Biomarkers Identification in the Microenvironment of Oral Squamous Cell Carcinoma: A Systematic Review of Proteomic Studies
Abstract
1. Introduction
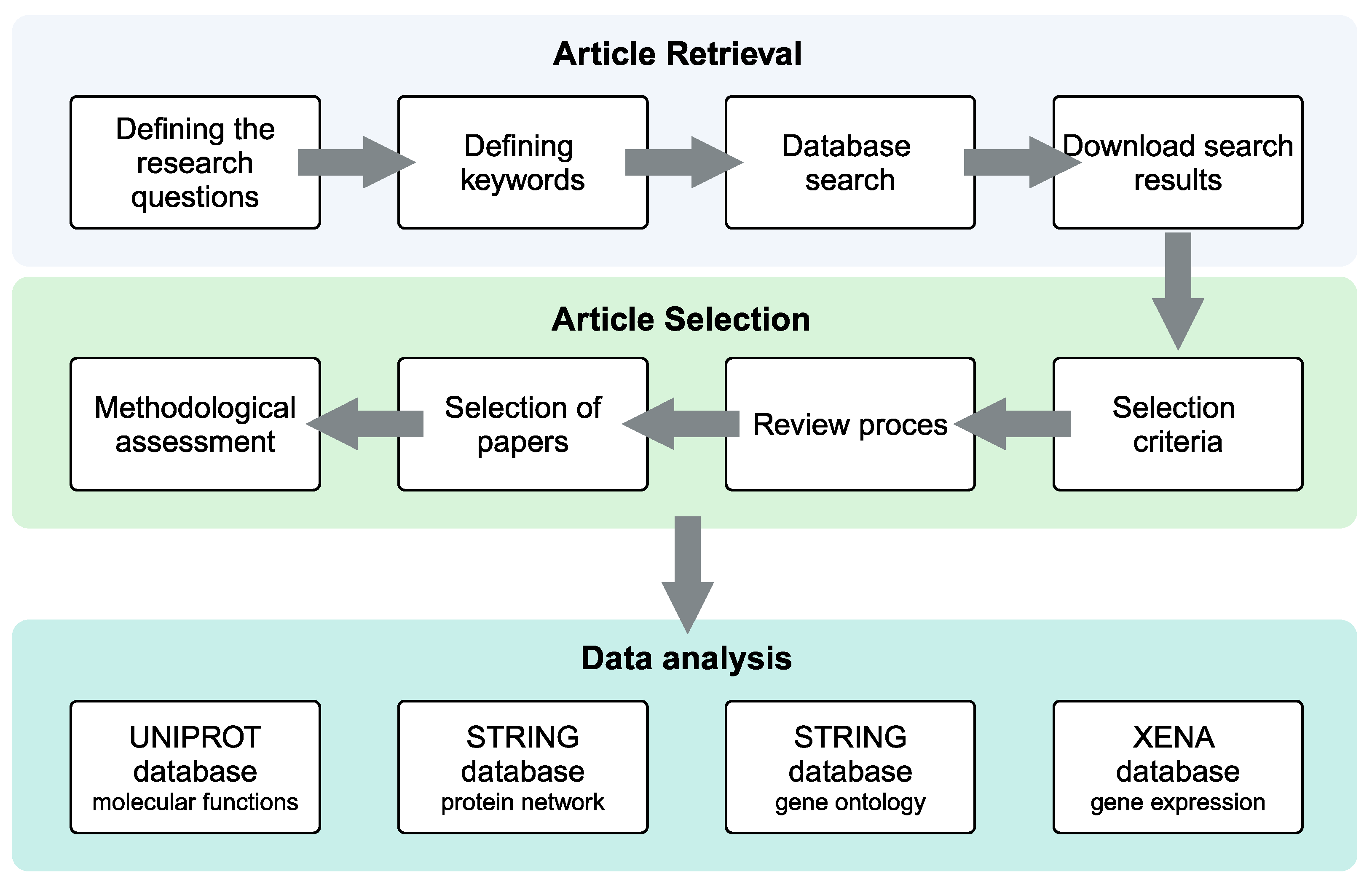
2. Materials and Methods
2.1. Systematic Review Protocol
2.2. Search Strategy and Eligibility Criteria
2.3. Methodological Quality Assessment and Data Extraction
3. Results
3.1. Systematic Review Analysis
| Refs | Cell Type | # Patient | # Control | Cohort of Patients | Sample Type | Technique |
|---|---|---|---|---|---|---|
| [31] | Tumor mass epithelium/stroma | 2 | 4 | Treatment-naïve patients | Proteome | Antibody microarray |
| [32] | Tumor mass | 5 | 5 | Treatment-naïve patients | Proteome | Imaging mass spectrometry associated with MALDI-TOF ultrafleXtreme |
| [33] | Tumor mass | 15 | / | Treatment-naïve patients | Proteome | Tandem mass tags LC-MS |
| [34] | Tumor mass center/adjacent epithelium | 3 | / | Treatment-naïve patients | Proteome | Dimethyl labeling associated LC-MS based on LTQ-Orbitrap Velos coupled with HPLC |
| [35] | Tumor mass | 5 | 5 | Treatment-naïve patients | Secretome | LC associated with nano-liquid MS |
| [30] | Tumor mass | 15 | 5 | All treatment-naïve patients, but one | Proteome/ Phosphoproteome | Tandem mass tags LC-MS |
| [36] | Tumor mass | 14 | 14 | Treatment-naïve patients | Proteome | LC-MS |
| [37] | Tumor mass | 20 | / | Treatment-naïve patients | Proteome | LC-MS |
| [38] | CAFs | 4 | 4 | Treatment-naïve patients | Secretome/ Proteome | Qexactive equipped with an EASY-Spray source tandem MS |
| [39] | CAFs | 1 | 1 | Treatment-naïve patients | Secretome | LC-MS |
| [40] | ECM | 6 | 3 | Treatment-naïve patients | Proteome | LC-MS |
| [41] | CAFs | 1 | 1 | Treatment-naïve patients | Secretome | MS based on LTQ-Orbitrap XL coupled to a nanoACQUITY Ultraperformance LC |
| [42] | CAFs | 1 | / | Treatment-naïve patients | Proteome | Phosphoproteomics by MS with Nano-LC |
| [43] | CAFs | 3 | 3 | Treatment-naïve patients | Proteome | Label-free LC-MS |
| [44] | MSCs | 1 | 2 | Treatment-naïve patients | Secretome | EASY-NLC 1000 Ultra-Performance LC system followed by MS in a Q Exactive TM Plus |
| [45] | TIF/NIF | 10 | 10 | Treatment-naïve patients | Proteome | In-gel digestion and GeLC-MS based on LTQ-Orbitrap Discovery coupled with HPLC |
| [46] | TIF/NIF | 2 | 2 | Treatment-naïve patients | Proteome | MS analysis on an LTQ-Orbitrap XL |
3.2. Proteomic Profiling of the Whole OSCC Mass
3.3. Proteomic Profiling of the OSSC Stromal Microenvironment
3.4. TME Protein–Protein Interaction and Association Network
3.4.1. Molecules Mediating Cell Adhesive Interactions with the ECM
- (1)
- (2)
- Chondroitin sulfate proteoglycan-4 (CSPG4), which consists of a transmembrane glycoprotein combined with a chondroitin sulfate proteoglycan, is expressed on the filopodia of migrating tumor cells, where it associates with the ECM-binding β1 integrins to favor tumor cell locomotion [55]. CSPG4 has been reported to be downregulated in CAFs and upregulated in TIFs when compared to their normal counterparts [41,45].
- (3)
- (4)
- (5)
- Fibulin-2 (FBLN2), another fibulin family member, is highly expressed by normal epithelial cells as compared to carcinoma cells and, upon binding to the β1 integrins, inactivates both the MAPK and the AKT pathways, thereby inhibiting cell proliferation, migration, and invasion [59]. FBLN2 levels have been found to be reduced in the ECM of OSCCs and increased in CAFs [40,41].
- (6)
- FGA represents the alpha chain of fibrinogen. The latter is a plasma protein whose cleavage to fibrin monomers is key to physiologic hemostasis [60], while in a tumor setting, it drives the formation of a fibrin matrix supporting cancer cell growth and metastasis [61]. FGA has been found to be downregulated in the ECM and upregulated in the mass of OSCCs [30,40].
- (7)
- (8)
- Heterochromatin protein 1 binding protein 3 (HP1BP3), a ubiquitously expressed nuclear protein belonging to the H1 histone family, plays an important role in cell growth and viability [62,63]. In cancer tissues, HP1BP3 is involved in the adhesion of malignant cells to collagen [64]. However, studies evaluating HP1BP3 expression levels in OSCCs produced conflicting results [36,37]. This is likely to depend on the disparate genetic, epigenetic, and metabolic alterations observed in OSCC tissues obtained from different patients [36,37].
- (9)
- (10)
- (11)
- (12)
- (13)
- (14)
- (15)
- (16)
- (17)
- (18)
3.4.2. Mediators of Glycolysis
- (1)
- (2)
- (3)
- CAV1 is a component of cell membrane caveolae that is upregulated and/or highly phosphorylated in tumor tissues, thereby increasing glycolysis, reducing oxidative phosphorylation, and promoting cancer cell invasion and cancer metastases [72]. CAV1 has been shown to be upregulated in the OSCC mass [33,35].
- (4)
- Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 (ENPP2) is a target of the Kirsten rat sarcoma viral oncogene homologue (KRAS), a transcriptional activator of key players in glucose metabolism [74]. A previous study reported that ENPP2 expression is upregulated in tumor cells following β4 integrins binding to ECM molecules that, in turn, promote cancer cell invasion [75]. These findings provide a link between cell–ECM adhesive interactions and glycolysis. Expression levels of ENPP2 have been reported to be reduced in CAFs [39,41].
- (5)
- (6)
- Pyruvate kinase M (PKM) is an enzyme that catalyzes the final step of glycolysis by converting phosphoenolpyruvate to pyruvate, leading to the origination of ATP, a crucial source of energy for the cells [77]. PKM is overexpressed in CAFs [44], while its phosphorylated form (Y175) is downregulated in the OSCC mass [30,43].
- (7)
- (8)
- (9)
4. Discussion
5. Conclusions and Future Perspective
- − Understand the biology of this tumor.
- − Individuate therapeutic targets and response markers.
- − Identify predictive and prognostic biomarkers for clinical use.
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- de Visser, K.E.; Joyce, J.A. The Evolving Tumor Microenvironment: From Cancer Initiation to Metastatic Outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef]
- Brassart-Pasco, S.; Brézillon, S.; Brassart, B.; Ramont, L.; Oudart, J.-B.; Monboisse, J.C. Tumor Microenvironment: Extracellular Matrix Alterations Influence Tumor Progression. Front. Oncol. 2020, 10, 397. [Google Scholar] [CrossRef]
- Park, S.J.; Min, H.J.; Yoon, C.; Kim, S.H.; Kim, J.H.; Lee, S.Y. Integrin Β1 Regulates the Perineural Invasion and Radioresistance of Oral Squamous Carcinoma Cells by Modulating Cancer Cell Stemness. Cell. Signal. 2023, 110, 110808. [Google Scholar] [CrossRef] [PubMed]
- Jang, T.-H.; Huang, W.-C.; Tung, S.-L.; Lin, S.-C.; Chen, P.-M.; Cho, C.-Y.; Yang, Y.-Y.; Yen, T.-C.; Lo, G.-H.; Chuang, S.-E.; et al. MicroRNA-485-5p Targets Keratin 17 to Regulate Oral Cancer Stemness and Chemoresistance via the Integrin/FAK/Src/ERK/β-Catenin Pathway. J. Biomed. Sci. 2022, 29, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Zhang, H.; Hu, G. Cancer and Microenvironment Plasticity: Double-Edged Swords in Metastasis. Trends Pharmacol. Sci. 2019, 40, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A Framework for Advancing Our Understanding of Cancer-Associated Fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Capello, M.; Katayama, H.; Hanash, S.M. Proteomic Profiling of the Tumor Microenvironment. Methods Mol. Biol. 2022, 2435, 157–167. [Google Scholar] [CrossRef]
- Thomas, S.N.; French, D.; Jannetto, P.J.; Rappold, B.A.; Clarke, W.A. Liquid Chromatography-Tandem Mass Spectrometry for Clinical Diagnostics. Nat. Rev. Methods Prim. 2022, 2, 96. [Google Scholar] [CrossRef]
- Haag, A.M. Mass Analyzers and Mass Spectrometers. Adv. Exp. Med. Biol. 2016, 919, 157–169. [Google Scholar] [CrossRef]
- Panarese, I.; Aquino, G.; Ronchi, A.; Longo, F.; Montella, M.; Cozzolino, I.; Roccuzzo, G.; Colella, G.; Caraglia, M.; Franco, R. Oral and Oropharyngeal Squamous Cell Carcinoma: Prognostic and Predictive Parameters in the Etiopathogenetic Route. Expert. Rev. Anticancer Ther. 2019, 19, 105–119. [Google Scholar] [CrossRef]
- Ahmad, W.M.A.W.; Yaqoob, M.A.; Noor, N.F.M.; Ghazali, F.M.M.; Rahman, N.A.; Tang, L.; Aleng, N.A.; Alam, M.K. The Predictive Model of Oral Squamous Cell Survival Carcinoma: A Methodology of Validation. Biomed. Res. Int. 2021, 2021, 5436894. [Google Scholar] [CrossRef]
- Du, E.; Mazul, A.L.; Farquhar, D.; Brennan, P.; Anantharaman, D.; Abedi-Ardekani, B.; Weissler, M.C.; Hayes, D.N.; Olshan, A.F.; Zevallos, J.P. Long-term Survival in Head and Neck Cancer: Impact of Site, Stage, Smoking, and Human Papillomavirus Status. Laryngoscope 2019, 129, 2506–2513. [Google Scholar] [CrossRef] [PubMed]
- Ciani, L.; Libonati, A.; Dri, M.; Pomella, S.; Campanella, V.; Barillari, G. About a Possible Impact of Endodontic Infections by Fusobacterium Nucleatum or Porphyromonas Gingivalis on Oral Carcinogenesis: A Literature Overview. Int. J. Mol. Sci. 2024, 25, 5083. [Google Scholar] [CrossRef] [PubMed]
- Kaminagakura, E.; Villa, L.L.; Andreoli, M.A.; Sobrinho, J.S.; Vartanian, J.G.; Soares, F.A.; Nishimoto, I.N.; Rocha, R.; Kowalski, L.P. High-Risk Human Papillomavirus in Oral Squamous Cell Carcinoma of Young Patients. Int. J. Cancer 2012, 130, 1726–1732. [Google Scholar] [CrossRef]
- Gondivkar, S.M.; Gadbail, A.R.; Sarode, S.C.; Hedaoo, A.; Dasgupta, S.; Sharma, B.; Sharma, A.; Gondivkar, R.S.; Yuwanati, M.; Patil, S.; et al. Oral and General Health-Related Quality of Life in Oral Squamous Cell Carcinoma Patients- Comparative Analysis of Different Treatment Regims. J. Oral Biol. Craniofacial Res. 2021, 11, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Pomella, S.; Cassandri, M.; Melaiu, O.; Marampon, F.; Gargari, M.; Campanella, V.; Rota, R.; Barillari, G. DNA Damage Response Gene Signature as Potential Treatment Markers for Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 2673. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef]
- Khanna, V.; Karjodkar, F.; Robbins, S.; Behl, M.; Arya, S.; Tripathi, A. Estimation of Serum Ferritin Level in Potentially Malignant Disorders, Oral Squamous Cell Carcinoma, and Treated Cases of Oral Squamous Cell Carcinoma. J. Cancer Res. Ther. 2017, 13, 550–555. [Google Scholar] [CrossRef]
- Chai, A.W.Y.; Lim, K.P.; Cheong, S.C. Translational Genomics and Recent Advances in Oral Squamous Cell Carcinoma. Semin. Cancer Biol. 2020, 61, 71–83. [Google Scholar] [CrossRef]
- Debela, D.T.; Muzazu, S.G.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New Approaches and Procedures for Cancer Treatment: Current Perspectives. SAGE Open Med. 2021, 9, 20503121211034370. [Google Scholar] [CrossRef] [PubMed]
- Shiravand, Y.; Khodadadi, F.; Kashani, S.M.A.; Hosseini-Fard, S.R.; Hosseini, S.; Sadeghirad, H.; Ladwa, R.; O’Byrne, K.; Kulasinghe, A. Immune Checkpoint Inhibitors in Cancer Therapy. Curr. Oncol. 2022, 29, 3044–3060. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, J.; Qian, H.; Zhuang, Q. Cancer-Associated Fibroblasts: From Basic Science to Anticancer Therapy. Exp. Mol. Med. 2023, 55, 1322–1332. [Google Scholar] [CrossRef]
- Li, X.; Bu, W.; Meng, L.; Liu, X.; Wang, S.; Jiang, L.; Ren, M.; Fan, Y.; Sun, H. CXCL12/CXCR4 Pathway Orchestrates CSC-like Properties by CAF Recruited Tumor Associated Macrophage in OSCC. Exp. Cell Res. 2019, 378, 131–138. [Google Scholar] [CrossRef]
- Chen, J.-H.; Wu, A.T.H.; Bamodu, O.A.; Yadav, V.K.; Chao, T.-Y.; Tzeng, Y.-M.; Mukhopadhyay, D.; Hsiao, M.; Lee, J.-C. Ovatodiolide Suppresses Oral Cancer Malignancy by Down-Regulating Exosomal Mir-21/STAT3/β-Catenin Cargo and Preventing Oncogenic Transformation of Normal Gingival Fibroblasts. Cancers 2019, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Cukura, A.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- McGuinness, L.A.; Higgins, J.P.T. Risk-of-bias VISualization (Robvis): An R Package and Shiny Web App for Visualizing Risk-of-bias Assessments. Res. Synth. Methods 2021, 12, 55–61. [Google Scholar] [CrossRef]
- Kaneko, T.; Zeng, P.Y.F.; Liu, X.; Abdo, R.; Barrett, J.W.; Zhang, Q.; Nichols, A.C.; Li, S.S.-C. Proteome and Phosphoproteome Signatures of Recurrence for HPV+ Head and Neck Squamous Cell Carcinoma. Commun. Med. 2022, 2, 95. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, V.; Leethanakul, C.; Bichsel, V.E.; Worth, J.M.; Prabhu, V.V.; Gutkind, J.S.; Liotta, L.A.; Munson, P.J.; Petricoin, E.F.; Krizman, D.B. Proteomic Profiling of the Cancer Microenvironment by Antibody Arrays. Proteomics 2001, 1, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Widlak, P.; Mrukwa, G.; Kalinowska, M.; Pietrowska, M.; Chekan, M.; Wierzgon, J.; Gawin, M.; Drazek, G.; Polanska, J. Detection of Molecular Signatures of Oral Squamous Cell Carcinoma and Normal Epithelium—Application of a Novel Methodology for Unsupervised Segmentation of Imaging Mass Spectrometry Data. Proteomics 2016, 16, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Routray, S.; Kumar, R.; Datta, K.K.; Puttamallesh, V.N.; Chatterjee, A.; Gowda, H.; Mohanty, N.; Dash, R.; Dixit, A. An Integrated Approach for Identification of a Panel of Candidate Genes Arbitrated for Invasion and Metastasis in Oral Squamous Cell Carcinoma. Sci. Rep. 2021, 11, 6208. [Google Scholar] [CrossRef]
- Alves, A.; Diel, L.; Ramos, G.; Pinto, A.; Bernardi, L.; Yates, J.; Lamers, M. Tumor Microenvironment and Oral Squamous Cell Carcinoma: A Crosstalk between the Inflammatory State and Tumor Cell Migration. Oral Oncol. 2021, 112, 105038. [Google Scholar] [CrossRef]
- Fraga, M.; Yáñez, M.; Sherman, M.; Llerena, F.; Hernandez, M.; Nourdin, G.; Álvarez, F.; Urrizola, J.; Rivera, C.; Lamperti, L.; et al. Immunomodulation of T Helper Cells by Tumor Microenvironment in Oral Cancer Is Associated With CCR8 Expression and Rapid Membrane Vitamin D Signaling Pathway. Front. Immunol. 2021, 12, 643298. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, M.; Bijnsdorp, I.V.; Böttger, F.; Piersma, S.R.; Pham, T.V.; Mumtaz, S.; Brakenhoff, R.H.; Akhtar, M.W.; Jimenez, C.R. Secreted Protein Markers in Oral Squamous Cell Carcinoma (OSCC). Clin. Proteomics 2022, 19, 4. [Google Scholar] [CrossRef]
- Miranda-Galvis, M.; Carneiro Soares, C.; Moretto Carnielli, C.; Ramalho Buttura, J.; Sales de Sá, R.; Kaminagakura, E.; Marchi, F.A.; Paes Leme, A.F.; Lópes Pinto, C.A.; Santos-Silva, A.R.; et al. New Insights into the Impact of Human Papillomavirus on Oral Cancer in Young Patients: Proteomic Approach Reveals a Novel Role for S100A8. Cells 2023, 12, 1323. [Google Scholar] [CrossRef] [PubMed]
- Principe, S.; Mejia-Guerrero, S.; Ignatchenko, V.; Sinha, A.; Ignatchenko, A.; Shi, W.; Pereira, K.; Su, S.; Huang, S.H.; O’Sullivan, B.; et al. Proteomic Analysis of Cancer-Associated Fibroblasts Reveals a Paracrine Role for MFAP5 in Human Oral Tongue Squamous Cell Carcinoma. J. Proteome Res. 2018, 17, 2045–2059. [Google Scholar] [CrossRef]
- Bagordakis, E.; Sawazaki-Calone, I.; Macedo, C.C.S.; Carnielli, C.M.; de Oliveira, C.E.; Rodrigues, P.C.; Rangel, A.L.C.A.; Dos Santos, J.N.; Risteli, J.; Graner, E.; et al. Secretome Profiling of Oral Squamous Cell Carcinoma-Associated Fibroblasts Reveals Organization and Disassembly of Extracellular Matrix and Collagen Metabolic Process Signatures. Tumour Biol. 2016, 37, 9045–9057. [Google Scholar] [CrossRef]
- He, Y.; Deng, P.; Yan, Y.; Zhu, L.; Chen, H.; Li, T.; Li, Y.; Li, J. Matrisome Provides a Supportive Microenvironment for Oral Squamous Cell Carcinoma Progression. J. Proteomics 2022, 253, 104454. [Google Scholar] [CrossRef]
- Álvarez-Teijeiro, S.; García-Inclán, C.; Villaronga, M.Á.; Casado, P.; Hermida-Prado, F.; Granda-Díaz, R.; Rodrigo, J.P.; Calvo, F.; Del-Río-Ibisate, N.; Gandarillas, A.; et al. Factors Secreted by Cancer-Associated Fibroblasts That Sustain Cancer Stem Properties in Head and Neck Squamous Carcinoma Cells as Potential Therapeutic Targets. Cancers 2018, 10, 334. [Google Scholar] [CrossRef]
- Prieto-Fernandez, L.; Villaronga, M.d.L.A.; Hermida-Prado, F.; Hijazi, M.; Montoro-Jimenez, I.; Pevida, M.; Llames, S.; Rodrigo, J.P.; Cutillas, P.; Calvo, F.; et al. Driving Role of Head and Neck Cancer Cell Secretome on the Invasion of Stromal Fibroblasts: Mechanistic Insights by Phosphoproteomics. Biomed. Pharmacother. 2023, 158, 114176. [Google Scholar] [CrossRef]
- Xiao, L.; Hu, Q.; Peng, Y.; Zheng, K.; Zhang, T.; Yang, L.; Wang, Z.; Tang, W.; Yu, J.; Xiao, Q.; et al. TRAP1 Suppresses Oral Squamous Cell Carcinoma Progression by Reducing Oxidative Phosphorylation Metabolism of Cancer-Associated Fibroblasts. BMC Cancer 2021, 21, 1329. [Google Scholar] [CrossRef]
- Li, S.; Han, Y.; Lu, M.; Liu, Z.; Jin, J.; Guo, Q.; Wang, Y.; Liu, H. Mesenchymal Stem Cell-Exosome-Mediated Matrix Metalloproteinase 1 Participates in Oral Leukoplakia and Carcinogenesis by Inducing Angiogenesis. J. Oral Pathol. Med. 2022, 51, 638–648. [Google Scholar] [CrossRef]
- Hsu, C.-W.; Chang, K.-P.; Huang, Y.; Liu, H.-P.; Hsueh, P.-C.; Gu, P.-W.; Yen, W.-C.; Wu, C.-C. Proteomic Profiling of Paired Interstitial Fluids Reveals Dysregulated Pathways and Salivary NID1 as a Biomarker of Oral Cavity Squamous Cell Carcinoma. Mol. Cell. Proteomics 2019, 18, 1939–1949. [Google Scholar] [CrossRef] [PubMed]
- Hardt, M.; Lam, D.K.; Dolan, J.C.; Schmidt, B.L. Surveying Proteolytic Processes in Human Cancer Microenvironments by Microdialysis and Activity-Based Mass Spectrometry. Proteomics Clin. Appl. 2011, 5, 636–643. [Google Scholar] [CrossRef]
- Routray, S. Caveolin-1 in Oral Squamous Cell Carcinoma Microenvironment: An Overview. Tumour Biol. 2014, 35, 9487–9495. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yan, M.; Zhang, J.; Wang, X.; Shen, Z.; Lv, Z.; Li, Z.; Wei, W.; Chen, W. TGFβ3-Mediated Induction of Periostin Facilitates Head and Neck Cancer Growth and Is Associated with Metastasis. Sci. Rep. 2016, 6, 20587. [Google Scholar] [CrossRef]
- Augsten, M. Cancer-Associated Fibroblasts as Another Polarized Cell Type of the Tumor Microenvironment. Front. Oncol. 2014, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Chu, H.-W.; Hsu, C.-W.; Chang, K.-P.; Liu, H.-P. Saliva Proteome Profiling Reveals Potential Salivary Biomarkers for Detection of Oral Cavity Squamous Cell Carcinoma. Proteomics 2015, 15, 3394–3404. [Google Scholar] [CrossRef]
- Singh, P.; Verma, J.K.; Singh, J.K. Validation of Salivary Markers, IL-1β, IL-8 and Lgals3bp for Detection of Oral Squamous Cell Carcinoma in an Indian Population. Sci. Rep. 2020, 10, 7365. [Google Scholar] [CrossRef]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in Inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef]
- Dzobo, K.; Senthebane, D.A.; Dandara, C. The Tumor Microenvironment in Tumorigenesis and Therapy Resistance Revisited. Cancers 2023, 15, 376. [Google Scholar] [CrossRef]
- Fucikova, J.; Spisek, R.; Kroemer, G.; Galluzzi, L. Calreticulin and Cancer. Cell Res. 2021, 31, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Price, M.A.; Wanshura, L.E.C.; He, J.; Yi, M.; Welch, D.R.; Li, G.; Conner, S.; Sachs, J.; Turley, E.A.; et al. Chondroitin Sulfate Proteoglycan 4 Enhanced Melanoma Motility and Growth Requires a Cysteine in the Core Protein Transmembrane Domain. Melanoma Res. 2019, 29, 365–375. [Google Scholar] [CrossRef]
- Nehls, C.; Böhling, A.; Podschun, R.; Schubert, S.; Grötzinger, J.; Schromm, A.; Fedders, H.; Leippe, M.; Harder, J.; Kaconis, Y.; et al. Influence of Disulfide Bonds in Human Beta Defensin-3 on Its Strain Specific Activity against Gram-Negative Bacteria. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183273. [Google Scholar] [CrossRef]
- Chang, H.-T.; Tsai, P.-W.; Huang, H.-H.; Liu, Y.-S.; Chien, T.-S.; Lan, C.-Y. LL37 and HBD-3 Elevate the β-1,3-Exoglucanase Activity of Candida Albicans Xog1p, Resulting in Reduced Fungal Adhesion to Plastic. Biochem. J. 2012, 441, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Song, E.; Hou, Y.; Yu, S.; Chen, S.; Huang, J.; Luo, T.; Kong, L.; Xu, J.; Wang, H. EFEMP1 Expression Promotes Angiogenesis and Accelerates the Growth of Cervical Cancer in Vivo. Gynecol. Oncol. 2011, 121, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Nenkov, M.; Schröder, D.C.; Abubrig, M.; Gassler, N.; Chen, Y. Fibulin 2 Is Hypermethylated and Suppresses Tumor Cell Proliferation through Inhibition of Cell Adhesion and Extracellular Matrix Genes in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 11834. [Google Scholar] [CrossRef]
- Vilar, R.; Fish, R.J.; Casini, A.; Neerman-Arbez, M. Fibrin(Ogen) in Human Disease: Both Friend and Foe. Haematologica 2020, 105, 284–296. [Google Scholar] [CrossRef]
- Yapijakis, C.; Bramos, A.; Nixon, A.M.; Ragos, V.; Vairaktaris, E. The Interplay between Hemostasis and Malignancy: The Oral Cancer Paradigm. Anticancer Res. 2012, 32, 1791–1800. [Google Scholar]
- Dutta, B.; Ren, Y.; Hao, P.; Sim, K.H.; Cheow, E.; Adav, S.; Tam, J.P.; Sze, S.K. Profiling of the Chromatin-Associated Proteome Identifies HP1BP3 as a Novel Regulator of Cell Cycle Progression. Mol. Cell. Proteomics 2014, 13, 2183–2197. [Google Scholar] [CrossRef] [PubMed]
- Garfinkel, B.P.; Melamed-Book, N.; Anuka, E.; Bustin, M.; Orly, J. HP1BP3 Is a Novel Histone H1 Related Protein with Essential Roles in Viability and Growth. Nucleic Acids Res. 2015, 43, 2074–2090. [Google Scholar] [CrossRef] [PubMed]
- Dutta, B.; Yan, R.; Lim, S.K.; Tam, J.P.; Sze, S.K. Quantitative Profiling of Chromatome Dynamics Reveals a Novel Role for HP1BP3 in Hypoxia-Induced Oncogenesis. Mol. Cell. Proteomics 2014, 13, 3236–3249. [Google Scholar] [CrossRef]
- Koshikawa, N.; Minegishi, T.; Nabeshima, K.; Seiki, M. Development of a New Tracking Tool for the Human Monomeric Laminin-Gamma 2 Chain in Vitro and in Vivo. Cancer Res. 2008, 68, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, C.; Li, N.; Wang, F.; Xu, Y.; Shen, Z.; Yang, L.; Li, Z.; He, C. The CircEPSTI1/Mir-942-5p/LTBP2 Axis Regulates the Progression of OSCC in the Background of OSF via EMT and the PI3K/Akt/MTOR Pathway. Cell Death Dis. 2020, 11, 682. [Google Scholar] [CrossRef]
- Pang, X.-F.; Lin, X.; Du, J.-J.; Zeng, D.-Y. LTBP2 Knockdown by SiRNA Reverses Myocardial Oxidative Stress Injury, Fibrosis and Remodelling during Dilated Cardiomyopathy. Acta Physiol. 2020, 228, e13377. [Google Scholar] [CrossRef] [PubMed]
- Kao, W.W.-Y.; Funderburgh, J.L.; Xia, Y.; Liu, C.-Y.; Conrad, G.W. Focus on Molecules: Lumican. Exp. Eye Res. 2006, 82, 3–4. [Google Scholar] [CrossRef]
- Dorafshan, S.; Razmi, M.; Safaei, S.; Gentilin, E.; Madjd, Z.; Ghods, R. Periostin: Biology and Function in Cancer. Cancer Cell Int. 2022, 22, 315. [Google Scholar] [CrossRef]
- Irigoyen, M.; Ansó, E.; Salvo, E.; de las Herrerías, J.D.; Martínez-Irujo, J.J.; Rouzaut, A. TGFβ-Induced Protein Mediates Lymphatic Endothelial Cell Adhesion to the Extracellular Matrix under Low Oxygen Conditions. Cell. Mol. Life Sci. 2008, 65, 2244–2255. [Google Scholar] [CrossRef]
- Abedsaeidi, M.; Hojjati, F.; Tavassoli, A.; Sahebkar, A. Biology of Tenascin C and Its Role in Physiology and Pathology. Curr. Med. Chem. 2023, 31, 2706–2731. [Google Scholar] [CrossRef]
- Díaz-Valdivia, N.; Simón, L.; Díaz, J.; Martinez-Meza, S.; Contreras, P.; Burgos-Ravanal, R.; Pérez, V.I.; Frei, B.; Leyton, L.; Quest, A.F.G. Mitochondrial Dysfunction and the Glycolytic Switch Induced by Caveolin-1 Phosphorylation Promote Cancer Cell Migration, Invasion, and Metastasis. Cancers 2022, 14, 2862. [Google Scholar] [CrossRef]
- Zhao, J.; Tian, M.; Zhang, S.; Delfarah, A.; Gao, R.; Rao, Y.; Savas, A.C.; Lu, A.; Bubb, L.; Lei, X.; et al. Deamidation Shunts RelA from Mediating Inflammation to Aerobic Glycolysis. Cell Metab. 2020, 31, 937–955.e7. [Google Scholar] [CrossRef]
- Ma, Z.; Li, Z.; Ma, Z.; Zhou, Z.; Zhuang, H.; Liu, C.; Huang, B.; Zou, Y.; Zheng, Z.; Yang, L.; et al. Development of a KRAS-Associated Metabolic Risk Model for Prognostic Prediction in Pancreatic Cancer. Biomed. Res. Int. 2021, 2021, 9949272. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Huang, H.; Cheepala, S.; Huang, S.; Chung, J. Curcumin Inhibition of Integrin (α6β4)-Dependent Breast Cancer Cell Motility and Invasion. Cancer Prev. Res. 2008, 1, 385–391. [Google Scholar] [CrossRef]
- Giudice, J.; Barcos, L.S.; Guaimas, F.F.; Penas-Steinhardt, A.; Giordano, L.; Jares-Erijman, E.A.; Coluccio Leskow, F. Insulin and Insulin like Growth Factor II Endocytosis and Signaling via Insulin Receptor B. Cell Commun. Signal. 2013, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Alquraishi, M.; Puckett, D.L.; Alani, D.S.; Humidat, A.S.; Frankel, V.D.; Donohoe, D.R.; Whelan, J.; Bettaieb, A. Pyruvate Kinase M2: A Simple Molecule with Complex Functions. Free Radic. Biol. Med. 2019, 143, 176–192. [Google Scholar] [CrossRef]
- Kang, Y.K.; Putluri, N.; Maity, S.; Tsimelzon, A.; Ilkayeva, O.; Mo, Q.; Lonard, D.; Michailidis, G.; Sreekumar, A.; Newgard, C.B.; et al. CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-ΚB-CMYC. PLoS Genet. 2015, 11, e1005116. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Huo, Y.-M.; Yang, M.-W.; Shen, Y.; Liu, D.-J.; Fu, X.-L.; Tao, L.-Y.; He, R.-Z.; Zhang, J.-F.; Hua, R.; et al. SF3B1 Mutation in Pancreatic Cancer Contributes to Aerobic Glycolysis and Tumor Growth through a PP2A-c-Myc Axis. Mol. Oncol. 2021, 15, 3076–3090. [Google Scholar] [CrossRef] [PubMed]
- Shoshan-Barmatz, V.; Israelson, A.; Brdiczka, D.; Sheu, S.S. The Voltage-Dependent Anion Channel (VDAC): Function in Intracellular Signalling, Cell Life and Cell Death. Curr. Pharm. Des. 2006, 12, 2249–2270. [Google Scholar] [CrossRef]
- Nelson, E.A.; Walker, S.R.; Li, W.; Liu, X.S.; Frank, D.A. Identification of Human STAT5-Dependent Gene Regulatory Elements Based on Interspecies Homology. J. Biol. Chem. 2006, 281, 26216–26224. [Google Scholar] [CrossRef]
- Talati, P.G.; Gu, L.; Ellsworth, E.M.; Girondo, M.A.; Trerotola, M.; Hoang, D.T.; Leiby, B.; Dagvadorj, A.; McCue, P.A.; Lallas, C.D.; et al. Jak2-Stat5a/b Signaling Induces Epithelial-to-Mesenchymal Transition and Stem-Like Cell Properties in Prostate Cancer. Am. J. Pathol. 2015, 185, 2505–2522. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, J.; Liu, Y.; Zhang, P.; Nie, J.; Zhao, R.; Shi, Q.; Sun, H.; Jiao, D.; Chen, Y.; et al. Mitochondrial STAT5A Promotes Metabolic Remodeling and the Warburg Effect by Inactivating the Pyruvate Dehydrogenase Complex. Cell Death Dis. 2021, 12, 634. [Google Scholar] [CrossRef]
- Banerjee, S. Empowering Clinical Diagnostics with Mass Spectrometry. ACS Omega 2020, 5, 2041–2048. [Google Scholar] [CrossRef] [PubMed]
- Kar, P.; Supakar, P.C. Expression of Stat5A in Tobacco Chewing-Mediated Oral Squamous Cell Carcinoma. Cancer Lett. 2006, 240, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Takenawa, T.; Ferdous, T.; Kuramitsu, Y.; Ueyama, Y. Calreticulin Is a Novel Independent Prognostic Factor for Oral Squamous Cell Carcinoma. Oncol. Lett. 2017, 13, 4857–4862. [Google Scholar] [CrossRef]
- Auzair, L.B.M.; Vincent-Chong, V.K.; Ghani, W.M.N.; Kallarakkal, T.G.; Ramanathan, A.; Lee, C.E.; Rahman, Z.A.A.; Ismail, S.M.; Abraham, M.T.; Zain, R.B. Caveolin 1 (Cav-1) and Actin-Related Protein 2/3 Complex, Subunit 1B (ARPC1B) Expressions as Prognostic Indicators for Oral Squamous Cell Carcinoma (OSCC). Eur. Arch. Oto-Rhino-Laryngol. 2016, 273, 1885–1893. [Google Scholar] [CrossRef]
- Lončar-Brzak, B.; Klobučar, M.; Veliki-Dalić, I.; Sabol, I.; Kraljević Pavelić, S.; Krušlin, B.; Mravak-Stipetić, M. Expression of Small Leucine-Rich Extracellular Matrix Proteoglycans Biglycan and Lumican Reveals Oral Lichen Planus Malignant Potential. Clin. Oral Investig. 2018, 22, 1071–1082. [Google Scholar] [CrossRef]
- Kurihara-Shimomura, M.; Sasahira, T.; Nakashima, C.; Kuniyasu, H.; Shimomura, H.; Kirita, T. The Multifarious Functions of Pyruvate Kinase M2 in Oral Cancer Cells. Int. J. Mol. Sci. 2018, 19, 1907. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Wang, X.; Zhang, Y.; Sun, Y. Periostin Serves an Important Role in the Pathogenesis of Oral Squamous Cell Carcinoma. Oncol. Lett. 2019, 17, 1292–1298. [Google Scholar] [CrossRef]
- Wang, B.-J.; Chi, K.-P.; Shen, R.-L.; Zheng, S.-W.; Guo, Y.; Li, J.-F.; Fei, J.; He, Y. TGFBI Promotes Tumor Growth and Is Associated with Poor Prognosis in Oral Squamous Cell Carcinoma. J. Cancer 2019, 10, 4902–4912. [Google Scholar] [CrossRef]
- Zhao, C.; Zhou, Y.; Ma, H.; Wang, J.; Guo, H.; Liu, H. A Four-Hypoxia-Genes-Based Prognostic Signature for Oral Squamous Cell Carcinoma. BMC Oral Health 2021, 21, 232. [Google Scholar] [CrossRef]
- Abdul Rahman, M.; Tan, M.L.; Johnson, S.P.; Hollows, R.J.; Chai, W.L.; Mansell, J.P.; Yap, L.F.; Paterson, I.C. Deregulation of Lysophosphatidic Acid Metabolism in Oral Cancer Promotes Cell Migration via the Up-Regulation of COX-2. PeerJ 2020, 8, e10328. [Google Scholar] [CrossRef] [PubMed]
- Gharat, L.; Rathod, G.P.; Kandalgaonkar, S. Quantitative Estimation of Serum Fibrinogen Degradation Product Levels in Oral Premalignant and Malignant Lesions. J. Int. Oral Health 2013, 5, 65–72. [Google Scholar]
- Cai, M.; Zheng, Z.; Bai, Z.; Ouyang, K.; Wu, Q.; Xu, S.; Huang, L.; Jiang, Y.; Wang, L.; Gao, J.; et al. Overexpression of Angiogenic Factors and Matrix Metalloproteinases in the Saliva of Oral Squamous Cell Carcinoma Patients: Potential Non-Invasive Diagnostic and Therapeutic Biomarkers. BMC Cancer 2022, 22, 530. [Google Scholar] [CrossRef]
- Spenlé, C.; Loustau, T.; Murdamoothoo, D.; Erne, W.; Beghelli-de la Forest Divonne, S.; Veber, R.; Petti, L.; Bourdely, P.; Mörgelin, M.; Brauchle, E.-M.; et al. Tenascin-C Orchestrates an Immune-Suppressive Tumor Microenvironment in Oral Squamous Cell Carcinoma. Cancer Immunol. Res. 2020, 8, 1122–1138. [Google Scholar] [CrossRef] [PubMed]
- Allon, I.; Pettesh, J.; Livoff, A.; Schlapobersky, M.; Nahlieli, O.; Michaeli, E. Voltage-Dependent Anion Channel 1 Expression in Oral Malignant and Premalignant Lesions. Diagnostics 2023, 13, 1225. [Google Scholar] [CrossRef]
- Guzman De Avila, J.; Silvera-Redondo, C.; Alviz-Amador, A. Bioinformatic Analysis of Plus Gene Expression Related to Progression from Leukoplakia to Oral Squamous Cell Carcinoma. Asian Pac. J. Cancer Prev. 2022, 23, 3833–3842. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Chang, Y.-S.; Chan, W.-L.; Yeh, K.-T.; Wei, R.-J.; Chang, J.-G. Detection of SF3B3 Gene Mutations in Oral Cancer by High Resolution Melting Analysis. Clin. Lab. 2014, 60, 2023–2029. [Google Scholar] [CrossRef]
- Tian, W.; Zhou, J.; Chen, M.; Qiu, L.; Li, Y.; Zhang, W.; Guo, R.; Lei, N.; Chang, L. Bioinformatics Analysis of the Role of Aldolase A in Tumor Prognosis and Immunity. Sci. Rep. 2022, 12, 11632. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Xu, L.; Zhang, J.; Cao, H. High Expression Levels of Pyrimidine Metabolic Rate-Limiting Enzymes Are Adverse Prognostic Factors in Lung Adenocarcinoma: A Study Based on The Cancer Genome Atlas and Gene Expression Omnibus Datasets. Purinergic Signal. 2020, 16, 347–366. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, K.; Wu, Q.; Kim, L.J.Y.; Morton, A.R.; Gimple, R.C.; Prager, B.C.; Shi, Y.; Zhou, W.; Bhargava, S.; et al. Targeting Pyrimidine Synthesis Accentuates Molecular Therapy Response in Glioblastoma Stem Cells. Sci. Transl. Med. 2019, 11, aau4972. [Google Scholar] [CrossRef] [PubMed]
- Drosouni, A.; Panagopoulou, M.; Aidinis, V.; Chatzaki, E. Autotaxin in Breast Cancer: Role, Epigenetic Regulation and Clinical Implications. Cancers 2022, 14, 5437. [Google Scholar] [CrossRef] [PubMed]
- Michalak, M.; Groenendyk, J.; Szabo, E.; Gold, L.I.; Opas, M. Calreticulin, a Multi-Process Calcium-Buffering Chaperone of the Endoplasmic Reticulum. Biochem. J. 2009, 417, 651–666. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Pantelis, A.; Kraus, D.; Reckenbeil, J.; Reich, R.; Jepsen, S.; Fischer, H.-P.; Allam, J.-P.; Novak, N.; Wenghoefer, M. Human α-Defensin (DEFA) Gene Expression Helps to Characterise Benign and Malignant Salivary Gland Tumours. BMC Cancer 2012, 12, 465. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, G.; Zhang, Y.; Cui, X.; Wang, S.; Gao, S.; Wang, Y.; Liu, Y.; Bae, J.H.; Yang, W.-H.; et al. Fibrinogen Alpha Chain Knockout Promotes Tumor Growth and Metastasis through Integrin-AKT Signaling Pathway in Lung Cancer. Mol. Cancer Res. 2020, 18, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, F.; Huang, Y.; Ke, K.; Zhao, B.; Chen, L.; Liao, N.; Wang, L.; Li, Q.; Liu, X.; et al. FGG Promotes Migration and Invasion in Hepatocellular Carcinoma Cells through Activating Epithelial to Mesenchymal Transition. Cancer Manag. Res. 2019, 11, 1653–1665. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhou, Y.; Wu, T.; Zhu, T.; Ji, X.; Kwon, Y.-S.; Zhang, C.; Yeo, G.; Black, D.L.; Sun, H.; et al. Genome-Wide Analysis of PTB-RNA Interactions Reveals a Strategy Used by the General Splicing Repressor to Modulate Exon Inclusion or Skipping. Mol. Cell 2009, 36, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Weng, L.; Wu, S.; Liu, B.; Yin, X.; Wang, Z.; Mao, A. HP1BP3 Promotes Tumor Growth and Metastasis by Upregulating MiR-23a to Target TRAF5 in Esophageal Squamous Cell Carcinoma. Am. J. Cancer Res. 2021, 11, 2928–2943. [Google Scholar]
- Liu, B.; Hu, Y.; Wan, L.; Wang, L.; Cheng, L.; Sun, H.; Liu, Y.; Wu, D.; Zhu, J.; Hong, X.; et al. Proteomics Analysis of Cancer Tissues Identifies IGF2R as a Potential Therapeutic Target in Laryngeal Carcinoma. Front. Endocrinol. 2022, 13, 1031210. [Google Scholar] [CrossRef]
- Takeda, T.; Komatsu, M.; Chiwaki, F.; Komatsuzaki, R.; Nakamura, K.; Tsuji, K.; Kobayashi, Y.; Tominaga, E.; Ono, M.; Banno, K.; et al. Upregulation of IGF2R Evades Lysosomal Dysfunction-Induced Apoptosis of Cervical Cancer Cells via Transport of Cathepsins. Cell Death Dis. 2019, 10, 876. [Google Scholar] [CrossRef]
- Zhang, Z.; Mou, Z.; Xu, C.; Wu, S.; Dai, X.; Chen, X.; Ou, Y.; Chen, Y.; Yang, C.; Jiang, H. Autophagy-Associated Circular RNA Hsa_circ_0007813 Modulates Human Bladder Cancer Progression via Hsa-MiR-361-3p/IGF2R Regulation. Cell Death Dis. 2021, 12, 778. [Google Scholar] [CrossRef]
- Wróblewska, J.P.; Dias-Santagata, D.; Ustaszewski, A.; Wu, C.-L.; Fujimoto, M.; Selim, M.A.; Biernat, W.; Ryś, J.; Marszalek, A.; Hoang, M.P. Prognostic Roles of BRAF, KIT, NRAS, IGF2R and SF3B1 Mutations in Mucosal Melanomas. Cells 2021, 10, 2216. [Google Scholar] [CrossRef]
- Xu, L.; Hou, Y.; Tu, G.; Chen, Y.; Du, Y.-E.; Zhang, H.; Wen, S.; Tang, X.; Yin, J.; Lang, L.; et al. Nuclear Drosha Enhances Cell Invasion via an EGFR-ERK1/2-MMP7 Signaling Pathway Induced by Dysregulated MiRNA-622/197 and Their Targets LAMC2 and CD82 in Gastric Cancer. Cell Death Dis. 2017, 8, e2642. [Google Scholar] [CrossRef] [PubMed]
- Kosanam, H.; Prassas, I.; Chrystoja, C.C.; Soleas, I.; Chan, A.; Dimitromanolakis, A.; Blasutig, I.M.; Rückert, F.; Gruetzmann, R.; Pilarsky, C.; et al. Laminin, Gamma 2 (LAMC2): A Promising New Putative Pancreatic Cancer Biomarker Identified by Proteomic Analysis of Pancreatic Adenocarcinoma Tissues. Mol. Cell. Proteomics 2013, 12, 2820–2832. [Google Scholar] [CrossRef]
- Shou, J.-Z.; Hu, N.; Takikita, M.; Roth, M.J.; Johnson, L.L.; Giffen, C.; Wang, Q.-H.; Wang, C.; Wang, Y.; Su, H.; et al. Overexpression of CDC25B and LAMC2 MRNA and Protein in Esophageal Squamous Cell Carcinomas and Premalignant Lesions in Subjects from a High-Risk Population in China. Cancer Epidemiol. Biomark. Prev. 2008, 17, 1424–1435. [Google Scholar] [CrossRef]
- Jiang, P.; He, S.; Li, Y.; Xu, Z. Identification of Therapeutic and Prognostic Biomarkers of Lamin C (LAMC) Family Members in Head and Neck Squamous Cell Carcinoma. Med. Sci. Monit. 2020, 26, e925735. [Google Scholar] [CrossRef]
- Zhong, P.; Liu, L.; Shen, A.; Chen, Z.; Hu, X.; Cai, Y.; Lin, J.; Wang, B.; Li, J.; Chen, Y.; et al. Five Extracellular Matrix-Associated Genes Upregulated in Oral Tongue Squamous Cell Carcinoma: An Integrated Bioinformatics Analysis. Oncol. Lett. 2019, 18, 5959–5967. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.; Aldridge, K.; Ensley, J.F.; Odell, E.; Boyd, A.; Jones, J.; Gutkind, J.S.; Yeudall, W.A. Laminin-Gamma2 Overexpression in Head-and-Neck Squamous Cell Carcinoma. Int. J. Cancer 2002, 99, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Lu, H.; Zhao, D.; Ou, Y.; Yu, K.; Gu, J.; Wang, L.; Jiang, S.; Chen, M.; Wang, J.; et al. LTPB2 Acts as a Prognostic Factor and Promotes Progression of Cervical Adenocarcinoma. Am. J. Transl. Res. 2015, 7, 1095–1105. [Google Scholar]
- Wang, J.; Liang, W.-J.; Min, G.-T.; Wang, H.-P.; Chen, W.; Yao, N. LTBP2 Promotes the Migration and Invasion of Gastric Cancer Cells and Predicts Poor Outcome of Patients with Gastric Cancer. Int. J. Oncol. 2018, 52, 1886–1898. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, G.; Zhao, C.; Geng, R.; Zhang, S.; Wang, W.; Chen, J.; Liu, H.; Wang, X. High Expression of LTBP2 Contributes to Poor Prognosis in Colorectal Cancer Patients and Correlates with the Mesenchymal Colorectal Cancer Subtype. Dis. Markers 2019, 2019, 5231269. [Google Scholar] [CrossRef]
- Turtoi, A.; Musmeci, D.; Wang, Y.; Dumont, B.; Somja, J.; Bevilacqua, G.; De Pauw, E.; Delvenne, P.; Castronovo, V. Identification of Novel Accessible Proteins Bearing Diagnostic and Therapeutic Potential in Human Pancreatic Ductal Adenocarcinoma. J. Proteome Res. 2011, 10, 4302–4313. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Tang, M.M.; Xu, X.; Jiang, B.; Huang, J.; Feng, X.; Qiang, J. LTBP2 Is a Prognostic Marker in Head and Neck Squamous Cell Carcinoma. Oncotarget 2016, 7, 45052–45059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tian, C.; Cheng, J.; Mao, W.; Li, M.; Chen, M. LTBP2 Inhibits Prostate Cancer Progression and Metastasis via the PI3K/AKT Signaling Pathway. Exp. Ther. Med. 2022, 24, 563. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Loustau, T.; Salomé, N.; Poilil Surendran, S.; Li, C.; Tucker, R.P.; Izzi, V.; Lamba, R.; Koch, M.; Orend, G. Advances on the Roles of Tenascin-C in Cancer. J. Cell Sci. 2022, 135, jcs260244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, W.; Zhang, N.; Chen, X.; Liu, W.; Zhang, L.; Liu, N. Systematic Pan-Cancer Analysis Identifies RBM39 as an Immunological and Prognostic Biomarker. J. Cell. Mol. Med. 2022, 26, 4859–4871. [Google Scholar] [CrossRef] [PubMed]
- Gökmen-Polar, Y.; Neelamraju, Y.; Goswami, C.P.; Gu, X.; Nallamothu, G.; Janga, S.C.; Badve, S. Expression Levels of SF3B3 Correlate with Prognosis and Endocrine Resistance in Estrogen Receptor-Positive Breast Cancer. Mod. Pathol. 2015, 28, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Halim, C.E.; Deng, S.; Ong, M.S.; Yap, C.T. Involvement of STAT5 in Oncogenesis. Biomedicines 2020, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Cabodi, S.; Tinnirello, A.; Di Stefano, P.; Bisarò, B.; Ambrosino, E.; Castellano, I.; Sapino, A.; Arisio, R.; Cavallo, F.; Forni, G.; et al. P130Cas as a New Regulator of Mammary Epithelial Cell Proliferation, Survival, and HER2-Neu Oncogene-Dependent Breast Tumorigenesis. Cancer Res. 2006, 66, 4672–4680. [Google Scholar] [CrossRef]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef]
- Suhaimi, S.A.; Chan, S.C.; Rosli, R. Matrix Metallopeptidase 3 Polymorphisms: Emerging Genetic Markers in Human Breast Cancer Metastasis. J. Breast Cancer 2020, 23, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Fernández, A.; Fueyo, A.; Folgueras, A.R.; Garabaya, C.; Pennington, C.J.; Pilgrim, S.; Edwards, D.R.; Holliday, D.L.; Jones, J.L.; Span, P.N.; et al. Matrix Metalloproteinase-8 Functions as a Metastasis Suppressor through Modulation of Tumor Cell Adhesion and Invasion. Cancer Res. 2008, 68, 2755–2763. [Google Scholar] [CrossRef] [PubMed]
- Hong, O.-Y.; Jang, H.-Y.; Lee, Y.-R.; Jung, S.H.; Youn, H.J.; Kim, J.-S. Inhibition of Cell Invasion and Migration by Targeting Matrix Metalloproteinase-9 Expression via Sirtuin 6 Silencing in Human Breast Cancer Cells. Sci. Rep. 2022, 12, 12125. [Google Scholar] [CrossRef]
- Krishnan, A.; Li, X.; Kao, W.-Y.; Viker, K.; Butters, K.; Masuoka, H.; Knudsen, B.; Gores, G.; Charlton, M. Lumican, an Extracellular Matrix Proteoglycan, Is a Novel Requisite for Hepatic Fibrosis. Lab. Investig. 2012, 92, 1712–1725. [Google Scholar] [CrossRef]
- Zahra, K.; Dey, T.; Ashish; Mishra, S.P.; Pandey, U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front. Oncol. 2020, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.M.; Lisanti, M.P. Caveolin-1 in Oncogenic Transformation, Cancer, and Metastasis. Am. J. Physiol. Physiol. 2005, 288, C494–C506. [Google Scholar] [CrossRef] [PubMed]
- Klingen, T.A.; Chen, Y.; Aas, H.; Wik, E.; Akslen, L.A. Fibulin-2 Expression Associates with Vascular Invasion and Patient Survival in Breast Cancer. PLoS ONE 2021, 16, e0249767. [Google Scholar] [CrossRef]
- Vaes, N.; Schonkeren, S.L.; Rademakers, G.; Holland, A.M.; Koch, A.; Gijbels, M.J.; Keulers, T.G.; de Wit, M.; Moonen, L.; Van der Meer, J.R.M.; et al. Loss of Enteric Neuronal Ndrg4 Promotes Colorectal Cancer via Increased Release of Nid1 and Fbln2. EMBO Rep. 2021, 22, e51913. [Google Scholar] [CrossRef] [PubMed]
- Corona, A.; Blobe, G.C. The Role of the Extracellular Matrix Protein TGFBI in Cancer. Cell. Signal. 2021, 84, 110028. [Google Scholar] [CrossRef]
- Fang, Y.; Liu, J.; Zhang, Q.; She, C.; Zheng, R.; Zhang, R.; Chen, Z.; Chen, C.; Wu, J. Overexpressed VDAC1 in Breast Cancer as a Novel Prognostic Biomarker and Correlates with Immune Infiltrates. World J. Surg. Oncol. 2022, 20, 211. [Google Scholar] [CrossRef]
- Wersäll, O.C.; Löfstedt, L.; Govorov, I.; Mints, M.; Gabrielson, M.; Shoshan, M. PGC1α and VDAC1 Expression in Endometrial Cancer. Mol. Clin. Oncol. 2021, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Rahmatpanah, F.; De Robles, G.; Lilly, M.; Keane, T.; Kumar, V.; Mercola, D.; Randhawa, P.; McClelland, M. RNA Expression Differences in Prostate Tumors and Tumor-Adjacent Stroma between Black and White Americans. Oncotarget 2021, 12, 1457–1469. [Google Scholar] [CrossRef]
- Kobayashi, N.; Kostka, G.; Garbe, J.H.O.; Keene, D.R.; Bächinger, H.P.; Hanisch, F.-G.; Markova, D.; Tsuda, T.; Timpl, R.; Chu, M.-L.; et al. A Comparative Analysis of the Fibulin Protein Family. Biochemical Characterization, Binding Interactions, and Tissue Localization. J. Biol. Chem. 2007, 282, 11805–11816. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Zhou, W.; Wang, X. Involvement of MiR-455 in the Protective Effect of H2S against Chemical Hypoxia-Induced Injury in BEAS-2B Cells. Pathol. Res. Pract. 2018, 214, 1804–1810. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, E.; Schmitt, B.M.; Menger, M.D.; Laschke, M.W. The Regulatory Mechanisms of NG2/CSPG4 Expression. Cell. Mol. Biol. Lett. 2017, 22, 4. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-H.; Kuo, C.-H.; Kuo, W.-W.; Ho, T.-J.; Pai, P.; Chen, W.-K.; Pan, L.-F.; Wang, C.-C.; Padma, V.V.; Huang, C.-Y. NFIL3 Suppresses Hypoxia-Induced Apoptotic Cell Death by Targeting the Insulin-like Growth Factor 2 Receptor. J. Cell. Biochem. 2015, 116, 1113–1120. [Google Scholar] [CrossRef]
- Jiang, J.; Jiang, Y.; Zhang, Y.-G.; Zhang, T.; Li, J.-H.; Huang, D.-L.; Hou, J.; Tian, M.-Y.; Sun, L.; Su, X.-M.; et al. The Effects of Hypoxia on Mitochondrial Function and Metabolism in Gastric Cancer Cells. Transl. Cancer Res. 2021, 10, 817–826. [Google Scholar] [CrossRef]
- Dai, C.; Lin, X.; Qi, Y.; Wang, Y.; Lv, Z.; Zhao, F.; Deng, Z.; Feng, X.; Zhang, T.; Pu, X. Vitamin D3 Improved Hypoxia-Induced Lung Injury by Inhibiting the Complement and Coagulation Cascade and Autophagy Pathway. BMC Pulm. Med. 2024, 24, 9. [Google Scholar] [CrossRef]
- Guarino, F.; Zinghirino, F.; Mela, L.; Pappalardo, X.G.; Ichas, F.; De Pinto, V.; Messina, A. NRF-1 and HIF-1α Contribute to Modulation of Human VDAC1 Gene Promoter during Starvation and Hypoxia in HeLa Cells. Biochim. Biophys. acta. Bioenerg. 2020, 1861, 148289. [Google Scholar] [CrossRef]
- Belton, M.; Brilha, S.; Manavaki, R.; Mauri, F.; Nijran, K.; Hong, Y.T.; Patel, N.H.; Dembek, M.; Tezera, L.; Green, J.; et al. Hypoxia and Tissue Destruction in Pulmonary TB. Thorax 2016, 71, 1145–1153. [Google Scholar] [CrossRef]
- Kim, E.-A.; Na, J.-M.; Kim, J.; Choi, S.Y.; Ahn, J.-Y.; Cho, S.-W. Neuroprotective Effect of 3-(Naphthalen-2-Yl(Propoxy)Methyl)Azetidine Hydrochloride on Brain Ischaemia/Reperfusion Injury. J. Neuroimmune Pharmacol. 2017, 12, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-C.; Zhou, W.; Shu, R.; Ni, J. Hypoxia Induces Apoptosis and Autophagic Cell Death in Human Periodontal Ligament Cells through HIF-1α PathSway. Cell Prolif. 2012, 45, 239–248. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Yan, L.; Du, W.; Zhang, M.; Chen, H.; Zhang, L.; Li, G.; Li, J.; Dong, Y.; et al. MMP-2 and MMP-9 Contribute to the Angiogenic Effect Produced by Hypoxia/15-HETE in Pulmonary Endothelial Cells. J. Mol. Cell. Cardiol. 2018, 121, 36–50. [Google Scholar] [CrossRef]
- Deng, C.; Deng, G.; Chu, H.; Chen, S.; Chen, X.; Li, X.; He, Y.; Sun, C.; Zhang, C. Construction of a Hypoxia-Immune-Related Prognostic Panel Based on Integrated Single-Cell and Bulk RNA Sequencing Analyses in Gastric Cancer. Front. Immunol. 2023, 14, 1140328. [Google Scholar] [CrossRef]
- Yang, L.; Cui, R.; Li, Y.; Liang, K.; Ni, M.; Gu, Y. Hypoxia-Induced TGFBI as a Serum Biomarker for Laboratory Diagnosis and Prognosis in Patients with Pancreatic Ductal Adenocarcinoma. Lab. Med. 2020, 51, 352–361. [Google Scholar] [CrossRef]
- Gonçalves, I.F.; Acar, E.; Costantino, S.; Szabo, P.L.; Hamza, O.; Tretter, E.V.; Klein, K.U.; Trojanek, S.; Abraham, D.; Paneni, F.; et al. Epigenetic Modulation of Tenascin C in the Heart: Implications on Myocardial Ischemia, Hypertrophy and Metabolism. J. Hypertens. 2019, 37, 1861–1870. [Google Scholar] [CrossRef]
- Niu, Y.; Lin, Z.; Wan, A.; Sun, L.; Yan, S.; Liang, H.; Zhan, S.; Chen, D.; Bu, X.; Liu, P.; et al. Loss-of-Function Genetic Screening Identifies Aldolase A as an Essential Driver for Liver Cancer Cell Growth Under Hypoxia. Hepatology 2021, 74, 1461–1479. [Google Scholar] [CrossRef]
- Verrier, E.R.; Weiss, A.; Bach, C.; Heydmann, L.; Turon-Lagot, V.; Kopp, A.; El Saghire, H.; Crouchet, E.; Pessaux, P.; Garcia, T.; et al. Combined Small Molecule and Loss-of-Function Screen Uncovers Estrogen Receptor Alpha and CAD as Host Factors for HDV Infection and Antiviral Targets. Gut 2020, 69, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Castillo Bennett, J.; Silva, P.; Martinez, S.; Torres, V.A.; Quest, A.F.G. Hypoxia-Induced Caveolin-1 Expression Promotes Migration and Invasion of Tumor Cells. Curr. Mol. Med. 2018, 18, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Joung, Y.-H.; Park, J.-H.; Park, T.; Lee, C.-S.; Kim, O.H.; Ye, S.-K.; Yang, U.M.; Lee, K.J.; Yang, Y.M. Hypoxia Activates Signal Transducers and Activators of Transcription 5 (STAT5) and Increases Its Binding Activity to the GAS Element in Mammary Epithelial Cells. Exp. Mol. Med. 2003, 35, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-H.; Hu, Y.; Yu, L.; Ke, C.; Vo, C.; Hsu, H.; Li, Z.; Di Donato, A.T.; Chaturbedi, A.; Hwang, J.W.; et al. Weaponizing Human EGF-Containing Fibulin-like Extracellular Matrix Protein 1 (EFEMP1) for 21st Century Cancer Therapeutics. Oncoscience 2016, 3, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Xu, X.; Zhang, Z.; Ma, C.; Xiang, R.; Liu, J.; Xu, W.; Wu, C.; Li, J.; Zhan, F.; et al. Hypoxia-Induced Long Non-Coding RNA DARS-AS1 Regulates RBM39 Stability to Promote Myeloma Malignancy. Haematologica 2020, 105, 1630–1640. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lee, Y.; Kang, Y.; Dai, B.; Perez, M.R.; Pratt, M.; Koay, E.J.; Kim, M.; Brekken, R.A.; Fleming, J.B. Hypoxia-Induced Autophagy of Stellate Cells Inhibits Expression and Secretion of Lumican into Microenvironment of Pancreatic Ductal Adenocarcinoma. Cell Death Differ. 2019, 26, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.P.; Harishankar, M.K.; Pillai, A.A.; Devi, A. Hypoxia Induced EMT: A Review on the Mechanism of Tumor Progression and Metastasis in OSCC. Oral Oncol. 2018, 80, 23–32. [Google Scholar] [CrossRef] [PubMed]
- DE Lima, P.O.; Jorge, C.C.; Oliveira, D.T.; Pereira, M.C. Hypoxic Condition and Prognosis in Oral Squamous Cell Carcinoma. Anticancer Res. 2014, 34, 605–612. [Google Scholar] [PubMed]
- Tan, Y.; Wang, Z.; Xu, M.; Li, B.; Huang, Z.; Qin, S.; Nice, E.C.; Tang, J.; Huang, C. Oral Squamous Cell Carcinomas: State of the Field and Emerging Directions. Int. J. Oral Sci. 2023, 15, 44. [Google Scholar] [CrossRef] [PubMed]
- Barillari, G.; Melaiu, O.; Gargari, M.; Pomella, S.; Bei, R.; Campanella, V. The Multiple Roles of CD147 in the Development and Progression of Oral Squamous Cell Carcinoma: An Overview. Int. J. Mol. Sci. 2022, 23, 8336. [Google Scholar] [CrossRef] [PubMed]
- Kelsen, S.G.; Duan, X.; Ji, R.; Perez, O.; Liu, C.; Merali, S. Cigarette Smoke Induces an Unfolded Protein Response in the Human Lung: A Proteomic Approach. Am. J. Respir. Cell Mol. Biol. 2008, 38, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, E.; Stinson, A.; Shan, L.; Yang, J.; Gietl, D.; Albino, A.P. Cigarette Smoke Induces Endoplasmic Reticulum Stress and the Unfolded Protein Response in Normal and Malignant Human Lung Cells. BMC Cancer 2008, 8, 229. [Google Scholar] [CrossRef]
- Jessie, K.; Pang, W.W.; Haji, Z.; Rahim, A.; Hashim, O.H. Proteomic Analysis of Whole Human Saliva Detects Enhanced Expression of Interleukin-1 Receptor Antagonist, Thioredoxin and Lipocalin-1 in Cigarette Smokers Compared to Non-Smokers. Int. J. Mol. Sci. 2010, 11, 4488–4505. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Chen, X.; Fang, X.; Chen, Q.; Hu, C. Caveolin-1 Aggravates Cigarette Smoke Extract-Induced MUC5AC Secretion in Human Airway Epithelial Cells. Int. J. Mol. Med. 2015, 35, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Mercer, B.A.; Wallace, A.M.; Brinckerhoff, C.E.; D’Armiento, J.M. Identification of a Cigarette Smoke-Responsive Region in the Distal MMP-1 Promoter. Am. J. Respir. Cell Mol. Biol. 2009, 40, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Foki, E.; Gangl, K.; Kranebitter, V.; Niederberger-Leppin, V.; Eckl-Dorna, J.; Wiebringhaus, R.; Thurnher, D.; Heiduschka, G. Early Effects of Cigarette Smoke Extract on Human Oral Keratinocytes and Carcinogenesis in Head and Neck Squamous Cell Carcinoma. Head Neck 2020, 42, 2348–2354. [Google Scholar] [CrossRef]
- Gupta, N.; Gupta, N.D.; Goyal, L.; Moin, S.; Khan, S.; Gupta, A.; Garg, S. The Influence of Smoking on the Levels of Matrix Metalloproteinase-8 and Periodontal Parameters in Smoker and Nonsmoker Patients with Chronic Periodontitis: A Clinicobiochemical Study. J. Oral Biol. Craniofacial Res. 2016, 6, S39–S43. [Google Scholar] [CrossRef]
- Sinha, I.; Modesto, J.; Krebs, N.M.; Stanley, A.E.; Walter, V.A.; Richie, J.P.; Muscat, J.E.; Sinha, R. Changes in Salivary Proteome before and after Cigarette Smoking in Smokers Compared to Sham Smoking in Nonsmokers: A Pilot Study. Tob. Induc. Dis. 2021, 19, 56. [Google Scholar] [CrossRef]
- Wang, X.-D.; Li, F.; Ma, D.-B.; Deng, X.; Zhang, H.; Gao, J.; Hao, L.; Liu, D.-D.; Wang, J. Periostin Mediates Cigarette Smoke Extract-Induced Proliferation and Migration in Pulmonary Arterial Smooth Muscle Cells. Biomed. Pharmacother. 2016, 83, 514–520. [Google Scholar] [CrossRef]
- Li, D.; Shen, C.; Liu, L.; Hu, J.; Qin, J.; Dai, L.; Gao, L.; Cheng, M.; Wang, D.; Bao, R.; et al. PKM2 Regulates Cigarette Smoke-Induced Airway Inflammation and Epithelial-to-Mesenchymal Transition via Modulating PINK1/Parkin-Mediated Mitophagy. Toxicology 2022, 477, 153251. [Google Scholar] [CrossRef]
- Luomanen, M.; Tiitta, O.; Heikinheimo, K.; Leimola-Virtanen, R.; Heinaro, I.; Happonen, R.P. Effect of Snuff and Smoking on Tenascin Expression in Oral Mucosa. J. Oral Pathol. Med. 1997, 26, 334–338. [Google Scholar] [CrossRef]
- Hoffman, J.L.; Faccidomo, S.; Kim, M.; Taylor, S.M.; Agoglia, A.E.; May, A.M.; Smith, E.N.; Wong, L.C.; Hodge, C.W. Alcohol Drinking Exacerbates Neural and Behavioral Pathology in the 3xTg-AD Mouse Model of Alzheimer’s Disease. Int. Rev. Neurobiol. 2019, 148, 169–230. [Google Scholar] [CrossRef]
- Ferrín, G.; Rodríguez-Perálvarez, M.; Aguilar-Melero, P.; Ranchal, I.; Llamoza, C.; Linares, C.I.; González-Rubio, S.; Muntané, J.; Briceño, J.; López-Cillero, P.; et al. Plasma Protein Biomarkers of Hepatocellular Carcinoma in HCV-Infected Alcoholic Patients with Cirrhosis. PLoS ONE 2015, 10, e0118527. [Google Scholar] [CrossRef] [PubMed]
- Sureshchandra, S.; Raus, A.; Jankeel, A.; Ligh, B.J.K.; Walter, N.A.R.; Newman, N.; Grant, K.A.; Messaoudi, I. Dose-Dependent Effects of Chronic Alcohol Drinking on Peripheral Immune Responses. Sci. Rep. 2019, 9, 7847. [Google Scholar] [CrossRef]
- Nong, F.-F.; Liang, Y.-Q.; Xing, S.-P.; Xiao, Y.-F.; Chen, H.-H.; Wen, B. Alcohol Promotes Epithelial Mesenchymal Transformation-Mediated Premetastatic Niche Formation of Colorectal Cancer by Activating Interaction between Laminin-Γ2 and Integrin-Β1. World J. Gastroenterol. 2022, 28, 5154–5174. [Google Scholar] [CrossRef] [PubMed]
- Zdanowicz, K.; Kowalczuk-Kryston, M.; Olanski, W.; Werpachowska, I.; Mielech, W.; Lebensztejn, D.M. Increase in Serum MMP-9 and TIMP-1 Concentrations during Alcohol Intoxication in Adolescents-A Preliminary Study. Biomolecules 2022, 12, 710. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Lee, B.C.; Ham, B.J.; Yang, B.-H.; Roh, S.; Choi, J.; Kang, T.-C.; Chai, Y.-G.; Choi, I.-G. Increased Transforming Growth Factor-Beta1 in Alcohol Dependence. J. Korean Med. Sci. 2009, 24, 941–944. [Google Scholar] [CrossRef]
- Rajendran, N.K.; Liu, W.; Cahill, P.A.; Redmond, E.M. Caveolin-1 Inhibition Mediates the Opposing Effects of Alcohol on γ-Secretase Activity in Arterial Endothelial and Smooth Muscle Cells. Physiol. Rep. 2023, 11, e15544. [Google Scholar] [CrossRef]
- Cassidy, L.L.; Dlugos, F.F.; Dlugos, C.A. Time Course of SERCA 2b and Calreticulin Expression in Purkinje Neurons of Ethanol-Fed Rats with Behavioral Correlates. Alcohol. Alcohol. 2013, 48, 667–678. [Google Scholar] [CrossRef]
- El Hajj, E.C.; El Hajj, M.C.; Voloshenyuk, T.G.; Mouton, A.J.; Khoutorova, E.; Molina, P.E.; Gilpin, N.W.; Gardner, J.D. Alcohol Modulation of Cardiac Matrix Metalloproteinases (MMPs) and Tissue Inhibitors of MMPs Favors Collagen Accumulation. Alcohol. Clin. Exp. Res. 2014, 38, 448–456. [Google Scholar] [CrossRef]
- Prystupa, A.; Szpetnar, M.; Boguszewska-Czubara, A.; Grzybowski, A.; Sak, J.; Załuska, W. Activity of MMP1 and MMP13 and Amino Acid Metabolism in Patients with Alcoholic Liver Cirrhosis. Med. Sci. Monit. 2015, 21, 1008–1014. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Steiner, J.L.; Pruznak, A.M.; Navaratnarajah, M.; Lang, C.H. Alcohol Differentially Alters Extracellular Matrix and Adhesion Molecule Expression in Skeletal Muscle and Heart. Alcohol. Clin. Exp. Res. 2015, 39, 1330–1340. [Google Scholar] [CrossRef]
- Kolch, W.; Mischak, H.; Pitt, A.R. The Molecular Make-up of a Tumour: Proteomics in Cancer Research. Clin. Sci. (Lond.) 2005, 108, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed]
- Chitturi Suryaprakash, R.T.; Shearston, K.; Farah, C.S.; Fox, S.A.; Iqbal, M.M.; Kadolsky, U.; Zhong, X.; Saxena, A.; Kujan, O. A Novel Preclinical In Vitro 3D Model of Oral Carcinogenesis for Biomarker Discovery and Drug Testing. Int. J. Mol. Sci. 2023, 24, 4096. [Google Scholar] [CrossRef] [PubMed]
- Mirnezami, R.; Nicholson, J.; Darzi, A. Preparing for Precision Medicine. N. Engl. J. Med. 2012, 366, 489–491. [Google Scholar] [CrossRef]
- Hyman, D.M.; Taylor, B.S.; Baselga, J. Implementing Genome-Driven Oncology. Cell 2017, 168, 584–599. [Google Scholar] [CrossRef]
- Chaves, P.; Garrido, M.; Oliver, J.; Pérez-Ruiz, E.; Barragan, I.; Rueda-Domínguez, A. Preclinical Models in Head and Neck Squamous Cell Carcinoma. Br. J. Cancer 2023, 128, 1819–1827. [Google Scholar] [CrossRef]
- Miserocchi, G.; Spadazzi, C.; Calpona, S.; De Rosa, F.; Usai, A.; De Vita, A.; Liverani, C.; Cocchi, C.; Vanni, S.; Calabrese, C.; et al. Precision Medicine in Head and Neck Cancers: Genomic and Preclinical Approaches. J. Pers. Med. 2022, 12, 854. [Google Scholar] [CrossRef]
- Tinhofer, I.; Braunholz, D.; Klinghammer, K. Preclinical Models of Head and Neck Squamous Cell Carcinoma for a Basic Understanding of Cancer Biology and Its Translation into Efficient Therapies. Cancers Head Neck 2020, 5, 9. [Google Scholar] [CrossRef]
- van Harten, A.M.; Poell, J.B.; Buijze, M.; Brink, A.; Wells, S.I.; René Leemans, C.; Wolthuis, R.M.F.; Brakenhoff, R.H. Characterization of a Head and Neck Cancer-Derived Cell Line Panel Confirms the Distinct TP53-Proficient Copy Number-Silent Subclass. Oral Oncol. 2019, 98, 53–61. [Google Scholar] [CrossRef]
- Cheng, H.; Yang, X.; Si, H.; Saleh, A.D.; Xiao, W.; Coupar, J.; Gollin, S.M.; Ferris, R.L.; Issaeva, N.; Yarbrough, W.G.; et al. Genomic and Transcriptomic Characterization Links Cell Lines with Aggressive Head and Neck Cancers. Cell Rep. 2018, 25, 1332–1345.e5. [Google Scholar] [CrossRef]
- Köpf-Maier, P.; Zimmermann, B. Organoid Reorganization of Human Tumors under in Vitro Conditions. Cell Tissue Res. 1991, 264, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Blattmann, P.; Aebersold, R. The Advent of Mass Spectrometry-Based Proteomics in Systems Biology Research. In Encyclopedia of Cell Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 166–176. [Google Scholar]
- Mani, D.R.; Krug, K.; Zhang, B.; Satpathy, S.; Clauser, K.R.; Ding, L.; Ellis, M.; Gillette, M.A.; Carr, S.A. Cancer Proteogenomics: Current Impact and Future Prospects. Nat. Rev. Cancer 2022, 22, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-C.; Chiang, C.-J.; Liu, T.-C.; Wu, C.-C.; Chen, Y.-T.; Chang, J.-G.; Shieh, G.S. Immunohistochemical Expression of Five Protein Combinations Revealed as Prognostic Markers in Asian Oral Cancer. Front. Genet. 2021, 12, 643461. [Google Scholar] [CrossRef] [PubMed]
- Pekarek, L.; Garrido-Gil, M.J.; Sánchez-Cendra, A.; Cassinello, J.; Pekarek, T.; Fraile-Martinez, O.; García-Montero, C.; Lopez-Gonzalez, L.; Rios-Parra, A.; Álvarez-Mon, M.; et al. Emerging Histological and Serological Biomarkers in Oral Squamous Cell Carcinoma: Applications in Diagnosis, Prognosis Evaluation and Personalized Therapeutics (Review). Oncol. Rep. 2023, 50, 213. [Google Scholar] [CrossRef]
- Gray, A.C.; Bradbury, A.; Dübel, S.; Knappik, A.; Plückthun, A.; Borrebaeck, C.A.K. Reproducibility: Bypass Animals for Antibody Production. Nature 2020, 581, 262. [Google Scholar] [CrossRef]


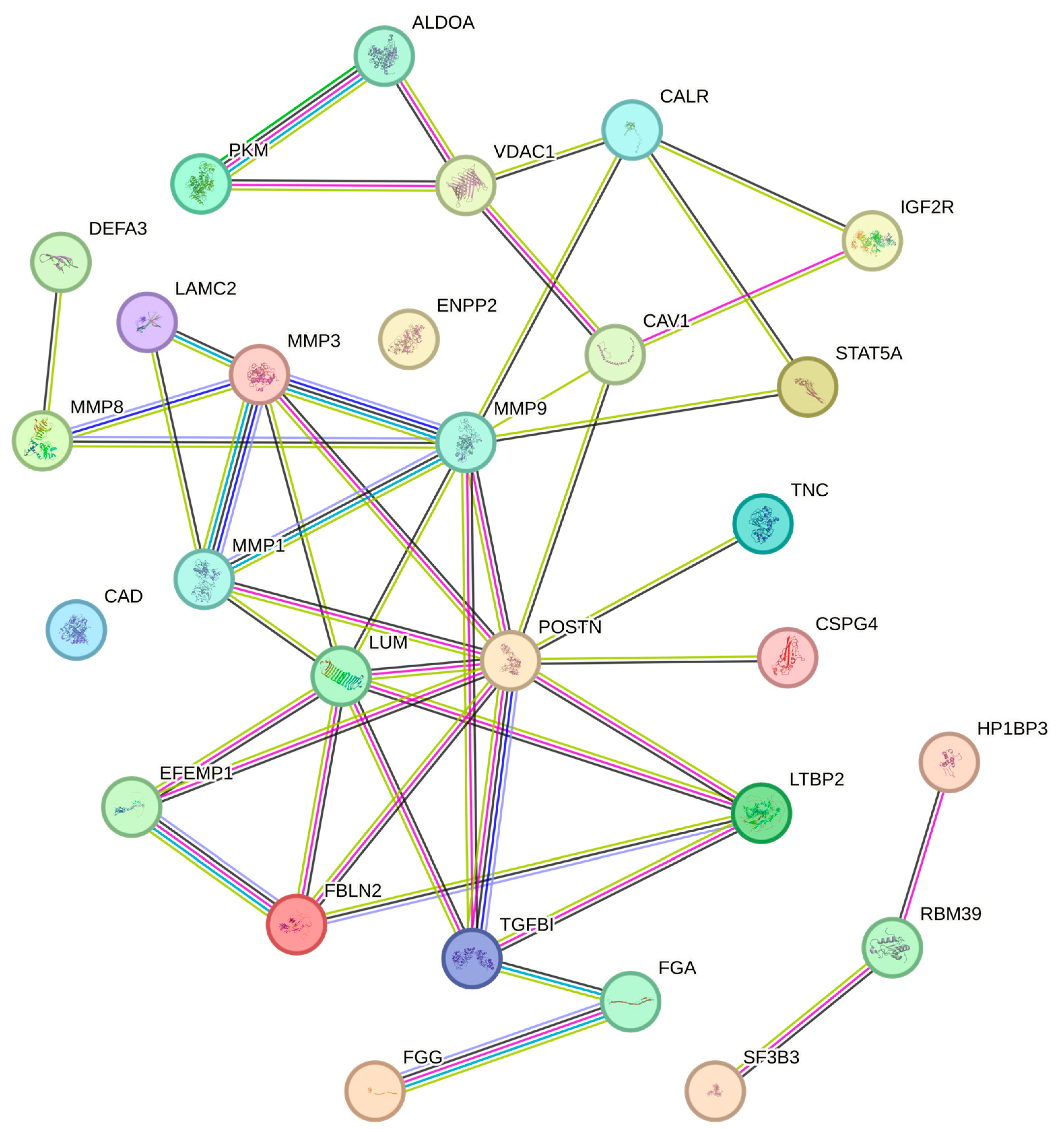
| Protein Name | Protein ID | Molecular Function | Variation | Validation | RNA Expression |
|---|---|---|---|---|---|
| Fructose-bisphosphate aldolase A | ALDOA | Actin binding Cadherin binding Cytoskeletal protein binding Fructose binding Fructose-bisphosphate aldolase activity Identical protein binding RNA binding Tubulin binding | Up CAF [43] Down MSC [44] | / | ns |
| CAD protein | CAD | Aspartate binding Aspartate carbamoyltransferase activity ATP binding Carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity Dihydroorotase activity Enzyme binding Identical protein binding Protein kinase activity Zinc ion binding” | Up tumor mass [36,37] | / | ns |
| Calreticulin | CALR | Calcium ion binding Carbohydrate binding Complement component C1q complex binding DNA binding Hormone binding Integrin binding Iron ion binding mRNA binding Nuclear androgen receptor binding Nuclear export signal receptor activity Peptide binding Protein-folding chaperone Protein-folding chaperone binding RNA binding Ubiquitin protein ligase binding Unfolded protein binding Zinc ion binding | Down CAF [38,41] | / | ns |
| Caveolin-1 | CAV1 | ATPase binding Cholesterol binding Enzyme binding Identical protein binding Inward rectifier potassium channel inhibitor activity Molecular adaptor activity Nitric oxide synthase binding Patched binding Peptidase activator activity Protein heterodimerization activity Protein kinase binding Protein sequestering activity Protein-containing complex binding Protein–macromolecule adaptor activity Signaling receptor binding small GTPase binding Transmembrane transporter binding | Up tumor mass [33,35] | IHC [33] | up |
| Chondroitin sulfate proteoglycan 4 | CSPG4 | Coreceptor activity Protein kinase binding | Down CAF [41] Up TIF [45] | / | ns |
| Neutrophil defensin 3 | DEFA3 | Protein homodimerization activity | Up tumor mass [30,34] | / | NA |
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | Calcium ion binding Epidermal growth factor receptor activity Epidermal growth factor receptor binding Growth factor activity | Down ECM [40] Up CAF [41] | / | ns |
| Ectonucleotide pyrophosphatase/ phosphodiesterase family member 2 | ENPP2 | Alkylglycerophosphoethanolamine phosphodiesterase activity Calcium ion binding Hydrolase activity Lysophospholipase activity Nucleic acid binding Phosphodiesterase I activity Polysaccharide binding Scavenger receptor activity Zinc ion binding | Down CAF [39,41] | / | ns |
| Fibulin-2 | FBLN2 | Calcium ion binding Extracellular matrix binding Extracellular matrix structural constituents Extracellular matrix constituents conferring elasticity | Down ECM [40] Down CAF [41] | / | ns |
| Fibrinogen alpha chain | FGA | Extracellular matrix structural constituents Metal ion binding Signaling receptor binding Structural molecule activity | Up tumor mass [30] Down ECM [40] | / | ns |
| Fibrinogen gamma chain | FGG | Cell adhesion molecule binding Extracellular matrix structural constituents Identical protein binding Metal ion binding Signaling receptor binding Structural molecule activity | Up tumor mass [30] Down ECM [40] | / | ns |
| Heterochromatin protein 1-binding protein 3 | HP1BP3 | DNA binding Nucleosome binding | Up tumor mass [36] Down tumor mass [37] | / | ns |
| Cation-independent mannose-6-phosphate receptor | IGF2R | Enzyme binding G-protein alpha-subunit binding Identical protein binding Insulin-like growth factor binding Insulin-like growth factor II binding Insulin-like growth factor receptor activity Mannose binding Phosphoprotein binding Retinoic acid binding Retromer complex binding Signaling receptor activity | Up tumor mass [36] Down MSC [44] | / | ns |
| Laminin subunit gamma-2 | LAMC2 | Cadherin binding Microtubule binding Microtubule plus end polymerase Microtubule plus-end binding Ribonucleoprotein complex binding | Up tumor mass [36] Up TIF [45] | / | up |
| Latent-transforming growth factor beta-binding protein 2 | LTBP2 | Calcium ion binding Growth factor binding Heparin binding Microfibril binding | Up tumor mass [36] Up CAF [39] | / | ns |
| Lumican | LUM | Collagen binding Extracellular matrix structural constituents confer compression resistance | Down CAF [38,41] | / | up |
| Interstitial collagenase | MMP-1 | Endopeptidase activity Metalloendopeptidase Peptidase activity Serine-type endopeptidase activity zinc ion binding | Down CAF [38] Up CAF [42] Up MSC [44] | / | up |
| Stromelysin-1 | MMP-3 | Endopeptidase activity Metalloendopeptidase activity Metallopeptidase activity Peptidase activity Serine-type endopeptidase activity zinc ion binding | Up CAF [39] Down CAF [41] | qPCR and ELISA [39] | up |
| Neutrophil collagenase | MMP-8 | Calcium ion binding Endopeptidase activity Metalloendopeptidase activity Metallopeptidase activity Peptidase activity Serine-type endopeptidase activity zinc ion binding | Up tumor mass [30] Up CAF [42] Up TIF [46] | IHC [46] | up |
| Matrix metalloproteinase 9 | MMP-9 | Collagen binding Endopeptidase activity Identical protein binding Metalloendopeptidase activity Metallopeptidase activity Peptidase activity Serine-type endopeptidase activity Zinc ion binding | Up tumor mass [30] Up CAF [42] Up TIF [46] | IHC [46] | up |
| Pyruvate kinase PKM | PKM | ATP binding Cadherin binding Histone H3T11 kinase activity Magnesium ion binding MHC class II protein complex binding mRNA binding Potassium ion binding Protein homodimerization activity Protein tyrosine kinase activity Pyruvate kinase activity RNA binding Transcription coactivator activity | Down tumor mass [30] Up CAF [43] | / | NA |
| Periostin | POSTN | Cell adhesion molecule binding Heparin binding Metal ion binding | Up tumor mass [33] Up TIF [45] | IHC [33] | up |
| RNA-binding protein 39 | RBM39 | RNA binding RS domain binding U1 snRNP binding | Up tumor mass [36] Up TIF [45] | / | ns |
| Splicing factor 3B subunit 3 | SF3B3 | Protein-containing complex binding U2 snRNA binding | Up tumor mass [36,37] | / | ns |
| Signal transducer and activator of transcription 5A | STAT5A | DNA-binding transcription factor activity DNA-binding transcription factor activity RNA polymerase II-specific DNA-binding transcription factor binding RNA polymerase II cis-regulatory region Sequence-specific DNA binding | Up tumor mass [30] Down epithelium [31] | / | ns |
| Transforming growth factor-beta-induced protein ig-h3 | TGFBI | Cell adhesion molecule binding Collagen binding Extracellular matrix binding Extracellular matrix structural constituents Identical protein binding Integrin binding | Up CAF [39] Down MSC [44] | / | up |
| Tenascin | TNC | Extracellular matrix structural constituent Syndecan binding | Up tumor mass [33,36] Up TIF [45] | IHC [33] | up |
| Voltage-dependent anion-selective channel protein 1 | VDAC1 | Ceramide binding Cholesterol binding Identical protein binding Phosphatidylcholine binding Porin activity Protein kinase binding Transmembrane transporter binding Voltage-gated monoatomic anion channel activity | Up CAF [38] Down CAF [43] | / | ns |
| #Term ID | Term Description | False Discovery Rate | Matching Proteins in Your Network |
|---|---|---|---|
| GO:0030198 | Extracellular matrix organization | 2.2 × 10−6 | MMP-8, LUM, 3, MMP-1, CAV1, MMP-9, POSTN, FBLN2, TGFBI |
| GO:0030574 | Collagen catabolic process | 1.8 × 10−3 | MMP-8, 3, MMP-1, MMP-9 |
| GO:0022617 | Extracellular matrix disassembly | 2.4 × 10−3 | MMP-8, 3, MMP-1, MMP-9 |
| GO:0071492 | Cellular response to UV-A | 2.4 × 10−3 | 3, MMP-1, MMP-9 |
| GO:0050789 | Regulation of biological processes | 1.5 × 10−2 | MMP-8, RBM39, ENPP2, LTBP2, LAMC2, TNC, LUM, 3, SF3B3, CSPG4, PKM, CALR, MMP-1, DEFA3, FGG, CAV1, STAT5A, IGF2R, MMP-9, POSTN, EFEMP1, VDAC1, FBLN2, HP1BP3, TGFBI, ALDOA, FGA |
| GO:0016043 | Cellular component organization | 1.8 × 10−2 | MMP-8, LTBP2, LAMC2, TNC, LUM, 3, SF3B3, CSPG4, CALR, MMP-1, FGG, CAV1, MMP-9, POSTN, FBLN2, HP1BP3, TGFBI, ALDOA, FGA |
| GO:0045907 | Positive regulation of vasoconstriction | 2.3 × 10−2 | FGG, CAV1, FGA |
| GO:0048522 | Positive regulation of the cellular process | 2.3 × 10−2 | MMP-8, ENPP2, LAMC2, TNC, LUM, 3, SF3B3, CSPG4, PKM, CALR, MMP-1, FGG, CAV1, STAT5A, IGF2R, MMP-9, VDAC1, FBLN2, FGA |
| GO:0150077 | Regulation of the neuroinflammatory response | 2.3 × 10−2 | MMP-8, MMP-3, MMP-9 |
| GO:0050794 | Regulation of the cellular process | 2.4 × 10−2 | MMP-8, RBM39, ENPP2, LTBP2, LAMC2, TNC, LUM, 3, SF3B3, CSPG4, PKM, CALR, MMP-1, DEFA3, FGG, CAV1, STAT5A, IGF2R, MMP-9, POSTN, EFEMP1, VDAC1, FBLN2, HP1BP3, TGFBI, FGA |
| GO:0010811 | Positive regulation of cell–substrate adhesion | 3.4 × 10−2 | CALR, FGG, FBLN2, FGA |
| GO:0032101 | Regulation of response to external stimulus | 3.7 × 10−2 | MMP-8, TNC, 3, CALR, FGG, CAV1, MMP-9, FGA |
| GO:1900026 | Positive regulation of substrate adhesion-dependent cell spreading | 3.7 × 10−2 | CALR, FGG, FGA |
| GO:0010634 | Positive regulation of epithelial cell migration | 5.0 × 10−2 | ENPP2, CALR, STAT5A, MMP-9 |
| #Term ID | Term Description | False Discovery Rate | Matching Proteins in Your Network |
|---|---|---|---|
| GO:0005201 | Extracellular matrix structural constituents | 1.6 × 10−4 | TNC, LUM, FGG, FBLN2, TGFBI, FGA |
| GO:0005488 | Binding | 1.5 × 10−2 | MMP-8, RBM39, ENPP2, LTBP2, LAMC2, CAD, TNC, LUM, 3, SF3B3, CSPG4, PKM, CALR, MMP-1, DEFA3, FGG, CAV1, STAT5A, IGF2R, MMP-9, POSTN, EFEMP1, VDAC1, FBLN2, HP1BP3, TGFBI, ALDOA, FGA |
| GO:0004222 | Metalloendopeptidase activity | 3.7 × 10−2 | MMP-8, 3, MMP-1, MMP-9 |
| #Term ID | Term Description | False Discovery Rate | Matching Proteins in Your Network |
|---|---|---|---|
| GO:0031012 | Extracellular matrix | 1.7 × 10−16 | MMP-8, LTBP2, LAMC2, TNC, LUM, 3, CSPG4, PKM, CALR, MMP-1, FGG, MMP-9, POSTN, EFEMP1, FBLN2, TGFBI, FGA |
| GO:0062023 | Collagen-containing extracellular matrix | 1.4 × 10−15 | MMP-8, LTBP2, LAMC2, TNC, LUM, CSPG4, PKM, CALR, FGG, MMP-9, POSTN, EFEMP1, FBLN2, TGFBI, FGA |
| GO:0005576 | Extracellular region | 5.4 × 10−9 | MMP-8, ENPP2, LTBP2, LAMC2, CAD, TNC, LUM, 3, CSPG4, PKM, CALR, MMP-1, DEFA3, FGG, IGF2R, MMP-9, POSTN, EFEMP1, VDAC1, FBLN2, TGFBI, ALDOA, FGA |
| GO:0005615 | Extracellular space | 5.6 × 10−9 | MMP-8, ENPP2, LTBP2, LAMC2, CAD, TNC, LUM, 3, CSPG4, PKM, CALR, DEFA3, FGG, IGF2R, MMP-9, POSTN, EFEMP1, VDAC1, TGFBI, ALDOA, FGA |
| GO:1903561 | Extracellular vesicle | 9.8 × 10−7 | LTBP2, CAD, LUM, CSPG4, PKM, CALR, DEFA3, FGG, IGF2R, MMP-9, EFEMP1, VDAC1, FBLN2, TGFBI, ALDOA, FGA |
| GO:0070062 | Extracellular exosome | 5.8 × 10−6 | LTBP2, CAD, LUM, CSPG4, PKM, CALR, DEFA3, FGG, IGF2R, MMP-9, EFEMP1, VDAC1, TGFBI, ALDOA, FGA |
| GO:0030141 | Secretory granule | 3.9 × 10−5 | MMP-8, PKM, CALR, DEFA3, FGG, CAV1, IGF2R, MMP-9, ALDOA, FGA |
| GO:0031982 | Vesicle | 8.7 × 10−5 | MMP-8, LTBP2, CAD, LUM, CSPG4, PKM, CALR, DEFA3, FGG, CAV1, IGF2R, MMP-9, EFEMP1, VDAC1, FBLN2, TGFBI, ALDOA, FGA |
| GO:0071944 | Cell periphery | 2.6 × 10−4 | MMP-8, ENPP2, LTBP2, LAMC2, TNC, LUM, 3, CSPG4, PKM, CALR, MMP-1, FGG, CAV1, IGF2R, MMP-9, POSTN, EFEMP1, VDAC1, FBLN2, TGFBI, FGA |
| GO:0034774 | Secretory granule lumen | 8.0 × 10−4 | MMP-8, PKM, DEFA3, FGG, ALDOA, FGA |
| GO:1904724 | Tertiary granule lumen | 9.0 × 10−3 | MMP-8, MMP-9, ALDOA |
| GO:0005577 | Fibrinogen complex | 9.9 × 10−3 | FGG, FGA |
| GO:0070013 | Intracellular organelle lumen | 1.1 × 10−2 | MMP-8, RBM39, CAD, TNC, LUM, SF3B3, CSPG4, PKM, CALR, DEFA3, FGG, STAT5A, IGF2R, MMP-9, VDAC1, HP1BP3, ALDOA, FGA |
| GO:0031093 | Platelet alpha granule lumen | 1.2 × 10−2 | FGG, ALDOA, FGA |
| GO:0005925 | Focal adhesion | 2.5 × 10−2 | TNC, CSPG4, CALR, CAV1, IGF2R |
| GO:0005604 | Basement membrane | 3.1 × 10−2 | LAMC2, TNC, TGFBI |
| GO:0005796 | Golgi lumen | 3.7 × 10−2 | LUM, CSPG4, DEFA3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pomella, S.; Melaiu, O.; Cifaldi, L.; Bei, R.; Gargari, M.; Campanella, V.; Barillari, G. Biomarkers Identification in the Microenvironment of Oral Squamous Cell Carcinoma: A Systematic Review of Proteomic Studies. Int. J. Mol. Sci. 2024, 25, 8929. https://doi.org/10.3390/ijms25168929
Pomella S, Melaiu O, Cifaldi L, Bei R, Gargari M, Campanella V, Barillari G. Biomarkers Identification in the Microenvironment of Oral Squamous Cell Carcinoma: A Systematic Review of Proteomic Studies. International Journal of Molecular Sciences. 2024; 25(16):8929. https://doi.org/10.3390/ijms25168929
Chicago/Turabian StylePomella, Silvia, Ombretta Melaiu, Loredana Cifaldi, Roberto Bei, Marco Gargari, Vincenzo Campanella, and Giovanni Barillari. 2024. "Biomarkers Identification in the Microenvironment of Oral Squamous Cell Carcinoma: A Systematic Review of Proteomic Studies" International Journal of Molecular Sciences 25, no. 16: 8929. https://doi.org/10.3390/ijms25168929
APA StylePomella, S., Melaiu, O., Cifaldi, L., Bei, R., Gargari, M., Campanella, V., & Barillari, G. (2024). Biomarkers Identification in the Microenvironment of Oral Squamous Cell Carcinoma: A Systematic Review of Proteomic Studies. International Journal of Molecular Sciences, 25(16), 8929. https://doi.org/10.3390/ijms25168929









