CATH | Protein | CATH | |
| LSU | eL15 | 3.40.1120 RP L15e | bL28 | 2.30.170 TP L24e; Chain T |
| eL21 | 2.30.30 SH3 type barrels | bL27 | 2.40.50 OB fold | |
| eL24 | 2.30.170 RP L24e; Chain T | bL19 | 2.30.30 SH3 type barrels | |
| eL31 | 3.10.440 RP L31e; Chain W | bL17 | 3.90.1030 50s RP L17; Chain A | |
| eL33 | 2.40.10 Thrombin, subunit H | bL20 | 1.10.1900 C-terminal domain of poly(a) binding protein | |
| eL40 | 4.10.1060 ZNF265 like | bL36 | 2.60.120 Jelly Rolls | |
| SSU | eS1 | 3.10.20 UB roll/3.30.479 Tetrahydropterin Synthase; Chain A | bS6/bS18 | 3.30.70 α-β Plaits/4.10.640 30s RP S18 |
| eS4 | 2.30.30 SH3 type barrels | bS16 | 3.30.1320 S16 RP; Chain A | |
| eS8 | 3.10.290 Structural Genomics Hypothetical 15.5Kd Protein In mrcA-pckA Intergenic Region; Chain A | bS20 | 1.20.58 Methane Monooxygenase Hydroxylase; Chain G, domain 1 | |
| eS17 | 1.10.60 Diphtheria Toxin Repressor; domain 2 | uS2-dom | 3.40.50 Rossmann fold |
| Urfolds/Metafolds | Universal 1 | B 2 | AE 3 | E 4 |
|---|---|---|---|---|
| SBBs | uL2, uL3, uL24, uS12, uS17 | bL19, bL21, bL27 | eL14, eL21, eL24, eL27, eL33, eS4, eS28 | eL6 |
| βαββαβ | uL1 (domain1), uL5, uL6, L7/L12, uL10, uL13, uL16, uL23, uS3, uS5 (C terminal domain), uS8, uS9, uS10 | bL9 (C terminal domain), bS6 | eL30, eL32, eS24 | eL22 |
| RLM | uL1 (domain 2), uL4, uS2 | |||
| Perpendicularly stacked α helices | uS13, uS2 (coiled coil domain) | bL20 | eS17 | |
| 3 parallel α helices | uL29, uS15 | bL34, bS20 | eL19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanoz, I.; Timsit, Y. Protein Fold Usages in Ribosomes: Another Glance to the Past. Int. J. Mol. Sci. 2024, 25, 8806. https://doi.org/10.3390/ijms25168806
Tanoz I, Timsit Y. Protein Fold Usages in Ribosomes: Another Glance to the Past. International Journal of Molecular Sciences. 2024; 25(16):8806. https://doi.org/10.3390/ijms25168806
Chicago/Turabian StyleTanoz, Inzhu, and Youri Timsit. 2024. "Protein Fold Usages in Ribosomes: Another Glance to the Past" International Journal of Molecular Sciences 25, no. 16: 8806. https://doi.org/10.3390/ijms25168806
APA StyleTanoz, I., & Timsit, Y. (2024). Protein Fold Usages in Ribosomes: Another Glance to the Past. International Journal of Molecular Sciences, 25(16), 8806. https://doi.org/10.3390/ijms25168806






