Temperature-Dependent tRNA Modifications in Bacillales
Abstract
1. Introduction
2. Results
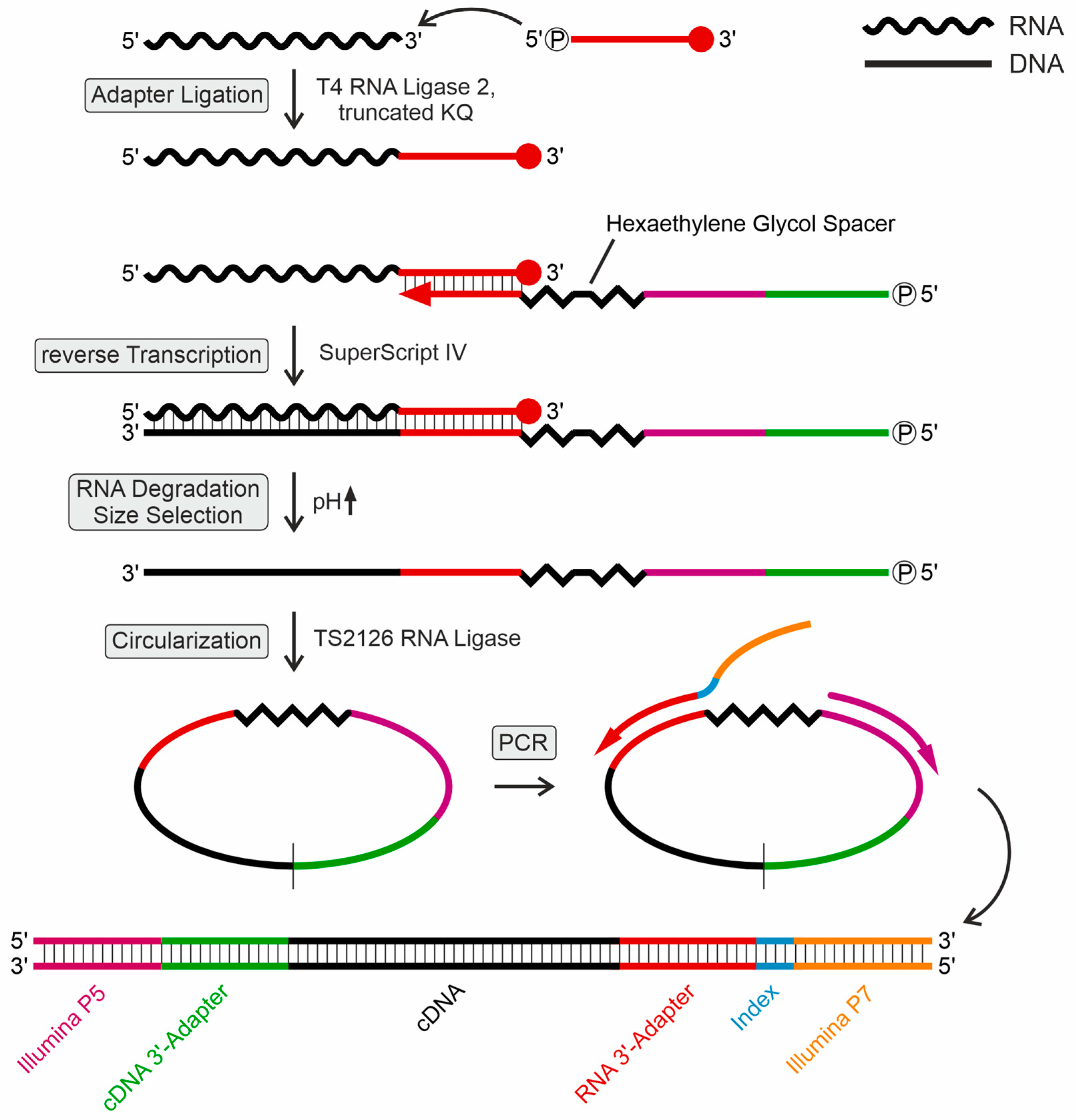
2.1. Specific Library Design for tRNA Sequencing and Subsequent Analysis of tRNA Modifications
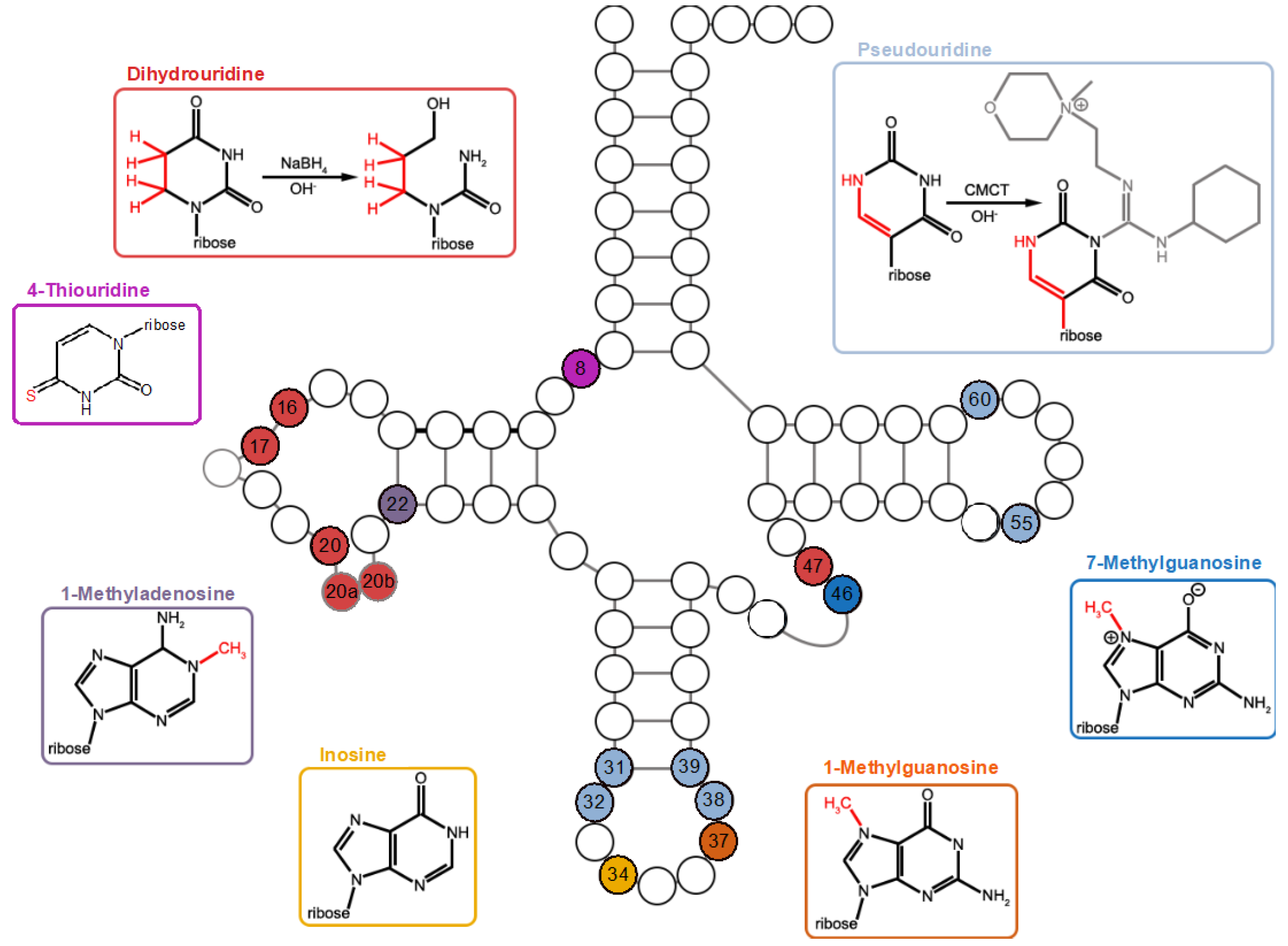
2.2. Variations in tRNA Modification Profiles among Closely Related Bacillales
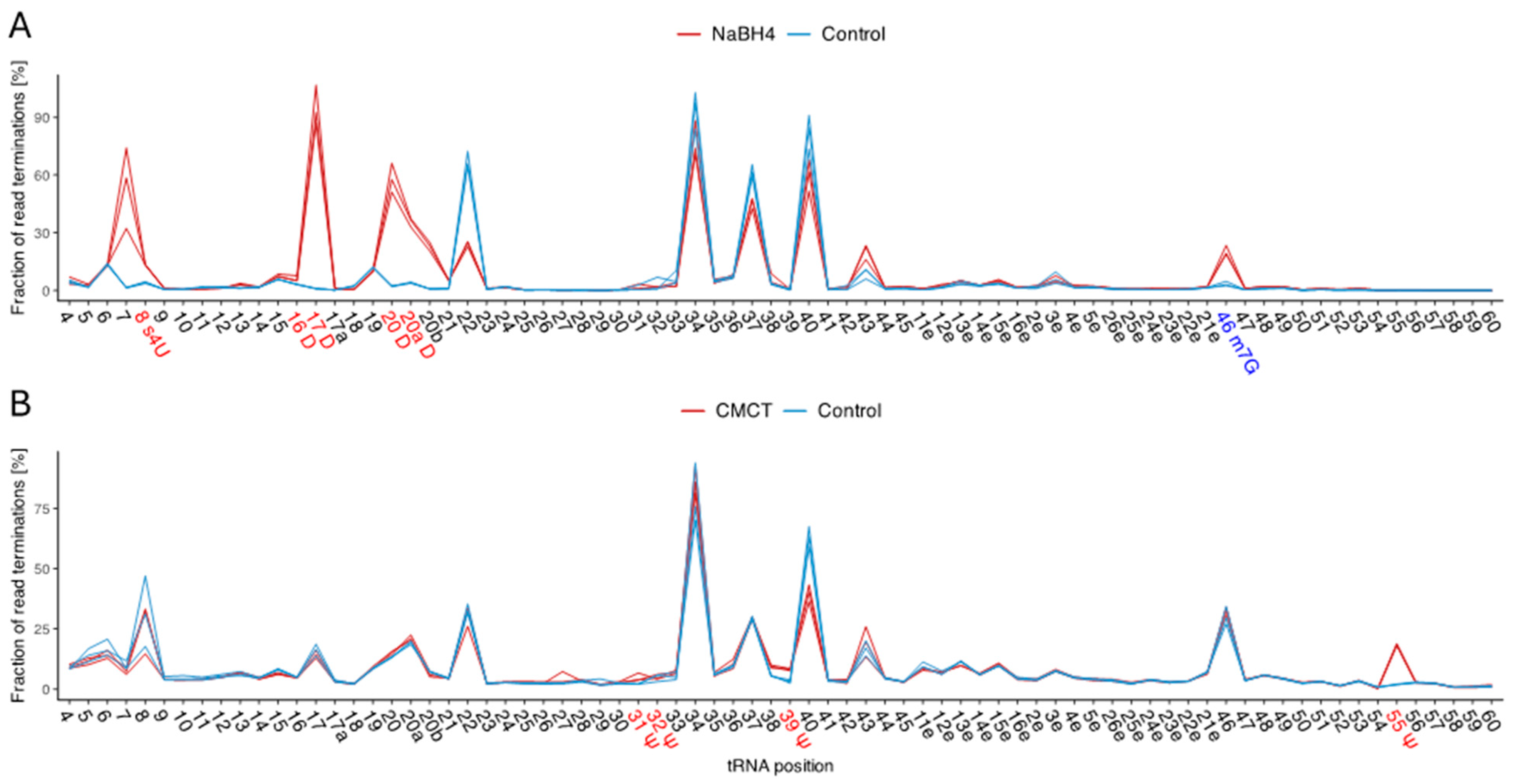
2.3. Specific tRNA Modifications Are Temperature-Dependent
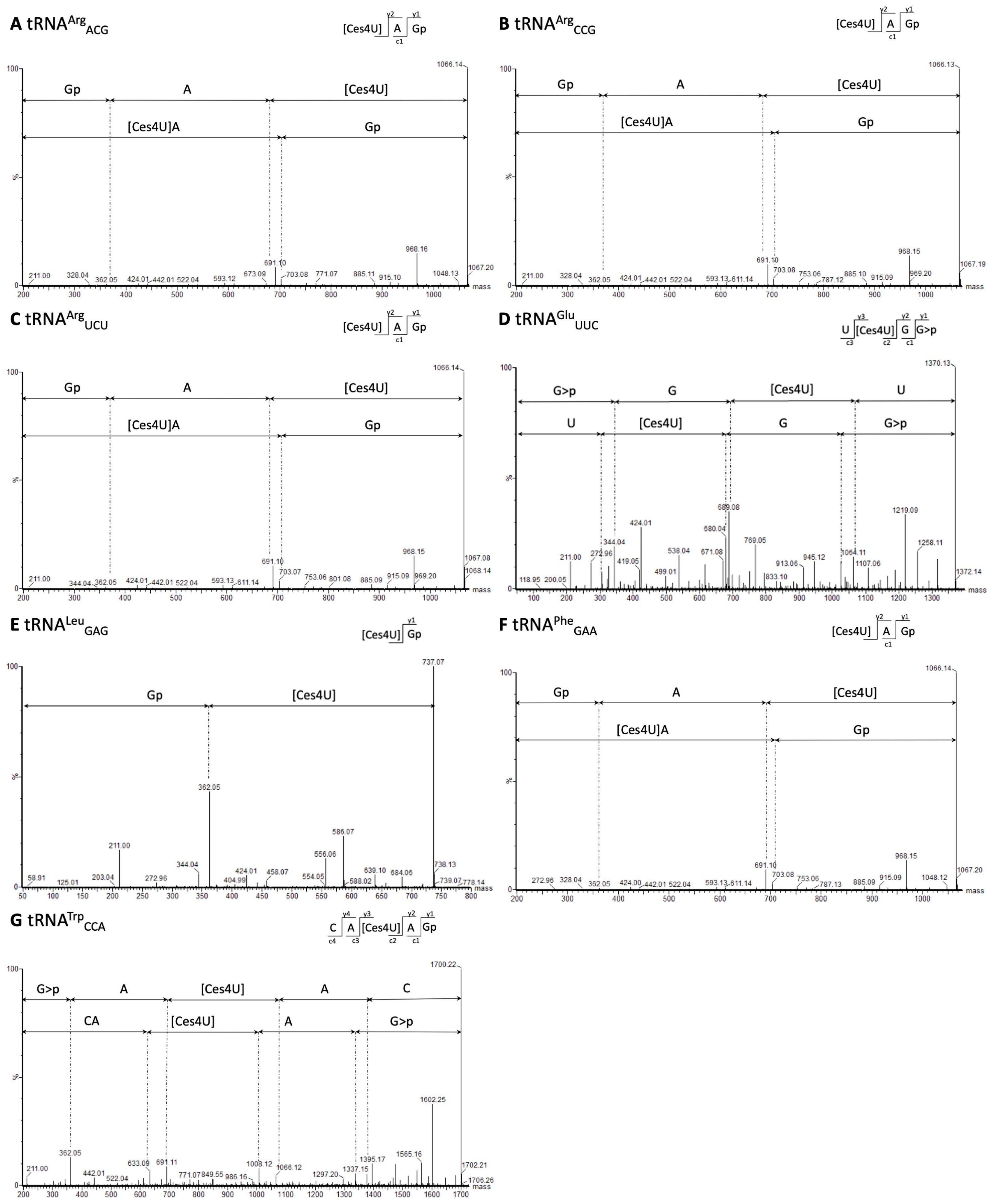
2.4. Performance Validation
3. Discussion
4. Materials and Methods
4.1. Cultivation of Bacteria
4.2. Isolation of Total RNA and Small RNA Enrichment
4.3. NaBH4-Treatment for the Detection of Dihydrouridine and 7-Methylguanosine
4.4. CMCT-Treatment for the Detection of Pseudouridine
4.5. Adenylation of Adapter Oligonucleotide and Ligation to Small RNA Samples
4.6. Reverse Transcription
4.7. Circularization, Amplification and Indexing of cDNA
4.8. High Throughput tRNA Sequencing
4.9. Annotation of tRNAs
4.10. Mapping of tRNA Reads
4.11. Library Normalization
4.12. Identification of Significant Base Misincorporation Sites
4.13. Profiling Position-Wise RT-Stops
4.14. Choice of Cutoff Values to Reduce Noise
- Fold change (FC): A sufficiently large effect size (FC) is required, i.e., position-specific increase of the chemically introduced RT-stops in treated compared to the untreated library.
- Total number of RT-stops: To reduce signals with an overestimated log2 FC, which can arise at positions with a low read coverage even at a few RT-stops.
4.15. Outcome Comparisons with mim-tRNAseq
4.16. MS/MS Analyses
4.16.1. tRNA Cyanoethylation
4.16.2. tRNA Isolation by 2D-Gel Electrophoresis
4.16.3. In-Gel RNase Digestion of tRNAs
4.16.4. Nano Liquid Chromatography-MS/MS of RNA Oligonucleotides and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA Modifications: Nature’s Combinatorial Chemistry Playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Kirino, Y.; Ikeuchi, Y.; Suzuki, T. Biosynthesis of Wybutosine, a Hyper-Modified Nucleoside in Eukaryotic Phenylalanine tRNA. EMBO J. 2006, 25, 2142–2154. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and Frequencies of Post-Transcriptional Modifications in tRNAs. RNA Biol. 2014, 11, 1619–1629. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Helm, M.; Motorin, Y. Use of Specific Chemical Reagents for Detection of Modified Nucleotides in RNA. J. Nucleic Acids 2011, 2011, 408053. [Google Scholar] [CrossRef]
- Findeiß, S.; Langenberger, D.; Stadler, P.F.; Hoffmann, S. Traces of Post-Transcriptional RNA Modifications in Deep Sequencing Data. Biol. Chem. 2011, 392, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, E.M.; Ehrenschwender, T.; Batista, P.J.; Chang, H.Y.; Kool, E.T. Identification of a Selective Polymerase Enables Detection of N6-Methyladenosine in RNA. J. Am. Chem. Soc. 2013, 135, 19079–19082. [Google Scholar] [CrossRef]
- Iida, K.; Jin, H.; Zhu, J.-K. Bioinformatics Analysis Suggests Base Modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genom. 2009, 10, 155. [Google Scholar] [CrossRef]
- Motorin, Y.; Marchand, V. Analysis of RNA Modifications by Second- and Third-Generation Deep Sequencing: 2020 Update. Genes 2021, 12, 278. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T.; Nishikawa, K.; Nemoto, F.; Kuchino, Y.; Nishimura, S.; Miyazawa, T.; Yokoyama, S. Codon and Amino-Acid Specificities of a Transfer RNA Are Both Converted by a Single Post-Transcriptional Modification. Nature 1988, 336, 179–181. [Google Scholar] [CrossRef]
- Muramatsu, T.; Yokoyama, S.; Horie, N.; Matsuda, A.; Ueda, T.; Yamaizumi, Z.; Kuchino, Y.; Nishimura, S.; Miyazawa, T. A Novel Lysine-Substituted Nucleoside in the First Position of the Anticodon of Minor Isoleucine tRNA from Escherichia coli. J. Biol. Chem. 1988, 263, 9261–9267. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-Wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, A.F.; Riordan, D.P.; Brown, P.O. Transcriptome-Wide Mapping of Pseudouridines: Pseudouridine Synthases Modify Specific mRNAs in S. Cerevisiae. PLoS ONE 2014, 9, e110799. [Google Scholar] [CrossRef]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine Profiling Reveals Regulated mRNA Pseudouridylation in Yeast and Human Cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Muller, S.; Behm-Ansmant, I.; Branlant, C. Identification of Modified Residues in RNAs by Reverse Transcription-Based Methods. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2007; Volume 425, pp. 21–53. ISBN 978-0-12-374155-4. [Google Scholar]
- Giege, R.; Sissler, M.; Florentz, C. Universal Rules and Idiosyncratic Features in tRNA Identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Duechler, M.; Leszczyńska, G.; Sochacka, E.; Nawrot, B. Nucleoside Modifications in the Regulation of Gene Expression: Focus on tRNA. Cell. Mol. Life Sci. 2016, 73, 3075–3095. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.; Lünse, C.E.; Mörl, M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. tRNA Stabilization by Modified Nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef]
- Voigts-Hoffmann, F.; Hengesbach, M.; Kobitski, A.Y.; Van Aerschot, A.; Herdewijn, P.; Nienhaus, G.U.; Helm, M. A Methyl Group Controls Conformational Equilibrium in Human Mitochondrial tRNALys. J. Am. Chem. Soc. 2007, 129, 13382–13383. [Google Scholar] [CrossRef] [PubMed]
- Du, X. Tertiary Structure Base Pairs between D- and TpsiC-Loops of Escherichia coli tRNALeu Play Important Roles in Both Aminoacylation and Editing. Nucleic Acids Res. 2003, 31, 2865–2872. [Google Scholar] [CrossRef][Green Version]
- Nobles, K.N.; Yarian, C.S.; Liu, G.; Guenther, R.H.; Agris, P.F. Highly Conserved Modified Nucleosides Influence Mg2+-Dependent tRNA Folding. Nucleic Acids Res. 2002, 30, 4751–4760. [Google Scholar] [CrossRef]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, Where, How, and Why. IUBMB Life 2000, 49, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Arnez, J.G.; Steitz, T.A. Crystal Structure of Unmodified tRNAGln Complexed with Glutaminyl-tRNA Synthetase and ATP Suggests a Possible Role for Pseudo-Uridines in Stabilization of RNA Structure. Biochemistry 1994, 33, 7560–7567. [Google Scholar] [CrossRef] [PubMed]
- Dalluge, J.J.; Hashizume, T.; Sopchik, A.E.; McCloskey, J.A.; Davis, D.R. Conformational Flexibility in RNA: The Role of Dihydrouridine. Nucleic Acids Res. 1996, 24, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Feller, G.; Gerday, C. Psychrophilic Enzymes: Hot Topics in Cold Adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Dalluge, J.J.; Hamamoto, T.; Horikoshi, K.; Morita, R.Y.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional Modification of tRNA in Psychrophilic Bacteria. J. Bacteriol. 1997, 179, 1918–1923. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, C.G.; Crain, P.F.; Gupta, R.; Hashizume, T.; Hocart, C.H.; Kowalak, J.A.; Pomerantz, S.C.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional Modification of tRNA in Thermophilic Archaea (Archaebacteria). J. Bacteriol. 1991, 173, 3138–3148. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Koh, H.; Söll, D. The Effect of Growth Temperatures on the in Vivo Ribose Methylation of Bacillus Stearothermophilus Transfer RNA. Arch. Biochem. Biophys. 1973, 154, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Kunibayashi, T.; Tomikawa, C.; Ochi, A.; Kanai, T.; Hirata, A.; Iwashita, C.; Hori, H. Pseudouridine at Position 55 in tRNA Controls the Contents of Other Modified Nucleotides for Low-Temperature Adaptation in the Extreme-Thermophilic Eubacterium Thermus Thermophilus. Nucleic Acids Res. 2011, 39, 2304–2318. [Google Scholar] [CrossRef] [PubMed]
- Cappannini, A.; Ray, A.; Purta, E.; Mukherjee, S.; Boccaletto, P.; Moafinejad, S.N.; Lechner, A.; Barchet, C.; Klaholz, B.P.; Stefaniak, F.; et al. MODOMICS: A Database of RNA Modifications and Related Information. 2023 Update. Nucleic Acids Res. 2024, 52, D239–D244. [Google Scholar] [CrossRef] [PubMed]
- De Crécy-Lagard, V.; Jaroch, M. Functions of Bacterial tRNA Modifications: From Ubiquity to Diversity. Trends Microbiol. 2021, 29, 41–53. [Google Scholar] [CrossRef]
- Pixa, G.; Dirheimer, G.; Keith, G. Sequence of tRNALeu from Bacillus Stearothermophilus CmAA. Biochem. Biophys. Res. Commun. 1983, 112, 578–585. [Google Scholar] [CrossRef]
- Finet, O.; Yague-Sanz, C.; Marchand, F.; Hermand, D. The Dihydrouridine Landscape from tRNA to mRNA: A Perspective on Synthesis, Structural Impact and Function. RNA Biol. 2022, 19, 735–750. [Google Scholar] [CrossRef]
- Tomasi, F.G.; Kimura, S.; Rubin, E.J.; Waldor, M.K. A tRNA Modification in Mycobacterium Tuberculosis Facilitates Optimal Intracellular Growth. eLife 2023, 12, RP87146. [Google Scholar] [CrossRef]
- Oerum, S.; Dégut, C.; Barraud, P.; Tisné, C. m1A Post-Transcriptional Modification in tRNAs. Biomolecules 2017, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Tomikawa, C. 7-Methylguanosine Modifications in Transfer RNA (tRNA). Int. J. Mol. Sci. 2018, 19, 4080. [Google Scholar] [CrossRef] [PubMed]
- De Crécy-Lagard, V.; Ross, R.L.; Jaroch, M.; Marchand, V.; Eisenhart, C.; Brégeon, D.; Motorin, Y.; Limbach, P.A. Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus Subtilis Sp Subtilis Strain 168. Biomolecules 2020, 10, 977. [Google Scholar] [CrossRef]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA Sequences and tRNA Genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef] [PubMed]
- Shigi, N. Biosynthesis and Degradation of Sulfur Modifications in tRNAs. Int. J. Mol. Sci. 2021, 22, 11937. [Google Scholar] [CrossRef]
- Yamagami, R.; Hori, H. Application of Mutational Profiling: New Functional Analyses Reveal the tRNA Recognition Mechanism of tRNA m1A22 Methyltransferase. J. Biol. Chem. 2023, 299, 102759. [Google Scholar] [CrossRef]
- Behrens, A.; Rodschinka, G.; Nedialkova, D.D. High-Resolution Quantitative Profiling of tRNA Abundance and Modification Status in Eukaryotes by Mim-tRNAseq. Mol. Cell 2021, 81, 1802–1815.e7. [Google Scholar] [CrossRef]
- Schwartz, S.; Motorin, Y. Next-Generation Sequencing Technologies for Detection of Modified Nucleotides in RNAs. RNA Biol. 2017, 14, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Verwilt, J.; Mestdagh, P.; Vandesompele, J. Artifacts and Biases of the Reverse Transcription Reaction in RNA Sequencing. RNA 2023, 29, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Sexton, A.N.; Wang, P.Y.; Rutenberg-Schoenberg, M.; Simon, M.D. Interpreting Reverse Transcriptase Termination and Mutation Events for Greater Insight into the Chemical Probing of RNA. Biochemistry 2017, 56, 4713–4721. [Google Scholar] [CrossRef] [PubMed]
- Wulff, B.-E.; Sakurai, M.; Nishikura, K. Elucidating the Inosinome: Global Approaches to Adenosine-to-Inosine RNA Editing. Nat. Rev. Genet. 2011, 12, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, L.; Li, X. Detection Technologies for RNA Modifications. Exp. Mol. Med. 2022, 54, 1601–1616. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed]
- Gamper, H.B.; Masuda, I.; Frenkel-Morgenstern, M.; Hou, Y.-M. Maintenance of Protein Synthesis Reading Frame by EF-P and m1G37-tRNA. Nat. Commun. 2015, 6, 7226. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Zheng, G.; Schwartz, M.H.; Clark, W.C.; Pan, T. Selective Enzymatic Demethylation of N2,N2-Dimethylguanosine in RNA and Its Application in High-Throughput tRNA Sequencing. Angew. Chem. Int. Ed. 2017, 56, 5017–5020. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.; Eyers, C.E. Analysis of Post-Translational Modifications by LC-MS/MS. In LC-MS/MS in Proteomics; Methods in Molecular Biology; Cutillas, P.R., Timms, J.F., Eds.; Humana Press: Totowa, NJ, USA, 2010; Volume 658, pp. 93–108. ISBN 978-1-60761-779-2. [Google Scholar]
- Panosyan, H.; Traube, F.R.; Brandmayr, C.; Wagner, M.; Carell, T. tRNA Modification Profiles in Obligate and Moderate Thermophilic Bacilli. Extremophiles 2022, 26, 11. [Google Scholar] [CrossRef]
- Lartigue, C.; Lebaudy, A.; Blanchard, A.; Yacoubi, B.E.; Rose, S.; Grosjean, H.; Douthwaite, S. The Flavoprotein Mcap0476 (RlmFO) Catalyzes m5U1939 Modification in Mycoplasma capricolum 23S rRNA. Nucleic Acids Res. 2014, 42, 8073–8082. [Google Scholar] [CrossRef][Green Version]
- Agris, P.F.; Vendeix, F.A.P.; Graham, W.D. tRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kramer, G.F.; Baker, J.C.; Ames, B.N. Near-UV Stress in Salmonella Typhimurium: 4-Thiouridine in tRNA, ppGpp, and ApppGpp as Components of an Adaptive Response. J. Bacteriol. 1988, 170, 2344–2351. [Google Scholar] [CrossRef] [PubMed]
- Griffey, R.H.; Davis, D.R.; Yamaizumi, Z.; Nishimura, S.; Hawkins, B.L.; Poulter, C.D. 15N-Labeled tRNA. Identification of 4-Thiouridine in Escherichia coli tRNASer1 and tRNATyr2 by 1H-15N Two-Dimensional NMR Spectroscopy. J. Biol. Chem. 1986, 261, 12074–12078. [Google Scholar] [CrossRef] [PubMed]
- Jamontas, R.; Laurynenas, A.; Povilaityte, D.; Meskys, R.; Aucynaite, A. RudS: Bacterial Desulfidase Responsible for tRNA 4-Thiouridine De-Modification. bioRxiv 2024. [Google Scholar] [CrossRef]
- Yared, M.-J.; Marcelot, A.; Barraud, P. Beyond the Anticodon: tRNA Core Modifications and Their Impact on Structure, Translation and Stress Adaptation. Genes 2024, 15, 374. [Google Scholar] [CrossRef] [PubMed]
- Yamagami, R.; Tomikawa, C.; Shigi, N.; Kazayama, A.; Asai, S.; Takuma, H.; Hirata, A.; Fourmy, D.; Asahara, H.; Watanabe, K.; et al. Folate-/FAD-dependent tRNA Methyltransferase from Thermus thermophilus Regulates Other Modifications in tRNA at Low Temperatures. Genes Cells 2016, 21, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Westhof, E. Pseudouridines or How to Draw on Weak Energy Differences. Biochem. Biophys. Res. Commun. 2019, 520, 702–704. [Google Scholar] [CrossRef] [PubMed]
- Noon, K.R.; Guymon, R.; Crain, P.F.; McCloskey, J.A.; Thomm, M.; Lim, J.; Cavicchioli, R. Influence of Temperature on tRNA Modification in Archaea: Methanococcoides burtonii (Optimum Growth Temperature [Topt], 23 °C) and Stetteria hydrogenophila (Topt, 95°C). J. Bacteriol. 2003, 185, 5483–5490. [Google Scholar] [CrossRef]
- Whitford, P.C.; Geggier, P.; Altman, R.B.; Blanchard, S.C.; Onuchic, J.N.; Sanbonmatsu, K.Y. Accommodation of Aminoacyl-tRNA into the Ribosome Involves Reversible Excursions along Multiple Pathways. RNA 2010, 16, 1196–1204. [Google Scholar] [CrossRef]
- Schmidt, L.; Werner, S.; Kemmer, T.; Niebler, S.; Kristen, M.; Ayadi, L.; Johe, P.; Marchand, V.; Schirmeister, T.; Motorin, Y.; et al. Graphical Workflow System for Modification Calling by Machine Learning of Reverse Transcription Signatures. Front. Genet. 2019, 10, 876. [Google Scholar] [CrossRef]
- Wintermeyer, W.; Schleich, H.-G.; Zachau, H.G. [4] Incorporation of Amines or Hydrazines into tRNA Replacing Wybutine or Dihydrouracil. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1979; Volume 59, pp. 110–121. ISBN 978-0-12-181959-0. [Google Scholar]
- Seidl, C.I.; Ryan, K. Circular Single-Stranded Synthetic DNA Delivery Vectors for MicroRNA. PLoS ONE 2011, 6, e16925. [Google Scholar] [CrossRef]
- Lama, L.; Ryan, K. Adenylylation of Small RNA Sequencing Adapters Using the TS2126 RNA Ligase I. RNA 2016, 22, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.; Weinberg, Z.; Vanderschuren, K.; Murdock, M.H.; Breaker, R.R. Natural Circularly Permuted Group II Introns in Bacteria Produce RNA Circles. iScience 2021, 24, 103431. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, D.H.; Rouskin, S.; Zhang, Y.; Li, G.-W.; Weissman, J.S.; Gross, C.A. Operon mRNAs Are Organized into ORF-Centric Structures That Predict Translation Efficiency. eLife 2017, 6, e22037. [Google Scholar] [CrossRef] [PubMed]
- Heyer, E.E.; Ozadam, H.; Ricci, E.P.; Cenik, C.; Moore, M.J. An Optimized Kit-Free Method for Making Strand-Specific Deep Sequencing Libraries from RNA Fragments. Nucleic Acids Res. 2015, 43, e2. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T. Genome-Wide Translational Profiling by Ribosome Footprinting. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 470, pp. 119–142. ISBN 978-0-12-375172-0. [Google Scholar]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sprinzl, M.; Vassilenko, K.S. Compilation of tRNA Sequences and Sequences of tRNA Genes. Nucleic Acids Res. 2004, 33, D139–D140. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Hoffmann, A.; Fallmann, J.; Vilardo, E.; Mörl, M.; Stadler, P.F.; Amman, F. Accurate Mapping of tRNA Reads. Bioinformatics 2018, 34, 1116–1124. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Hood, M.; Scott, L.; Peng, Q.; Mukherjee, S.; Tung, J.; Zhou, X. Differential Expression Analysis for RNAseq Using Poisson Mixed Models. Nucleic Acids Res. 2017, 45, e106. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Heiss, M.; Kellner, S. Detection of Nucleic Acid Modifications by Chemical Reagents. RNA Biol. 2017, 14, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, A.; Limbach, P.A. Mass Spectrometry of the Fifth Nucleoside: A Review of the Identification of Pseudouridine in Nucleic Acids. Anal. Chim. Acta 2008, 623, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Wolff, P.; Villette, C.; Zumsteg, J.; Heintz, D.; Antoine, L.; Chane-Woon-Ming, B.; Droogmans, L.; Grosjean, H.; Westhof, E. Comparative Patterns of Modified Nucleotides in Individual tRNA Species from a Mesophilic and Two Thermophilic Archaea. RNA 2020, 26, 1957–1975. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Sugawara, H.; Shumway, M.; on behalf of the International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]


| P. halocryophilus | E. sibiricum | B. subtilis | G. stearothermophilus | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 °C | 20 °C | 30 °C | 10 °C | 20 °C | 30 °C | 20 °C | 30 °C | 37 °C | 40 °C | 55 °C | 70 °C | |
| s4U8 | 1 | 3 | 2 | 3 | - | 1 | 4 | 4 | 4 | 30 | 32 | 24 |
| D16 | 5 | 6 | 5 | 3 | 3 | 3 | 7 | 4 | 4 | - | 3 | 3 |
| D17 | 14 | 14 | 15 | 15 | 16 | 16 | 20 | 21 | 21 | 1 | 12 | 13 |
| D20 | 17 | 17 | 17 | 25 | 23 | 24 | 21 | 21 | 19 | 6 | 18 | 19 |
| D20a | 12 | 12 | 12 | 11 | 11 | 11 | 14 | 14 | 14 | 2 | 10 | 11 |
| D20b | - | - | - | - | 1 | - | - | - | - | - | - | - |
| Ψ31 | 1 | 1 | 1 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 2 | 5 |
| Ψ32 | - | - | 1 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 2 | 2 |
| Ψ38 | 2 | 2 | 2 | 1 | 1 | 1 | - | - | - | - | - | - |
| Ψ39 | 11 | 11 | 11 | 13 | 13 | 14 | 13 | 13 | 13 | 9 | 13 | 14 |
| m7G46 | 17 | 17 | 17 | 18 | 18 | 18 | 22 | 22 | 23 | 18 | 18 | 18 |
| D47 | - | 3 | 1 | 14 | 4 | 7 | - | - | 10 | 2 | 2 | 5 |
| Ψ55 | 24 | 20 | 21 | 20 | 31 | 31 | 27 | 29 | 25 | 9 | 22 | 29 |
| Ψ60 | - | 5 | 13 | - | - | 4 | - | 12 | - | - | 12 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffmann, A.; Lorenz, C.; Fallmann, J.; Wolff, P.; Lechner, A.; Betat, H.; Mörl, M.; Stadler, P.F. Temperature-Dependent tRNA Modifications in Bacillales. Int. J. Mol. Sci. 2024, 25, 8823. https://doi.org/10.3390/ijms25168823
Hoffmann A, Lorenz C, Fallmann J, Wolff P, Lechner A, Betat H, Mörl M, Stadler PF. Temperature-Dependent tRNA Modifications in Bacillales. International Journal of Molecular Sciences. 2024; 25(16):8823. https://doi.org/10.3390/ijms25168823
Chicago/Turabian StyleHoffmann, Anne, Christian Lorenz, Jörg Fallmann, Philippe Wolff, Antony Lechner, Heike Betat, Mario Mörl, and Peter F. Stadler. 2024. "Temperature-Dependent tRNA Modifications in Bacillales" International Journal of Molecular Sciences 25, no. 16: 8823. https://doi.org/10.3390/ijms25168823
APA StyleHoffmann, A., Lorenz, C., Fallmann, J., Wolff, P., Lechner, A., Betat, H., Mörl, M., & Stadler, P. F. (2024). Temperature-Dependent tRNA Modifications in Bacillales. International Journal of Molecular Sciences, 25(16), 8823. https://doi.org/10.3390/ijms25168823






