Is Exon Skipping a Viable Therapeutic Approach for Vascular Ehlers–Danlos Syndrome with Mutations in COL3A1 Exon 10 or 15?
Abstract
1. Introduction
2. Results
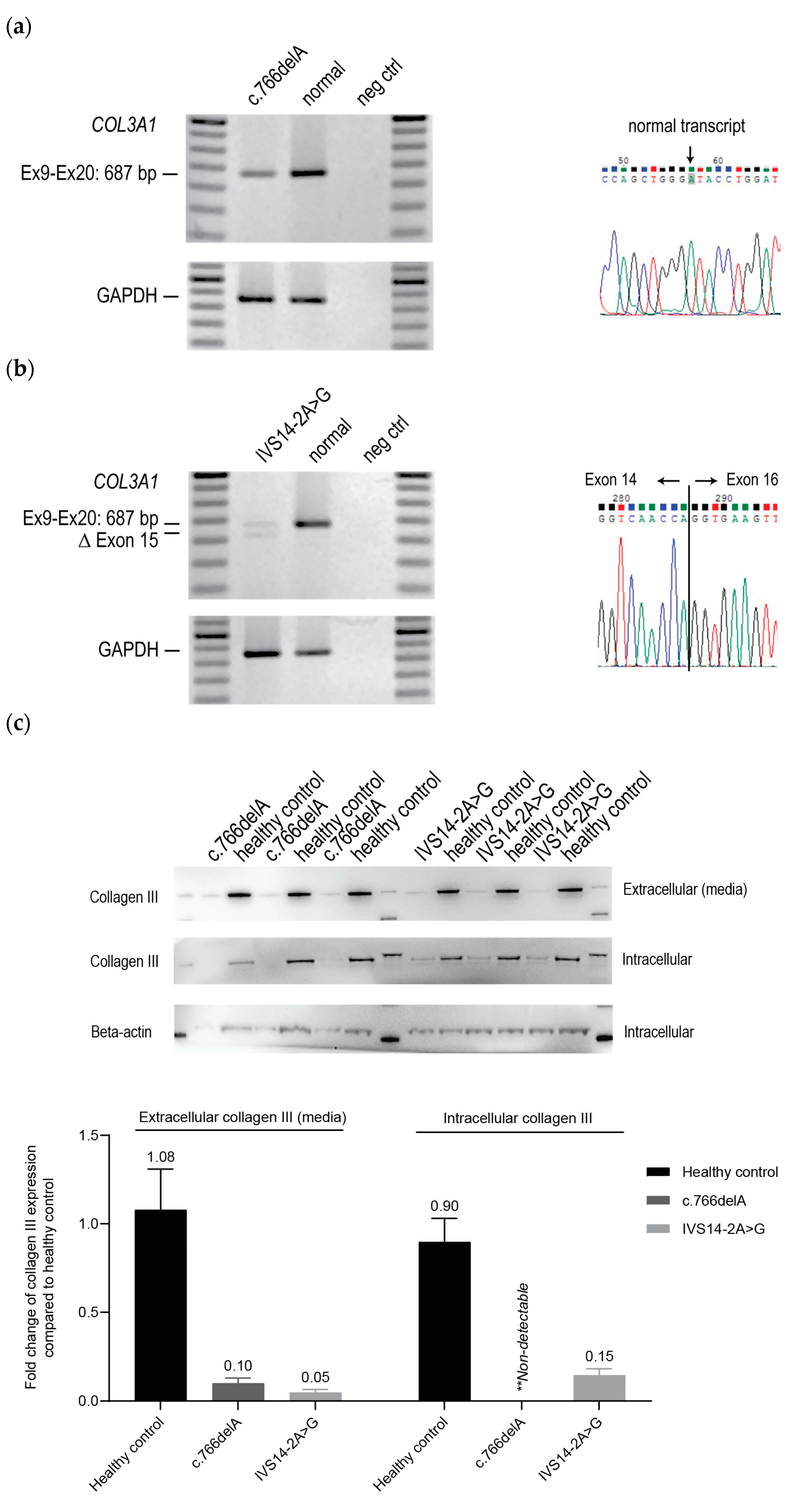
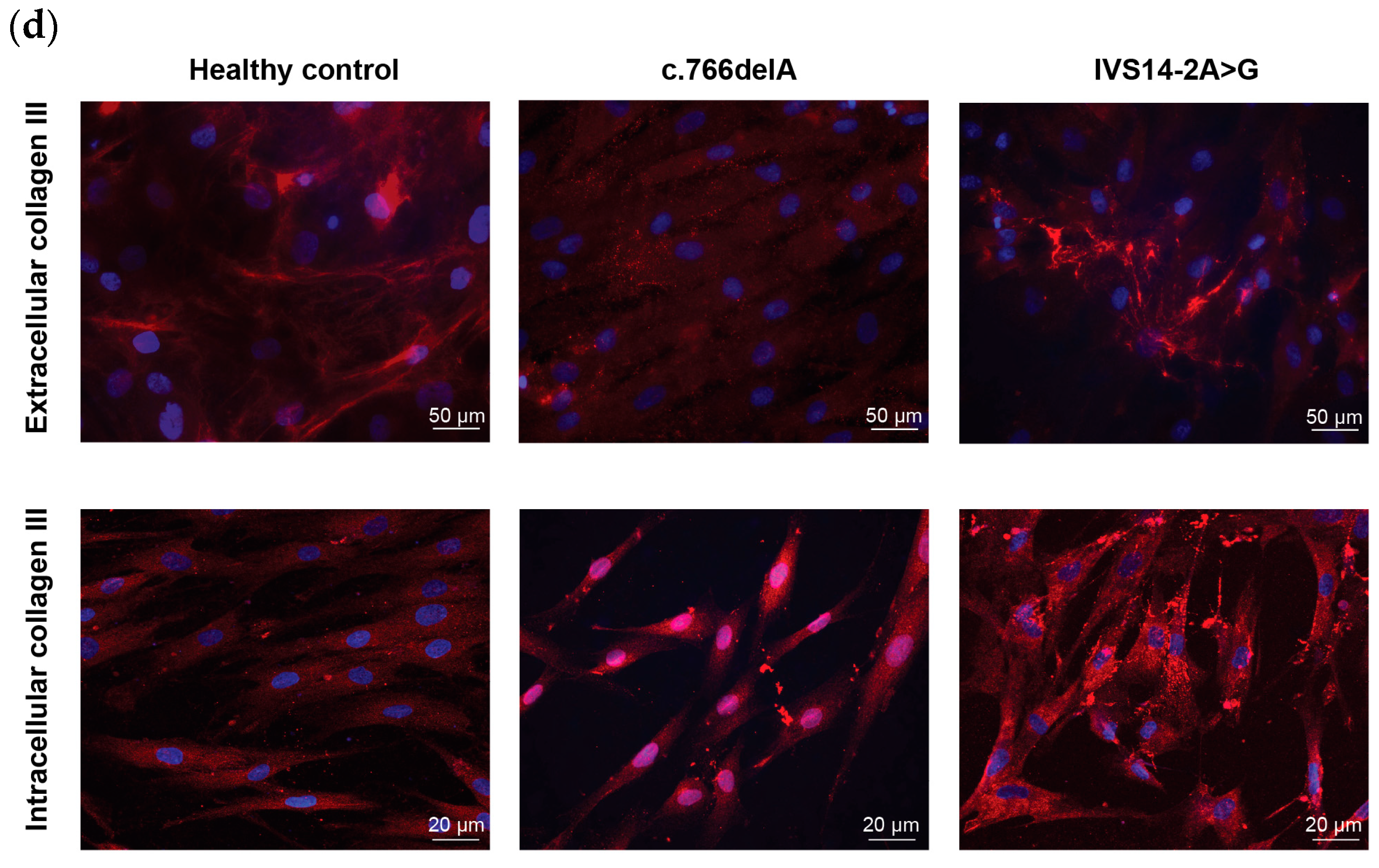
2.1. Cellular Phenotypes of vEDS Patient Fibroblasts Harbouring COL3A1 Mutations
2.2. ASO-Mediated Exon Skipping to Reframe COL3A1 Transcripts and Assessment of Collagen III Expression in vEDS Patient-Derived Fibroblasts Carrying COL3A1 c.766delA
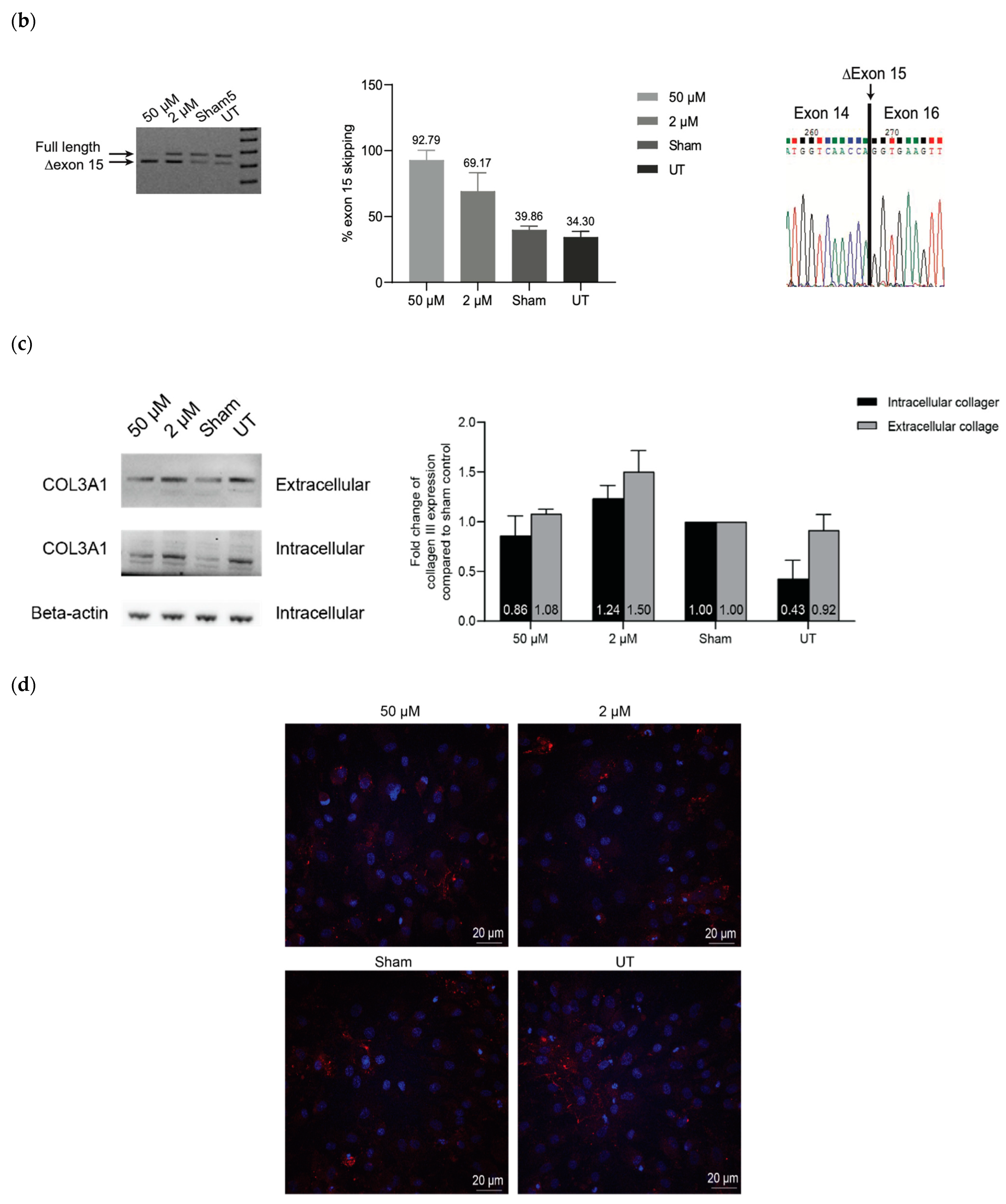
2.3. ASO-Mediated Exon Skipping to Reframe COL3A1 Transcripts and Assessment of Collagen III Expression in vEDS Patient-Derived Fibroblasts Carrying COL3A1 IVS14-2A>G
3. Discussion
4. Materials and Methods
4.1. Antisense Oligonucleotide Design and Synthesis
4.2. Cell Culture and Transfection
4.3. RNA Harvest and RT-PCR
4.4. Protein Harvest and Western Blot
4.4.1. Cell Lysate Preparation
4.4.2. Spent Media Preparation
4.4.3. Western Blot
4.5. Fibroblast-Derived Extracellular Matrix
4.6. Immunofluorescence Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boot-Handford, R.P.; Tuckwell, D.S.; Plumb, D.A.; Rock, C.F.; Poulsom, R. A novel and highly conserved collagen (pro(alpha)1(XXVII)) with a unique expression pattern and unusual molecular characteristics establishes a new clade within the vertebrate fibrillar collagen family. J. Biol. Chem. 2003, 278, 31067–31077. [Google Scholar] [CrossRef] [PubMed]
- Exposito, J.-Y.; Cluzel, C.; Garrone, R.; Lethias, C. Evolution of collagens. Anat. Rec. 2002, 268, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Laub, F.; Zhou, P.; Hahn, R.A.; Tanaka, S.; Burgeson, R.E.; Gerecke, D.R.; Ramirez, F.; Gordon, M.K. Collagen XXIV, a vertebrate fibrillar collagen with structural features of invertebrate collagens: Selective expression in developing cornea and bone. J. Biol. Chem. 2003, 278, 43236–43244. [Google Scholar] [CrossRef] [PubMed]
- Pace, J.M.; Corrado, M.; Missero, C.; Byers, P.H. Identification, characterization and expression analysis of a new fibrillar collagen gene, COL27A1. Matrix Biol. 2003, 22, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef]
- Cheung, D.T.; Benya, P.D.; Perelman, N.; DiCesare, P.E.; Nimni, M.E. A highly specific and quantitative method for determining type III/I collagen ratios in tissues. Matrix 1990, 10, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Kuivaniemi, H.; Tromp, G. Type III collagen (COL3A1): Gene and protein structure, tissue distribution, and associated diseases. Gene 2019, 707, 151–171. [Google Scholar] [CrossRef]
- Ramshaw, J.A.; Shah, N.K.; Brodsky, B. Gly-X-Y tripeptide frequencies in collagen: A context for host-guest triple-helical peptides. J. Struct. Biol. 1998, 122, 86–91. [Google Scholar] [CrossRef]
- Epstein, D.; Elias-Bishko, S.; Hershko, A. Requirement for protein synthesis in the regulation of protein breakdown in cultured hepatoma cells. Biochemistry 1975, 14, 5199–5204. [Google Scholar] [CrossRef]
- van Kuppevelt, T.H.; Veerkamp, J.H.; Timmermans, J.A. Immunoquantification of type I, III, IV and V collagen in small samples of human lung parenchyma. Int. J. Biochem. Cell Biol. 1995, 27, 775–782. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sumiyoshi, H.; Nakagami, K.; Tahara, E. Distribution of collagen types I, III, and V in fibrotic and neoplastic human liver. Acta Pathol. Jpn. 1984, 34, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Hurst, P.R.; Gibbs, R.D.; Clark, D.E.; Myers, D.B. Temporal changes to uterine collagen types I, III and V in relation to early pregnancy in the rat. Reprod. Fertil. Dev. 1994, 6, 669–677. [Google Scholar] [CrossRef] [PubMed]
- McCullagh, K.A.; Balian, G. Collagen characterisation and cell transformation in human atherosclerosis. Nature 1975, 258, 73–75. [Google Scholar] [CrossRef]
- Kaplan, E.P.; Richier, J.C.; Howard, P.S.; Ewalt, D.H.; Lin, V.K. Type III collagen messenger RNA is modulated in non-compliant human bladder tissue. J. Urol. 1997, 157, 2366–2369. [Google Scholar] [CrossRef] [PubMed]
- Weber, K.T. Cardiac interstitium in health and disease: The fibrillar collagen network. J. Am. Coll. Cardiol. 1989, 13, 1637–1652. [Google Scholar] [CrossRef] [PubMed]
- Asgari, M.; Latifi, N.; Heris, H.K.; Vali, H.; Mongeau, L. In vitro fibrillogenesis of tropocollagen type III in collagen type I affects its relative fibrillar topology and mechanics. Sci. Rep. 2017, 7, 1392. [Google Scholar] [CrossRef] [PubMed]
- Malfait, F. Vascular aspects of the Ehlers-Danlos Syndromes. Matrix Biol. 2018, 71–72, 380–395. [Google Scholar] [CrossRef]
- Germain, D.P. Ehlers-Danlos syndrome type IV. Orphanet. J. Rare Dis. 2007, 2, 32. [Google Scholar] [CrossRef] [PubMed]
- Malfait, F.; Francomano, C.; Byers, P.; Belmont, J.; Berglund, B.; Black, J.; Bloom, L.; Bowen, J.M.; Brady, A.F.; Burrows, N.P.; et al. The 2017 international classification of the Ehlers–Danlos syndromes. Am. J. Med. Genet. Part C Semin. Med. Genet. 2017, 175, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.; Bartlett, M.; Baker, D.; Berlin, C.; Bowen, J.; Cummings, C.; Fallows, C.; Green, C.; Griffin, J.; Julier, K.; et al. An exemplary model of genetic counselling for highly specialised services. J. Community Genet. 2023, 14, 115–119. [Google Scholar] [CrossRef]
- Murray, M.L.; Pepin, M.; Peterson, S.; Byers, P.H. Pregnancy-related deaths and complications in women with vascular Ehlers-Danlos syndrome. Genet. Med. 2014, 16, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Matthew, J.E. Arterial complications of vascular Ehlers-Danlos syndrome. J. Vasc. Surg. 2016, 64, 1869–1880. [Google Scholar]
- Schwarze, U.; Schievink, W.I.; Petty, E.; Jaff, M.R.; Babovic-Vuksanovic, D.; Cherry, K.J.; Pepin, M.; Byers, P.H. Haploinsufficiency for one COL3A1 allele of type III procollagen results in a phenotype similar to the vascular form of Ehlers-Danlos syndrome, Ehlers-Danlos syndrome type IV. Am. J. Hum. Genet. 2001, 69, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Leistritz, D.F.; Pepin, M.G.; Schwarze, U.; Byers, P.H. COL3A1 haploinsufficiency results in a variety of Ehlers-Danlos syndrome type IV with delayed onset of complications and longer life expectancy. Genet. Med. 2011, 13, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.; Albuisson, J.; Ranque, B.; Golmard, L.; Mazzella, J.-M.; Bal-Theoleyre, L.; Fauret, A.-L.; Mirault, T.; Denarié, N.; Mousseaux, E.; et al. The type of variants at the COL3A1 gene associates with the phenotype and severity of vascular Ehlers–Danlos syndrome. Eur. J. Hum. Genet. 2015, 23, 1657–1664. [Google Scholar] [CrossRef]
- Shalhub, S.; Black, J.H., 3rd; Cecchi, A.C.; Xu, Z.; Griswold, B.F.; Safi, H.J.; Milewicz, D.M.; McDonnell, N.B. Molecular diagnosis in vascular Ehlers-Danlos syndrome predicts pattern of arterial involvement and outcomes. J. Vasc. Surg. 2014, 60, 160–169. [Google Scholar] [CrossRef]
- Pichavant, C.; Aartsma-Rus, A.; Clemens, P.R.; Davies, K.E.; Dickson, G.; Takeda, S.; Wilton, S.D.; Wolff, J.A.; Wooddell, C.I.; Xiao, X.; et al. Current status of pharmaceutical and genetic therapeutic approaches to treat DMD. Mol. Ther. 2011, 19, 830–840. [Google Scholar] [CrossRef]
- Li, D.; Adams, A.M.; Johnsen, R.D.; Fletcher, S.; Wilton, S.D. Morpholino Oligomer-Induced Dystrophin Isoforms to Map the Functional Domains in the Dystrophin Protein. Mol. Ther. Nucleic Acids 2020, 22, 263–272. [Google Scholar] [CrossRef]
- Pepin, M.G.; Schwarze, U.; Rice, K.M.; Liu, M.; Leistritz, D.; Byers, P.H. Survival is affected by mutation type and molecular mechanism in vascular Ehlers-Danlos syndrome (EDS type IV). Genet. Med. 2014, 16, 881–888. [Google Scholar] [CrossRef]
- Cale, J.M.; Greer, K.; Fletcher, S.; Wilton, S.D. Proof-of-Concept: Antisense Oligonucleotide Mediated Skipping of Fibrillin-1 Exon 52. Int. J. Mol. Sci. 2021, 22, 3479. [Google Scholar] [CrossRef]
- Bremer, J.; Bornert, O.; Nyström, A.; Gostynski, A.; Jonkman, M.F.; Aartsma-Rus, A.; van den Akker, P.C.; Pasmooij, A.M. Antisense Oligonucleotide-mediated Exon Skipping as a Systemic Therapeutic Approach for Recessive Dystrophic Epidermolysis Bullosa. Mol. Ther. Nucleic Acids 2016, 5, e379. [Google Scholar] [CrossRef] [PubMed]
- Turczynski, S.; Titeux, M.; Tonasso, L.; Décha, A.; Ishida-Yamamoto, A.; Hovnanian, A. Targeted Exon Skipping Restores Type VII Collagen Expression and Anchoring Fibril Formation in an In Vivo RDEB Model. J. Investig. Dermatol. 2016, 136, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, T.; Horinouchi, T.; Adachi, T.; Terakawa, M.; Takaoka, Y.; Omachi, K.; Takasato, M.; Takaishi, K.; Shoji, T.; Onishi, Y.; et al. Development of an exon skipping therapy for X-linked Alport syndrome with truncating variants in COL4A5. Nat. Commun. 2020, 11, 2777. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.K.; Taga, Y.; Sakai, T.; Ito, S.; Hattori, S.; Nagata, K.; Koide, T. Lowering the culture temperature corrects collagen abnormalities caused by HSP47 gene knockout. Sci. Rep. 2019, 9, 17433. [Google Scholar] [CrossRef] [PubMed]
- Muntoni, F.; Torelli, S.; Ferlini, A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003, 2, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Mays, P.K.; Bishop, J.E.; Laurent, G.J. Age-related changes in the proportion of types I and III collagen. Mech. Ageing Dev. 1988, 45, 203–212. [Google Scholar] [CrossRef]
- Cheng, W.; Rong, Y.-H.; Ning, F.-G.; Zhang, G.-A. The content and ratio of type I and III collagen in skin differ with age and injury. Afr. J. Biotechnol. 2011, 10, 2524–2529. [Google Scholar]
- Khoshnoodi, J.; Pedchenko, V.; Hudson, B.G. Mammalian collagen IV. Microsc. Res. Tech. 2008, 71, 357–370. [Google Scholar] [CrossRef]
- Bekheirnia, M.R.; Reed, B.; Gregory, M.C.; McFann, K.; Shamshirsaz, A.A.; Masoumi, A.; Schrier, R.W. Genotype-phenotype correlation in X-linked Alport syndrome. J. Am. Soc. Nephrol. 2010, 21, 876–883. [Google Scholar] [CrossRef]
- Berg, R.A.; Prockop, D.J. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem. Biophys. Res. Commun. 1973, 52, 115–120. [Google Scholar] [CrossRef]
- Sakakibara, S.; Inouye, K.; Shudo, K.; Kishida, Y.; Kobayashi, Y.; Prockop, D.J. Synthesis of (Pro-Hyp-Gly) n of defined molecular weights. Evidence for the stabilization of collagen triple helix by hydroxypyroline. Biochim. Biophys. Acta 1973, 303, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.D.; Page, A.P. Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol. Cell. Biol. 2000, 20, 4084–4093. [Google Scholar] [CrossRef] [PubMed]
- Jürgensen, H.J.; Madsen, D.H.; Ingvarsen, S.; Melander, M.C.; Gårdsvoll, H.; Patthy, L.; Engelholm, L.H.; Behrendt, N. A novel functional role of collagen glycosylation: Interaction with the endocytic collagen receptor uparap/ENDO180. J. Biol. Chem. 2011, 286, 32736–32748. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Zhu, Y.; Xu, W.; Ye, S.; Zhang, R.; Lu, L.; Jiang, S. Characterization by high-resolution crystal structure analysis of a triple-helix region of human collagen type III with potent cell adhesion activity. Biochem. Biophys. Res. Commun. 2019, 508, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.N.; Brandts, J.F. Determination of cis-trans proline isomerization by trypsin proteolysis. Application to a model pentapeptide and to oxidized ribonuclease A. Biochemistry 1983, 22, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Boudko, S.; Engel, J.; Bächinger, H.P. Vascular Ehlers-Danlos syndrome mutations in type III collagen differently stall the triple helical folding. J. Biol. Chem. 2013, 288, 19166–19176. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Mizuno, K.; Bächinger, H.P. Ziploc-ing the structure 2.0: Endoplasmic reticulum-resident peptidyl prolyl isomerases show different activities toward hydroxyproline. J. Biol. Chem. 2017, 292, 9273–9282. [Google Scholar] [CrossRef] [PubMed]
- Aung-Htut, M.T.; McIntosh, C.S.; West, K.A.; Fletcher, S.; Wilton, S.D. In Vitro Validation of Phosphorodiamidate Morpholino Oligomers. Molecules 2019, 24, 2922. [Google Scholar] [CrossRef] [PubMed]
- Wilton, S.D.; Lim, L.; Dye, D.; Laing, N. Bandstab: A PCR-based alternative to cloning PCR products. Biotechniques 1997, 22, 642–645. [Google Scholar] [CrossRef]
- Kumar, P.; Satyam, A.; Fan, X.; Collin, E.; Rochev, Y.; Rodriguez, B.J.; Gorelov, A.; Dillon, S.; Joshi, L.; Raghunath, M.; et al. Macromolecularly crowded in vitro microenvironments accelerate the production of extracellular matrix-rich supramolecular assemblies. Sci. Rep. 2015, 5, 8729. [Google Scholar] [CrossRef]


| Name | ASO Sequence 5′ --> 3′ | Length (bp) | Target COL3A1 Exon |
|---|---|---|---|
| COL3A1_H10A(−13+12) | ACCUUUGAUACCCUAAAAAUAAUGA | 25 | 10 |
| COL3A1_H10A(+15+39) | ACCAGGGAAUCCAGGUAUCCCAGCU | 25 | 10 |
| COL3A1_H10D(+12−13) | AAACUCUACUUACUCUGUGUCCUUU | 25 | 10 |
| COL3A1_H15A(−43−19) | AUUUCAAGAGCUCUUUUUCAACUAA | 25 | 15 |
| COL3A1_H15A(−15+10) | CAGGAGGGCCCCGAAAAAAUUAAAU | 25 | 15 |
| COL3A1_H15A(+20+44) | GGGGAUCCAGGGAAUCCGGCAGUUC | 25 | 15 |
| COL3A1_H15D(+6−19) | UAUAGAAACACAUGUUUACCUUAGC | 25 | 15 |
| COL3A1_H15A(+25+49) | CACCAGGGGAUCCAGGGAAUCCGGC | 25 | 15 |
| COL3A1_H15A(+15+39) | UCCAGGGAAUCCGGCAGUUCCAGGA | 25 | 15 |
| COL3A1_H15A(+20+39) | UCCAGGGAAUCCGGCAGUUC | 20 | 15 |
| COL3A1_H15A(+25+44) | GGGGAUCCAGGGAAUCCGGC | 20 | 15 |
| Control * | CCTCTTACCTCAGTTACAATTTATA | 25 | None |
| Name | Sequence 5′ --> 3′ | Amplicon Size (bp) | Optimized Condition |
|---|---|---|---|
| COL3A1_Ex1F | GCAGGGAACAACTTGATGGTGC | 1079 | 58 °C 30 min |
| 94 °C 2 min | |||
| 94 °C 10 s | |||
| COL3A1_Ex14R | GGTTGACCATCACTGCCTCGA | 58 °C 10 s | |
| 68 °C 1 min | |||
| for 20 cycles | |||
| COL3A1_Ex9F | GACCTGGAGAGCGAGGATTG | 687 | 58 °C 30 min |
| 94 °C 2 min | |||
| 94 °C 10 s | |||
| COL3A1_Ex20R | CCTTGCCATCTTCGCCTTTA | 58 °C 10 s | |
| 68 °C 1 min | |||
| for 20 cycles | |||
| hGAPDH Ex1F | ATGCTGGCGCTGAGTACGT | 874 | 94 °C 2 min |
| 94 °C 30 s | |||
| hGAPDH Ex3R | AGGGGTCTACATGGCAACTG | 58 °C 30 s | |
| 68 °C 1 min | |||
| for 20 cycles |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Utama, S.; Cale, J.M.; Mitrpant, C.; Fletcher, S.; Wilton, S.D.; Aung-Htut, M.T. Is Exon Skipping a Viable Therapeutic Approach for Vascular Ehlers–Danlos Syndrome with Mutations in COL3A1 Exon 10 or 15? Int. J. Mol. Sci. 2024, 25, 8816. https://doi.org/10.3390/ijms25168816
Utama S, Cale JM, Mitrpant C, Fletcher S, Wilton SD, Aung-Htut MT. Is Exon Skipping a Viable Therapeutic Approach for Vascular Ehlers–Danlos Syndrome with Mutations in COL3A1 Exon 10 or 15? International Journal of Molecular Sciences. 2024; 25(16):8816. https://doi.org/10.3390/ijms25168816
Chicago/Turabian StyleUtama, Sasiwimon, Jessica M. Cale, Chalermchai Mitrpant, Sue Fletcher, Steve D. Wilton, and May T. Aung-Htut. 2024. "Is Exon Skipping a Viable Therapeutic Approach for Vascular Ehlers–Danlos Syndrome with Mutations in COL3A1 Exon 10 or 15?" International Journal of Molecular Sciences 25, no. 16: 8816. https://doi.org/10.3390/ijms25168816
APA StyleUtama, S., Cale, J. M., Mitrpant, C., Fletcher, S., Wilton, S. D., & Aung-Htut, M. T. (2024). Is Exon Skipping a Viable Therapeutic Approach for Vascular Ehlers–Danlos Syndrome with Mutations in COL3A1 Exon 10 or 15? International Journal of Molecular Sciences, 25(16), 8816. https://doi.org/10.3390/ijms25168816






