The Ubiquitin Ligase Adaptor NDFIP1 Interacts with TRESK and Negatively Regulates the Background K+ Current
Abstract
1. Introduction
2. Results
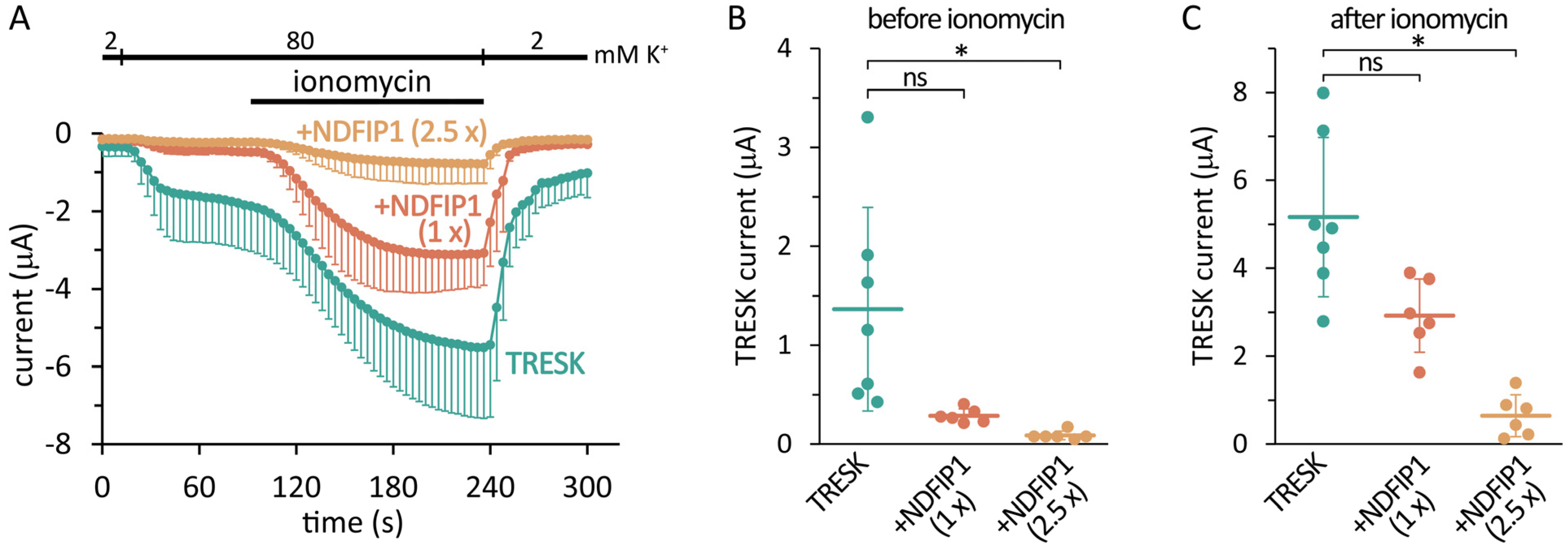
2.1. The Coexpression of NDFIP1 with TRESK Reduces the Background K+ Current
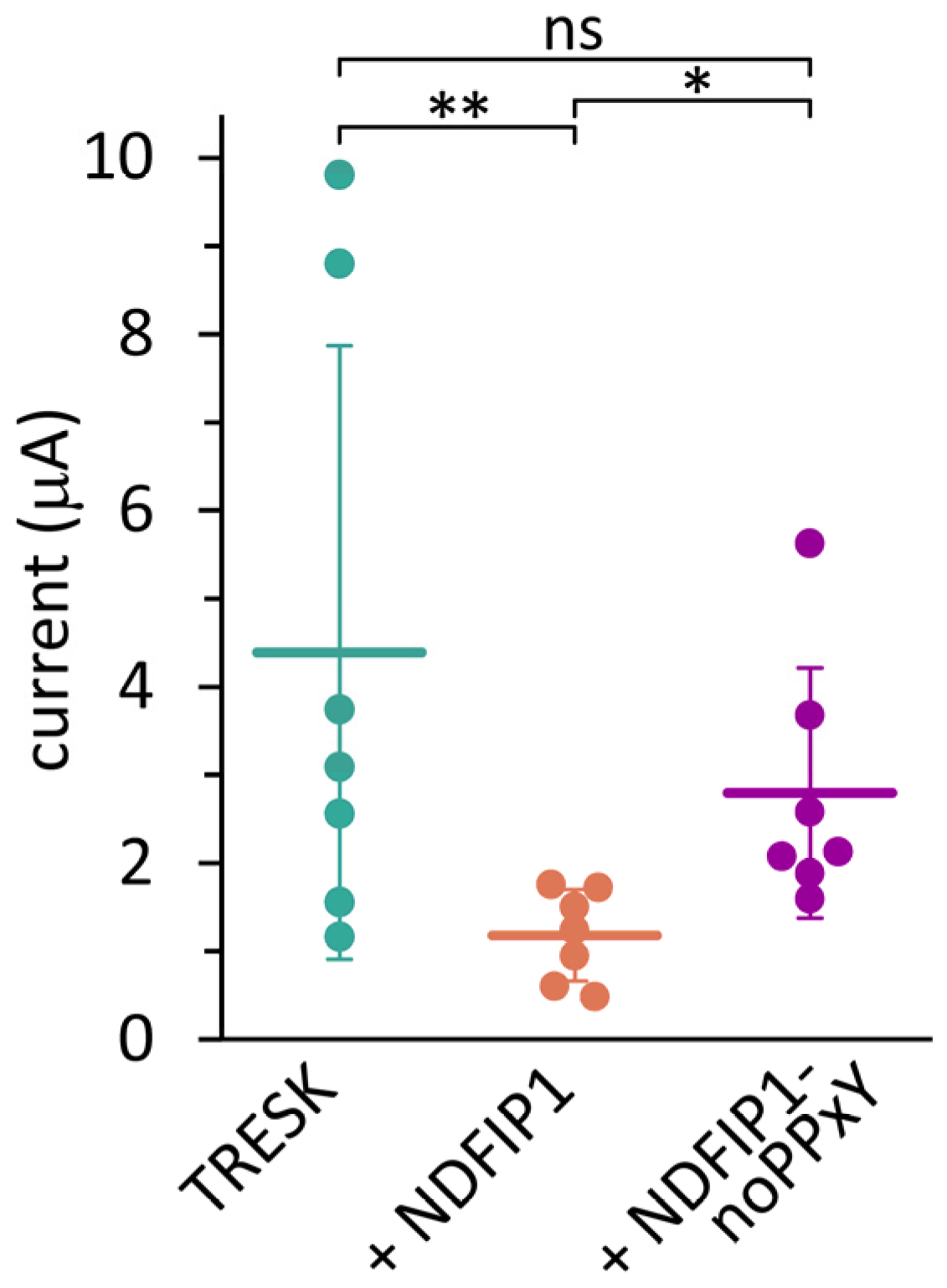
2.2. The Effect of NDFIP1 on TRESK Is Mediated by E3 Ubiquitin Ligases
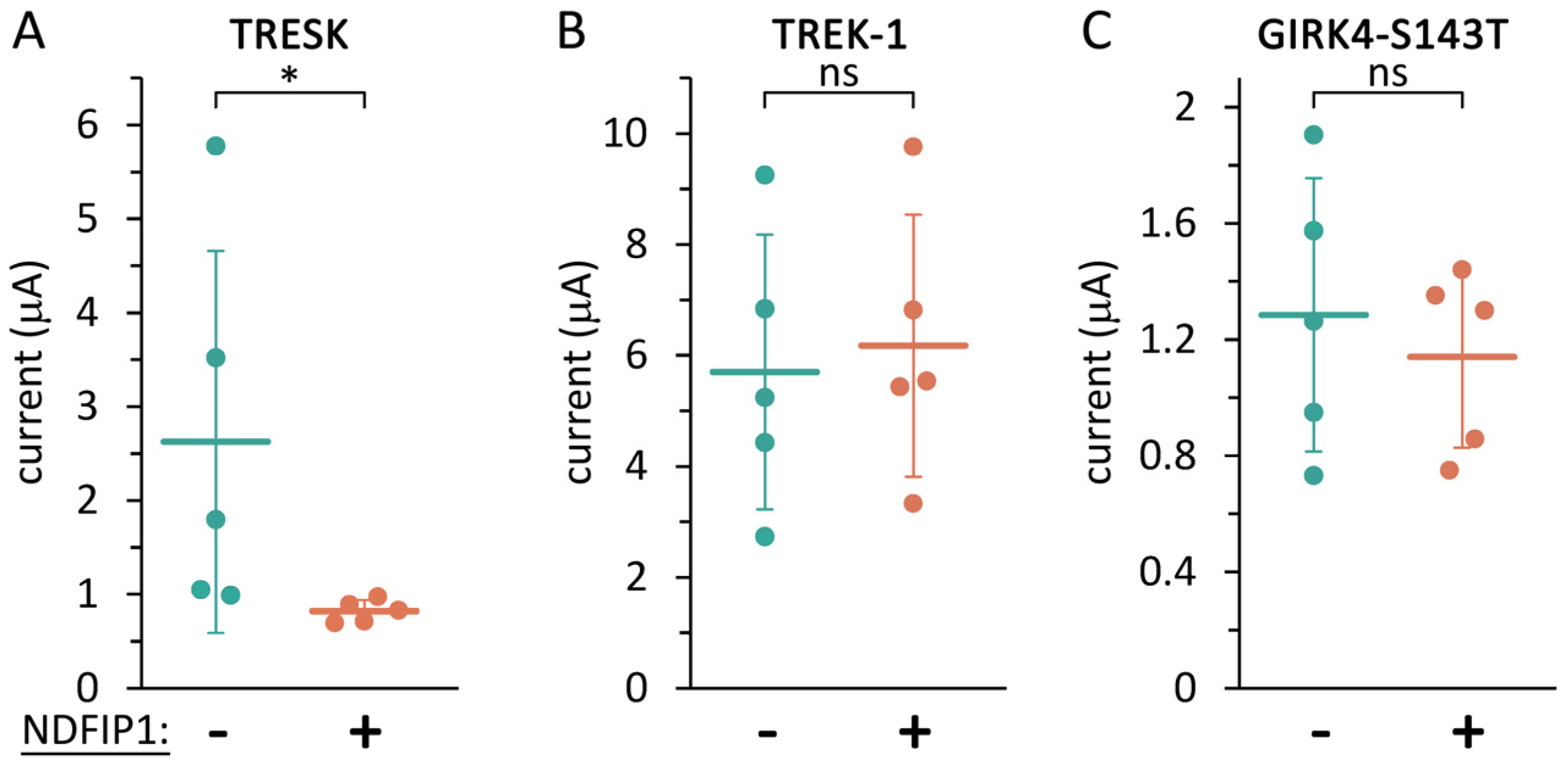
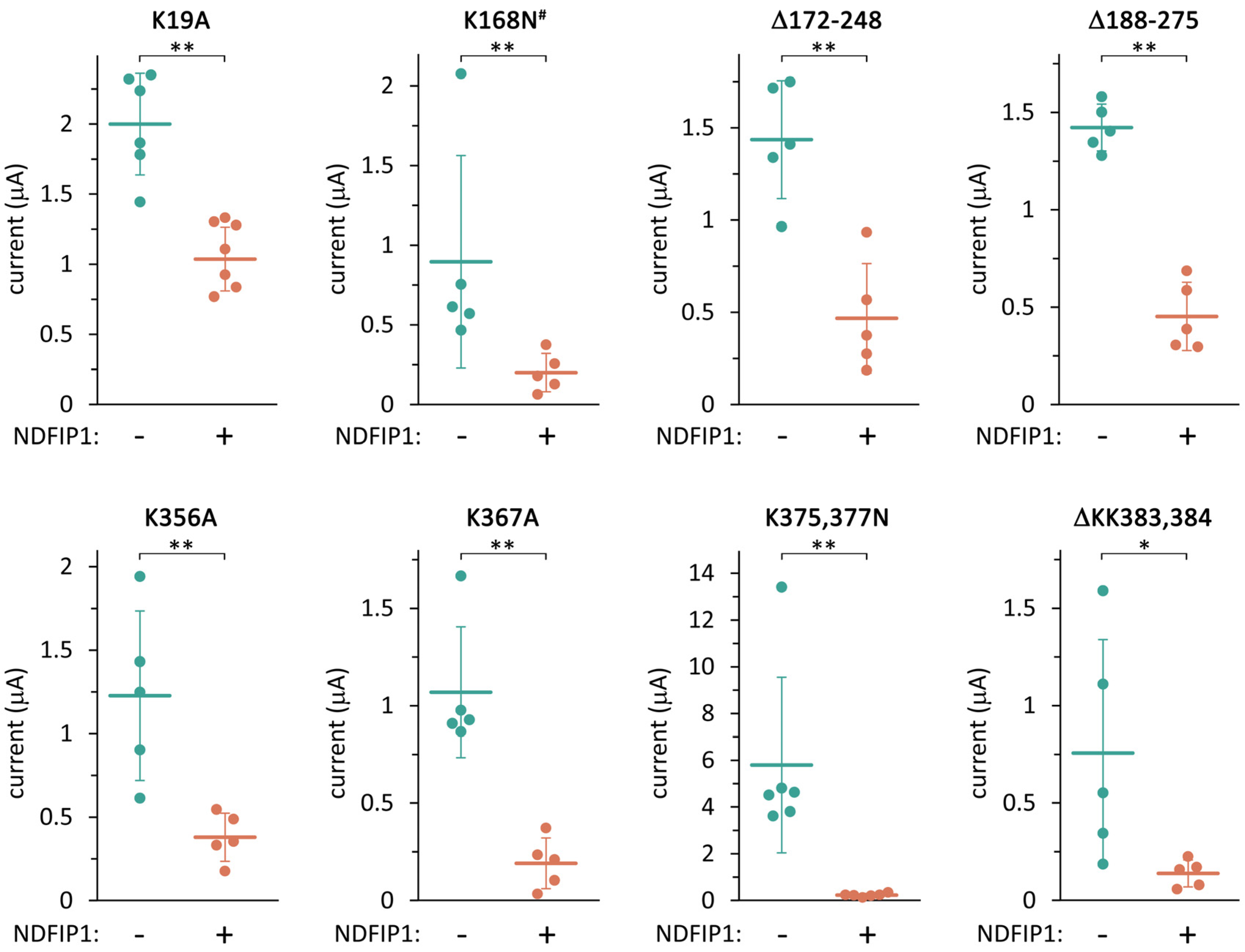
2.3. TRESK Regulation by NDFIP1 Does Not Depend on the Ubiquitination of a Single Lysine Residue in the Channel
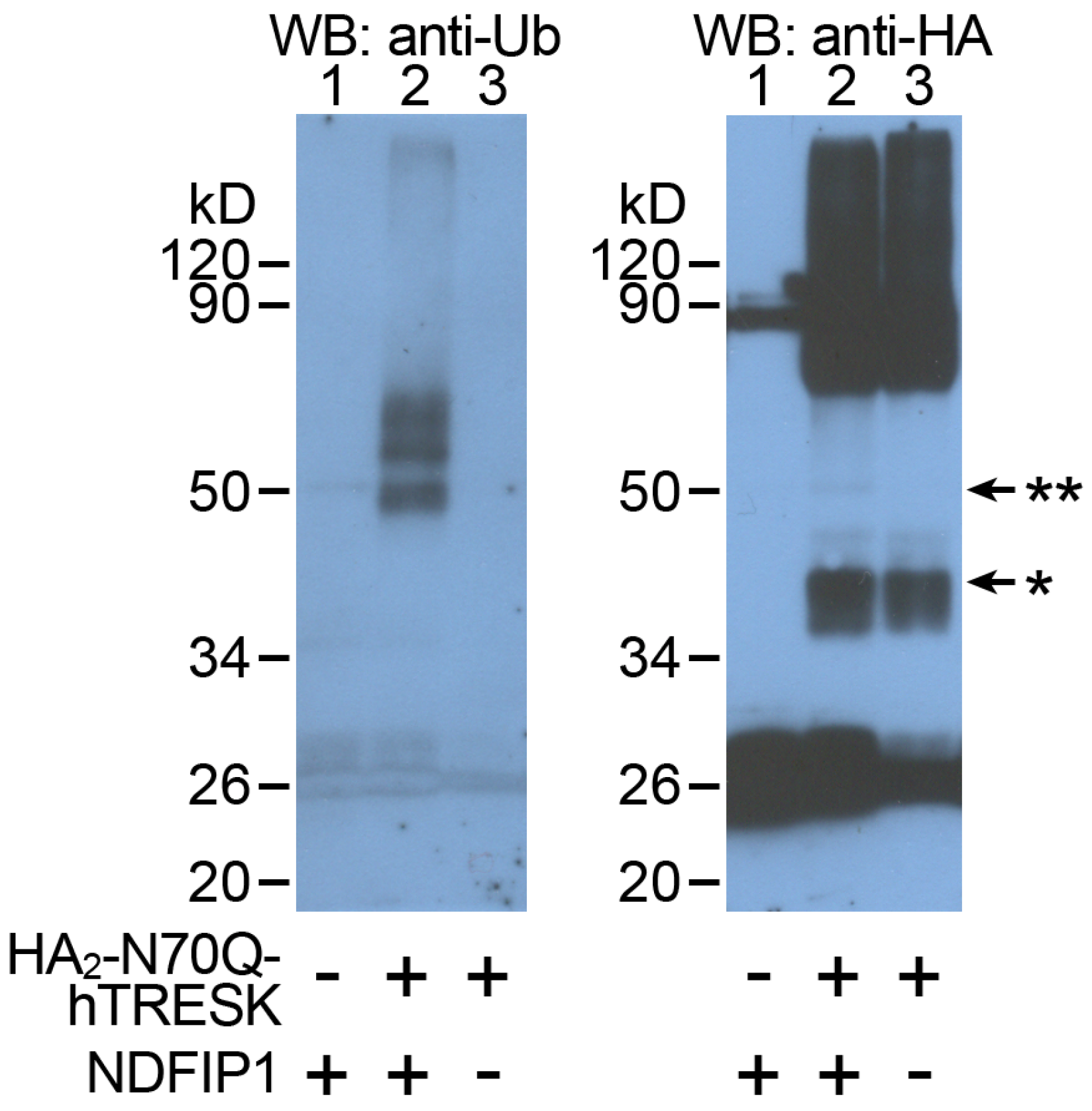
2.4. TRESK Is Ubiquitinated When NDFIP1 Is Coexpressed
2.5. Protein–Protein Interaction between TRESK and NDFIP1
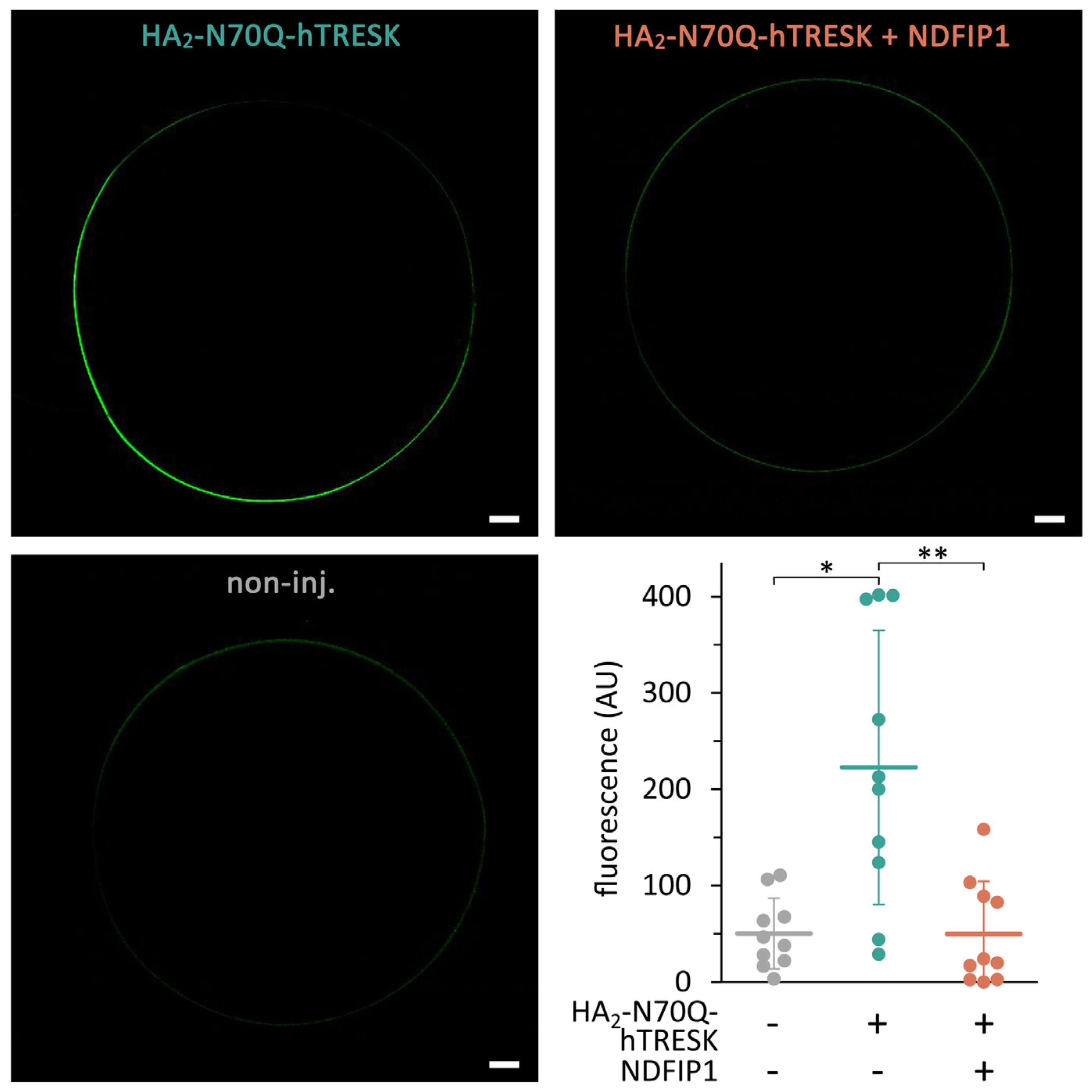
2.6. Coexpression of NDFIP1 Reduces the Quantity of TRESK Protein on the Oocyte Surface
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Molecular Biology
4.3. Two-Electrode Voltage Clamp
4.4. (Co-)Immunoprecipitation
4.5. Western Blot
4.6. Confocal Microscopy
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schreiber, J.A.; Dufer, M.; Seebohm, G. The Special One: Architecture, Physiology and Pharmacology of the TRESK Channel. Cell. Physiol. Biochem. 2022, 56, 663–684. [Google Scholar] [PubMed]
- Andres-Bilbe, A.; Castellanos, A.; Pujol-Coma, A.; Callejo, G.; Comes, N.; Gasull, X. The Background K+ Channel TRESK in Sensory Physiology and Pain. Int. J. Mol. Sci. 2020, 21, 5206. [Google Scholar] [CrossRef]
- Sano, Y.; Inamura, K.; Miyake, A.; Mochizuki, S.; Kitada, C.; Yokoi, H.; Nozawa, K.; Okada, H.; Matsushime, H.; Furuichi, K. A novel two-pore domain K+ channel, TRESK, is localized in the spinal cord. J. Biol. Chem. 2003, 278, 27406–27412. [Google Scholar] [CrossRef]
- Czirják, G.; Tóth, Z.E.; Enyedi, P. The two-pore domain K+ channel, TRESK, is activated by the cytoplasmic calcium signal through calcineurin. J. Biol. Chem. 2004, 279, 18550–18558. [Google Scholar] [CrossRef]
- Kang, D.; Mariash, E.; Kim, D. Functional expression of TRESK-2, a new member of the tandem-pore K+ channel family. J. Biol. Chem. 2004, 279, 28063–28070. [Google Scholar] [CrossRef]
- Kang, D.; Kim, D. TREK-2 (K2P10.1) and TRESK (K2P18.1) are major background K+ channels in dorsal root ganglion neurons. Am. J. Physiol. Cell Physiol. 2006, 291, C138–C146. [Google Scholar] [CrossRef]
- Dobler, T.; Springauf, A.; Tovornik, S.; Weber, M.; Schmitt, A.; Sedlmeier, R.; Wischmeyer, E.; Doring, F. TRESK two-pore-domain K+ channels constitute a significant component of background potassium currents in murine dorsal root ganglion neurones. J. Physiol. 2007, 585 Pt 3, 867–879. [Google Scholar] [CrossRef]
- Usoskin, D.; Furlan, A.; Islam, S.; Abdo, H.; Lonnerberg, P.; Lou, D.; Hjerling-Leffler, J.; Haeggstrom, J.; Kharchenko, O.; Kharchenko, P.V.; et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015, 18, 145–153. [Google Scholar] [CrossRef] [PubMed]
- LaPaglia, D.M.; Sapio, M.R.; Burbelo, P.D.; Thierry-Mieg, J.; Thierry-Mieg, D.; Raithel, S.J.; Ramsden, C.E.; Iadarola, M.J.; Mannes, A.J. RNA-Seq investigations of human post-mortem trigeminal ganglia. Cephalalgia 2018, 38, 912–932. [Google Scholar] [CrossRef]
- Tulleuda, A.; Cokic, B.; Callejo, G.; Saiani, B.; Serra, J.; Gasull, X. TRESK channel contribution to nociceptive sensory neurons excitability: Modulation by nerve injury. Mol. Pain 2011, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, C.X.; Zhong, J.Y.; Wang, H.B. Intrathecal TRESK gene recombinant adenovirus attenuates spared nerve injury-induced neuropathic pain in rats. Neuroreport 2013, 24, 131–136. [Google Scholar] [CrossRef]
- Weir, G.A.; Pettingill, P.; Wu, Y.; Duggal, G.; Ilie, A.S.; Akerman, C.J.; Cader, M.Z. The Role of TRESK in Discrete Sensory Neuron Populations and Somatosensory Processing. Front. Mol. Neurosci. 2019, 12, 170. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.T.; Siregar, A.S.; Kim, E.J.; Lee, E.S.; Nyiramana, M.M.; Woo, M.S.; Hah, Y.S.; Han, J.; Kang, D. Upregulation of TRESK Channels Contributes to Motor and Sensory Recovery after Spinal Cord Injury. Int. J. Mol. Sci. 2020, 21, 8997. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, A.; Pujol-Coma, A.; Andres-Bilbe, A.; Negm, A.; Callejo, G.; Soto, D.; Noel, J.; Comes, N.; Gasull, X. TRESK background K+ channel deletion selectively uncovers enhanced mechanical and cold sensitivity. J. Physiol. 2020, 598, 1017–1038. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiao, H.; Ren, W.; Ren, F. TRESK alleviates trigeminal neuralgia induced by infraorbital nerve chronic constriction injury in rats. Mol. Pain 2019, 15, 1744806919882511. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.P.; Jing, H.B.; Xi, K.; Zhang, Z.X.; Jin, Z.R.; Cai, S.Q.; Tian, Y.; Cai, J.; Xing, G.G. Contribution of TRESK two-pore domain potassium channel to bone cancer-induced spontaneous pain and evoked cutaneous pain in rats. Mol. Pain 2021, 17, 17448069211023230. [Google Scholar] [CrossRef] [PubMed]
- Lafreniere, R.G.; Cader, M.Z.; Poulin, J.F.; Andres-Enguix, I.; Simoneau, M.; Gupta, N.; Boisvert, K.; Lafreniere, F.; McLaughlan, S.; Dube, M.P.; et al. A dominant-negative mutation in the TRESK potassium channel is linked to familial migraine with aura. Nat. Med. 2010, 16, 1157–1160. [Google Scholar] [CrossRef]
- Guo, Z.; Qiu, C.S.; Jiang, X.; Zhang, J.; Li, F.; Liu, Q.; Dhaka, A.; Cao, Y.Q. TRESK K+ Channel Activity Regulates Trigeminal Nociception and Headache. eNeuro 2019, 6, ENEURO.0236-19.2019. [Google Scholar] [CrossRef]
- Royal, P.; Andres-Bilbe, A.; Avalos Prado, P.; Verkest, C.; Wdziekonski, B.; Schaub, S.; Baron, A.; Lesage, F.; Gasull, X.; Levitz, J.; et al. Migraine-Associated TRESK Mutations Increase Neuronal Excitability through Alternative Translation Initiation and Inhibition of TREK. Neuron 2019, 101, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Czirják, G.; Enyedi, P. TRESK background K+ channel is inhibited by phosphorylation via two distinct pathways. J. Biol. Chem. 2010, 285, 14549–14557. [Google Scholar] [CrossRef]
- Braun, G.; Nemcsics, B.; Enyedi, P.; Czirják, G. TRESK background K+ channel is inhibited by PAR-1/MARK microtubule affinity-regulating kinases in Xenopus oocytes. PLoS ONE 2011, 6, e28119. [Google Scholar] [CrossRef] [PubMed]
- Czirják, G.; Enyedi, P. Targeting of calcineurin to an NFAT-like docking site is required for the calcium-dependent activation of the background K+ channel, TRESK. J. Biol. Chem. 2006, 281, 14677–14682. [Google Scholar] [CrossRef] [PubMed]
- Czirják, G.; Enyedi, P. The LQLP calcineurin docking site is a major determinant of the calcium-dependent activation of human TRESK background K+ channel. J. Biol. Chem. 2014, 289, 29506–29518. [Google Scholar] [CrossRef] [PubMed]
- Imbrici, P.; Nematian-Ardestani, E.; Hasan, S.; Pessia, M.; Tucker, S.J.; D’Adamo, M.C. Altered functional properties of a missense variant in the TRESK K+ channel (KCNK18) associated with migraine and intellectual disability. Pflug. Arch. 2020, 472, 923–930. [Google Scholar] [CrossRef]
- Pavinato, L.; Nematian-Ardestani, E.; Zonta, A.; De Rubeis, S.; Buxbaum, J.; Mancini, C.; Bruselles, A.; Tartaglia, M.; Pessia, M.; Tucker, S.J.; et al. KCNK18 Biallelic Variants Associated with Intellectual Disability and Neurodevelopmental Disorders Alter TRESK Channel Activity. Int. J. Mol. Sci. 2021, 22, 6064. [Google Scholar] [CrossRef]
- Kollert, S.; Dombert, B.; Doring, F.; Wischmeyer, E. Activation of TRESK channels by the inflammatory mediator lysophosphatidic acid balances nociceptive signalling. Sci. Rep. 2015, 5, 12548. [Google Scholar] [CrossRef]
- Lalic, T.; Steponenaite, A.; Wei, L.; Vasudevan, S.R.; Mathie, A.; Peirson, S.N.; Lall, G.S.; Cader, M.Z. TRESK is a key regulator of nocturnal suprachiasmatic nucleus dynamics and light adaptive responses. Nat. Commun. 2020, 11, 4614. [Google Scholar] [CrossRef]
- Huang, W.; Ke, Y.; Zhu, J.; Liu, S.; Cong, J.; Ye, H.; Guo, Y.; Wang, K.; Zhang, Z.; Meng, W.; et al. TRESK channel contributes to depolarization-induced shunting inhibition and modulates epileptic seizures. Cell Rep. 2021, 36, 109404. [Google Scholar] [CrossRef]
- Ruck, T.; Bock, S.; Pfeuffer, S.; Schroeter, C.B.; Cengiz, D.; Marciniak, P.; Lindner, M.; Herrmann, A.; Liebmann, M.; Kovac, S.; et al. K2P18.1 translates T cell receptor signals into thymic regulatory T cell development. Cell Res. 2022, 32, 72–88. [Google Scholar] [CrossRef]
- Wright, P.D.; Weir, G.; Cartland, J.; Tickle, D.; Kettleborough, C.; Cader, M.Z.; Jerman, J. Cloxyquin (5-chloroquinolin-8-ol) is an activator of the two-pore domain potassium channel TRESK. Biochem. Biophys. Res. Commun. 2013, 441, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, M.; Dobolyi, A.; Czirják, G.; Enyedi, P. Selective and state-dependent activation of TRESK (K2P18.1) background potassium channel by cloxyquin. Br. J. Pharmacol. 2017, 174, 2102–2113. [Google Scholar] [CrossRef]
- Schreiber, J.A.; Derksen, A.; Goerges, G.; Schutte, S.; Sorgel, J.; Kiper, A.K.; Strutz-Seebohm, N.; Ruck, T.; Meuth, S.G.; Decher, N.; et al. Cloxyquin activates hTRESK by allosteric modulation of the selectivity filter. Commun. Biol. 2023, 6, 745. [Google Scholar] [CrossRef]
- Lengyel, M.; Hajdu, D.; Dobolyi, A.; Rosta, J.; Czirják, G.; Dux, M.; Enyedi, P. TRESK background potassium channel modifies the TRPV1-mediated nociceptor excitability in sensory neurons. Cephalalgia 2021, 41, 827–838. [Google Scholar] [CrossRef]
- Pettingill, P.; Weir, G.A.; Wei, T.; Wu, Y.; Flower, G.; Lalic, T.; Handel, A.; Duggal, G.; Chintawar, S.; Cheung, J.; et al. A causal role for TRESK loss of function in migraine mechanisms. Brain 2019, 142, 3852–3867. [Google Scholar] [CrossRef] [PubMed]
- Dilek, M.; Kilinc, Y.B.; Kilinc, E.; Torun, I.E.; Saylan, A.; Duzcu, S.E. Activation of TRESK background potassium channels by cloxyquin exerts protective effects against excitotoxic-induced brain injury and neuroinflammation in neonatal rats. J. Neuroimmunol. 2022, 368, 577894. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kim, G.T.; Kim, E.J.; La, J.H.; Lee, J.S.; Lee, E.S.; Park, J.Y.; Hong, S.G.; Han, J. Lamotrigine inhibits TRESK regulated by G-protein coupled receptor agonists. Biochem. Biophys. Res. Commun. 2008, 367, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Walsh, Y.; Leach, M.; Veale, E.L.; Mathie, A. Block of TREK and TRESK K2P channels by lamotrigine and two derivatives sipatrigine and CEN-092. Biochem. Biophys. Rep. 2021, 26, 101021. [Google Scholar] [CrossRef]
- Bautista, D.M.; Sigal, Y.M.; Milstein, A.D.; Garrison, J.L.; Zorn, J.A.; Tsuruda, P.R.; Nicoll, R.A.; Julius, D. Pungent agents from Szechuan peppers excite sensory neurons by inhibiting two-pore potassium channels. Nat. Neurosci. 2008, 11, 772–779. [Google Scholar] [CrossRef]
- Castellanos, A.; Andres, A.; Bernal, L.; Callejo, G.; Comes, N.; Gual, A.; Giblin, J.P.; Roza, C.; Gasull, X. Pyrethroids inhibit K2P channels and activate sensory neurons: Basis of insecticide-induced paraesthesias. Pain 2018, 159, 92–105. [Google Scholar] [CrossRef]
- Lengyel, M.; Erdélyi, F.; Pergel, E.; Balint-Polonka, A.; Dobolyi, A.; Bozsaki, P.; Dux, M.; Király, K.; Hegedűs, T.; Czirják, G.; et al. Chemically Modified Derivatives of the Activator Compound Cloxyquin Exert Inhibitory Effect on TRESK (K2P18.1) Background Potassium Channel. Mol. Pharmacol. 2019, 95, 652–660. [Google Scholar] [CrossRef]
- Harvey, K.F.; Shearwin-Whyatt, L.M.; Fotia, A.; Parton, R.G.; Kumar, S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J. Biol. Chem. 2002, 277, 9307–9317. [Google Scholar] [CrossRef] [PubMed]
- Mund, T.; Pelham, H.R. Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 2009, 10, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Oliver, P.M.; Cao, X.; Worthen, G.S.; Shi, P.; Briones, N.; MacLeod, M.; White, J.; Kirby, P.; Kappler, J.; Marrack, P.; et al. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 2006, 25, 929–940. [Google Scholar] [CrossRef]
- Howitt, J.; Putz, U.; Lackovic, J.; Doan, A.; Dorstyn, L.; Cheng, H.; Yang, B.; Chan-Ling, T.; Silke, J.; Kumar, S.; et al. Divalent metal transporter 1 (DMT1) regulation by Ndfip1 prevents metal toxicity in human neurons. Proc. Natl. Acad. Sci. USA 2009, 106, 15489–15494. [Google Scholar] [CrossRef]
- Foot, N.J.; Leong, Y.A.; Dorstyn, L.E.; Dalton, H.E.; Ho, K.; Zhao, L.; Garrick, M.D.; Yang, B.; Hiwase, D.; Kumar, S. Ndfip1-deficient mice have impaired DMT1 regulation and iron homeostasis. Blood 2011, 117, 638–646. [Google Scholar] [CrossRef]
- Low, L.H.; Chow, Y.L.; Li, Y.; Goh, C.P.; Putz, U.; Silke, J.; Ouchi, T.; Howitt, J.; Tan, S.S. Nedd4 family interacting protein 1 (Ndfip1) is required for ubiquitination and nuclear trafficking of BRCA1-associated ATM activator 1 (BRAT1) during the DNA damage response. J. Biol. Chem. 2015, 290, 7141–7150. [Google Scholar] [CrossRef]
- Trimpert, C.; Wesche, D.; de Groot, T.; Pimentel Rodriguez, M.M.; Wong, V.; van den Berg, D.T.M.; Cheval, L.; Ariza, C.A.; Doucet, A.; Stagljar, I.; et al. NDFIP allows NEDD4/NEDD4L-induced AQP2 ubiquitination and degradation. PLoS ONE 2017, 12, e0183774. [Google Scholar] [CrossRef] [PubMed]
- Hettema, E.H.; Valdez-Taubas, J.; Pelham, H.R. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004, 23, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Weber, J.; Polo, S.; Maspero, E. HECT E3 Ligases: A Tale with Multiple Facets. Front. Physiol. 2019, 10, 370. [Google Scholar] [CrossRef]
- Vivaudou, M.; Chan, K.W.; Sui, J.L.; Jan, L.Y.; Reuveny, E.; Logothetis, D.E. Probing the G-protein regulation of GIRK1 and GIRK4, the two subunits of the KACh channel, using functional homomeric mutants. J. Biol. Chem. 1997, 272, 31553–31560. [Google Scholar] [CrossRef]
- Pergel, E.; Lengyel, M.; Enyedi, P.; Czirják, G. TRESK (K2P18.1) Background Potassium Channel Is Activated by Novel-Type Protein Kinase C via Dephosphorylation. Mol. Pharmacol. 2019, 95, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Egenberger, B.; Polleichtner, G.; Wischmeyer, E.; Doring, F. N-linked glycosylation determines cell surface expression of two-pore-domain K+ channel TRESK. Biochem. Biophys. Res. Commun. 2010, 391, 1262–1267. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 2021, 184, 3022–3040. [Google Scholar] [CrossRef]
- Kang, Y.; Guo, J.; Yang, T.; Li, W.; Zhang, S. Regulation of the human ether-a-go-go-related gene (hERG) potassium channel by Nedd4 family interacting proteins (Ndfips). Biochem. J. 2015, 472, 71–82. [Google Scholar] [CrossRef]
- Shearwin-Whyatt, L.M.; Brown, D.L.; Wylie, F.G.; Stow, J.L.; Kumar, S. N4WBP5A (Ndfip2), a Nedd4-interacting protein, localizes to multivesicular bodies and the Golgi, and has a potential role in protein trafficking. J. Cell Sci. 2004, 117 Pt 16, 3679–3689. [Google Scholar] [CrossRef] [PubMed]
- Howitt, J.; Lackovic, J.; Low, L.H.; Naguib, A.; Macintyre, A.; Goh, C.P.; Callaway, J.K.; Hammond, V.; Thomas, T.; Dixon, M.; et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. J. Cell Biol. 2012, 196, 29–36. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, X.; Ye, X. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J. Immunol. 2012, 189, 5304–5313. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Supek, F.; Nelson, N.; Culotta, V.C. Negative control of heavy metal uptake by the Saccharomyces cerevisiae BSD2 gene. J. Biol. Chem. 1997, 272, 11763–11769. [Google Scholar] [CrossRef]
- Xia, M.; Liang, S.; Li, S.; Ji, M.; Chen, B.; Zhang, M.; Dong, C.; Chen, B.; Gong, W.; Wen, G.; et al. Iatrogenic Iron Promotes Neurodegeneration and Activates Self-Protection of Neural Cells against Exogenous Iron Attacks. Function 2021, 2, zqab003. [Google Scholar] [CrossRef]
- Tang, Y.L.; Zhu, L.; Yang, H.F.; Tao, Y.; Lu, W.; Cheng, H. Research progress on the role of Ndfip1 (Nedd4 family interacting protein 1) in immune cells. Allergol. Immunopathol. 2023, 51, 77–83. [Google Scholar] [CrossRef]
- Field, N.S.; Moser, E.K.; Oliver, P.M. Itch regulation of innate and adaptive immune responses in mice and humans. J. Leukoc. Biol. 2020, 108, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Beal, A.M.; Ramos-Hernandez, N.; Riling, C.R.; Nowelsky, E.A.; Oliver, P.M. TGF-beta induces the expression of the adaptor Ndfip1 to silence IL-4 production during iTreg cell differentiation. Nat. Immunol. 2011, 13, 77–85. [Google Scholar] [CrossRef]
- Layman, A.A.K.; Deng, G.; O’Leary, C.E.; Tadros, S.; Thomas, R.M.; Dybas, J.M.; Moser, E.K.; Wells, A.D.; Doliba, N.M.; Oliver, P.M. Ndfip1 restricts mTORC1 signalling and glycolysis in regulatory T cells to prevent autoinflammatory disease. Nat. Commun. 2017, 8, 15677. [Google Scholar] [CrossRef]
- Mund, T.; Pelham, H.R. Regulation of PTEN/Akt and MAP kinase signaling pathways by the ubiquitin ligase activators Ndfip1 and Ndfip2. Proc. Natl. Acad. Sci. USA 2010, 107, 11429–11434. [Google Scholar] [CrossRef]
- Foot, N.; Henshall, T.; Kumar, S. Ubiquitination and the Regulation of Membrane Proteins. Physiol. Rev. 2017, 97, 253–281. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.M.; Bilodeau, P.S.; Zhdankina, O.; Winistorfer, S.C.; Hauglund, M.J.; Allaman, M.M.; Kearney, W.R.; Robertson, A.D.; Boman, A.L.; Piper, R.C. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 2004, 6, 252–259. [Google Scholar] [CrossRef]
- Gorla, M.; Santiago, C.; Chaudhari, K.; Layman, A.A.K.; Oliver, P.M.; Bashaw, G.J. Ndfip Proteins Target Robo Receptors for Degradation and Allow Commissural Axons to Cross the Midline in the Developing Spinal Cord. Cell Rep. 2019, 26, 3298–3312. [Google Scholar] [CrossRef] [PubMed]
- Kamynina, E.; Debonneville, C.; Hirt, R.P.; Staub, O. Liddle’s syndrome: A novel mouse Nedd4 isoform regulates the activity of the epithelial Na+ channel. Kidney Int. 2001, 60, 466–471. [Google Scholar] [CrossRef]
- Fombonne, J.; Bissey, P.A.; Guix, C.; Sadoul, R.; Thibert, C.; Mehlen, P. Patched dependence receptor triggers apoptosis through ubiquitination of caspase-9. Proc. Natl. Acad. Sci. USA 2012, 109, 10510–10515. [Google Scholar] [CrossRef]
- Czirják, G.; Enyedi, P. Formation of functional heterodimers between the TASK-1 and TASK-3 two-pore domain potassium channel subunits. J. Biol. Chem. 2002, 277, 5426–5432. [Google Scholar] [CrossRef] [PubMed]
- Pergel, E.; Veres, I.; Csigi, G.I.; Czirják, G. Translocation of TMEM175 Lysosomal Potassium Channel to the Plasma Membrane by Dynasore Compounds. Int. J. Mol. Sci. 2021, 22, 10515. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pergel, E.; Tóth, D.J.; Baukál, D.; Veres, I.; Czirják, G. The Ubiquitin Ligase Adaptor NDFIP1 Interacts with TRESK and Negatively Regulates the Background K+ Current. Int. J. Mol. Sci. 2024, 25, 8879. https://doi.org/10.3390/ijms25168879
Pergel E, Tóth DJ, Baukál D, Veres I, Czirják G. The Ubiquitin Ligase Adaptor NDFIP1 Interacts with TRESK and Negatively Regulates the Background K+ Current. International Journal of Molecular Sciences. 2024; 25(16):8879. https://doi.org/10.3390/ijms25168879
Chicago/Turabian StylePergel, Enikő, Dániel J. Tóth, Dóra Baukál, Irén Veres, and Gábor Czirják. 2024. "The Ubiquitin Ligase Adaptor NDFIP1 Interacts with TRESK and Negatively Regulates the Background K+ Current" International Journal of Molecular Sciences 25, no. 16: 8879. https://doi.org/10.3390/ijms25168879
APA StylePergel, E., Tóth, D. J., Baukál, D., Veres, I., & Czirják, G. (2024). The Ubiquitin Ligase Adaptor NDFIP1 Interacts with TRESK and Negatively Regulates the Background K+ Current. International Journal of Molecular Sciences, 25(16), 8879. https://doi.org/10.3390/ijms25168879





