Activation of the G Protein-Coupled Bile Acid Receptor TGR5 Modulates the HCP5/miR-139-5p/DDIT4 Axis to Antagonize Cervical Cancer Progression
Abstract
1. Introduction
2. Results
2.1. TGR5 Is Downregulated in CC, and Its Deficiency Promotes Cervical Inflammation Response
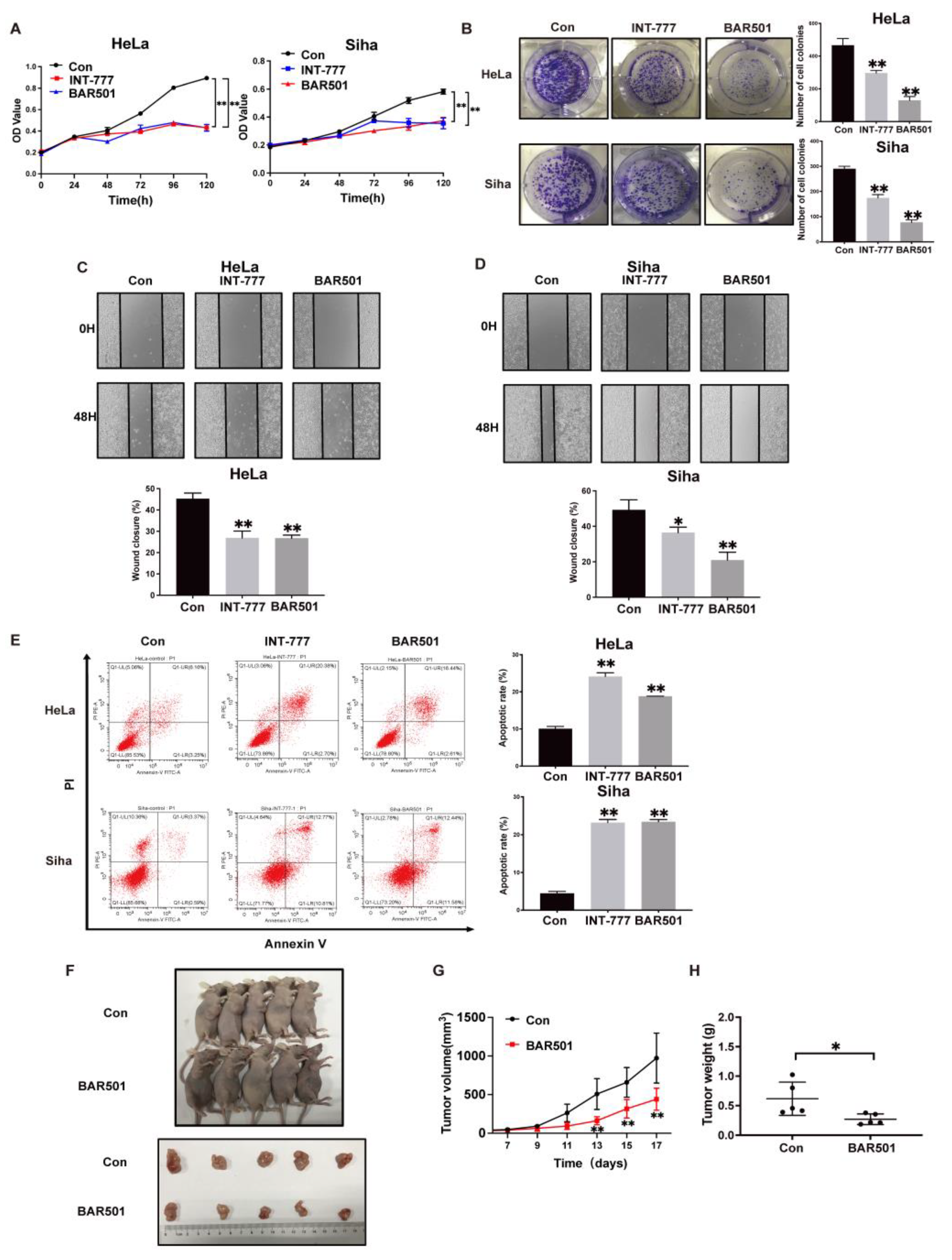
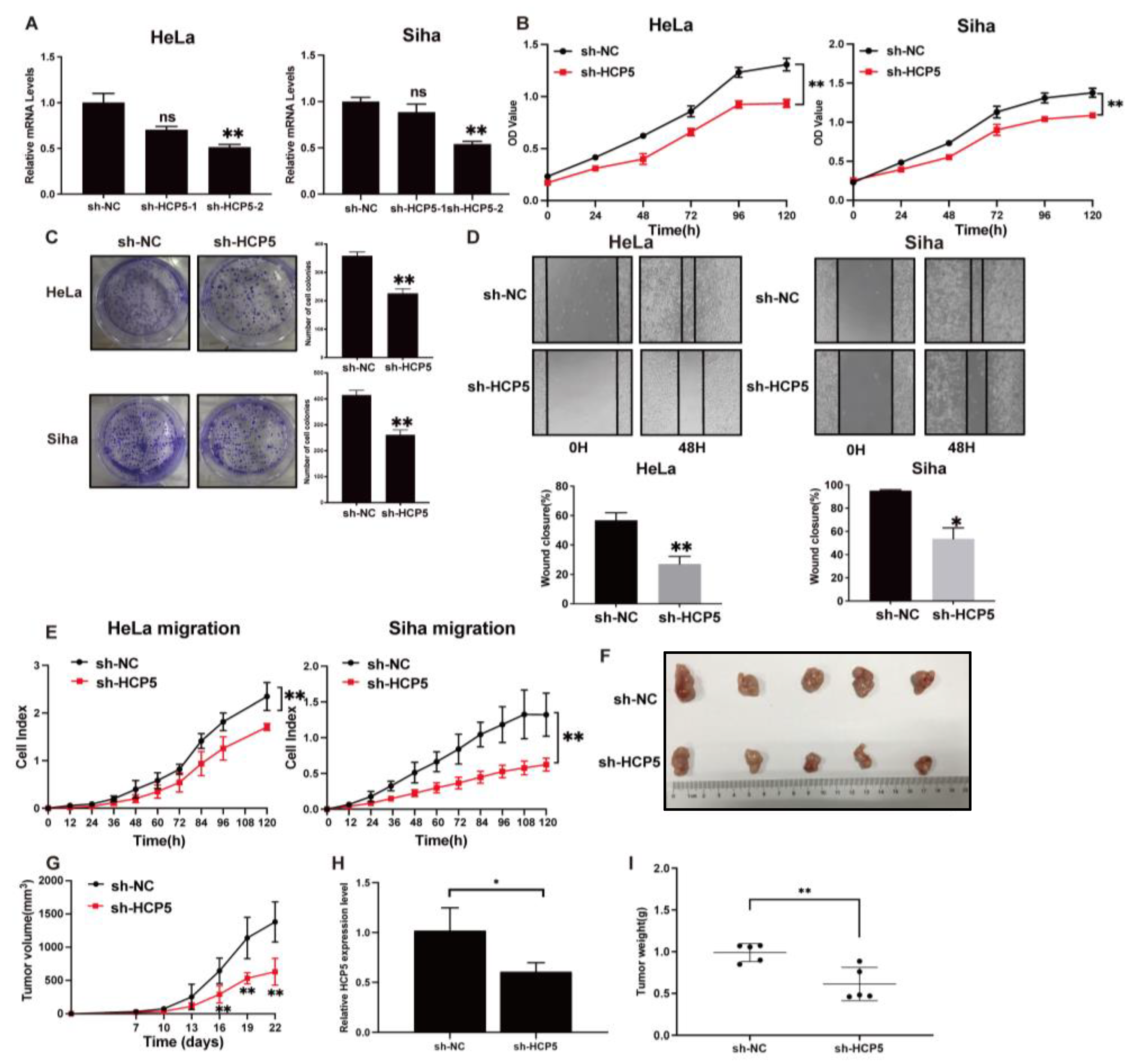
2.2. TGR5 Activation Suppresses Cervical Cancer Progression through the Downregulation of HCP5 Expression
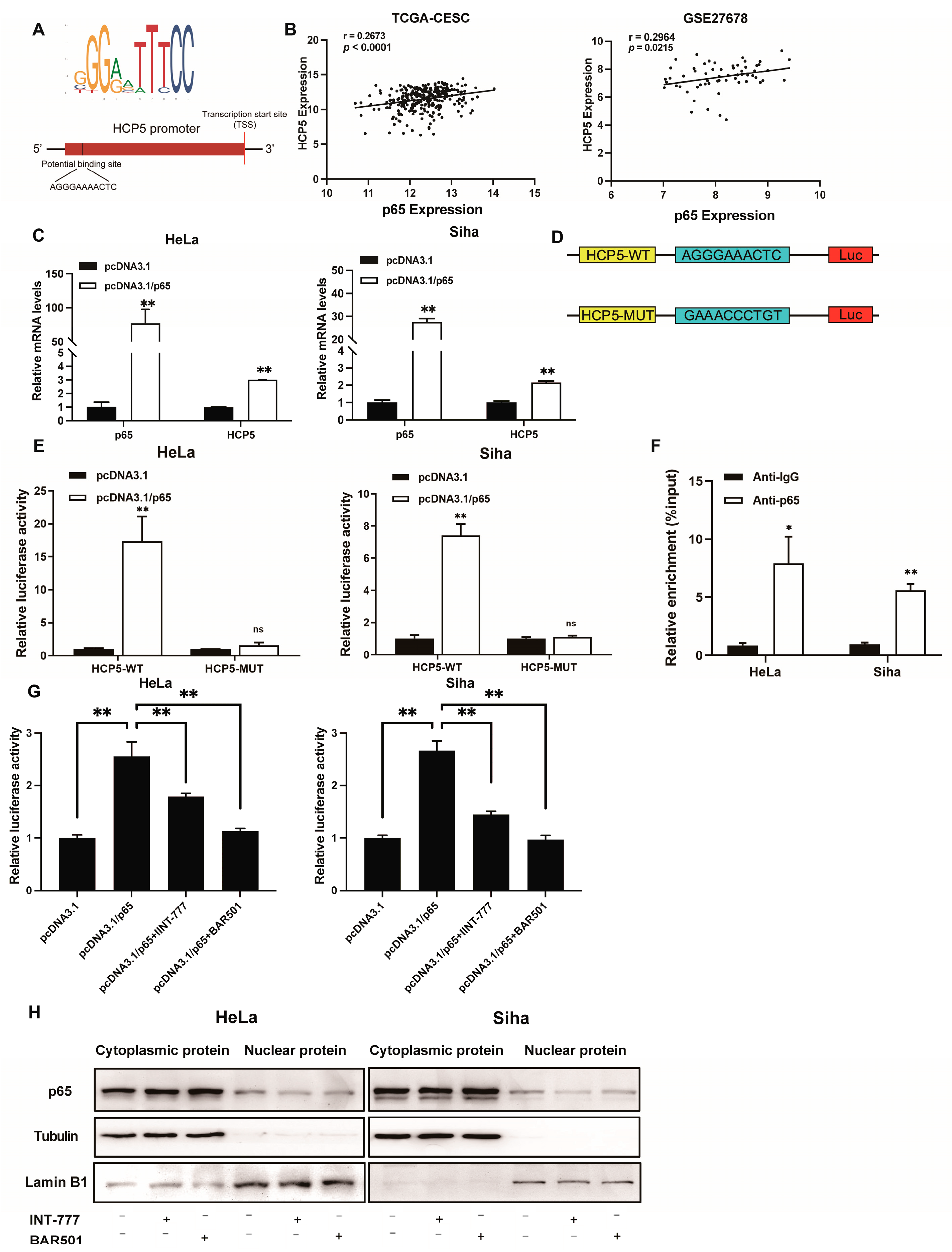
2.3. TGR5 Suppressed HCP5 Expression Level through Transcriptional Inactivation
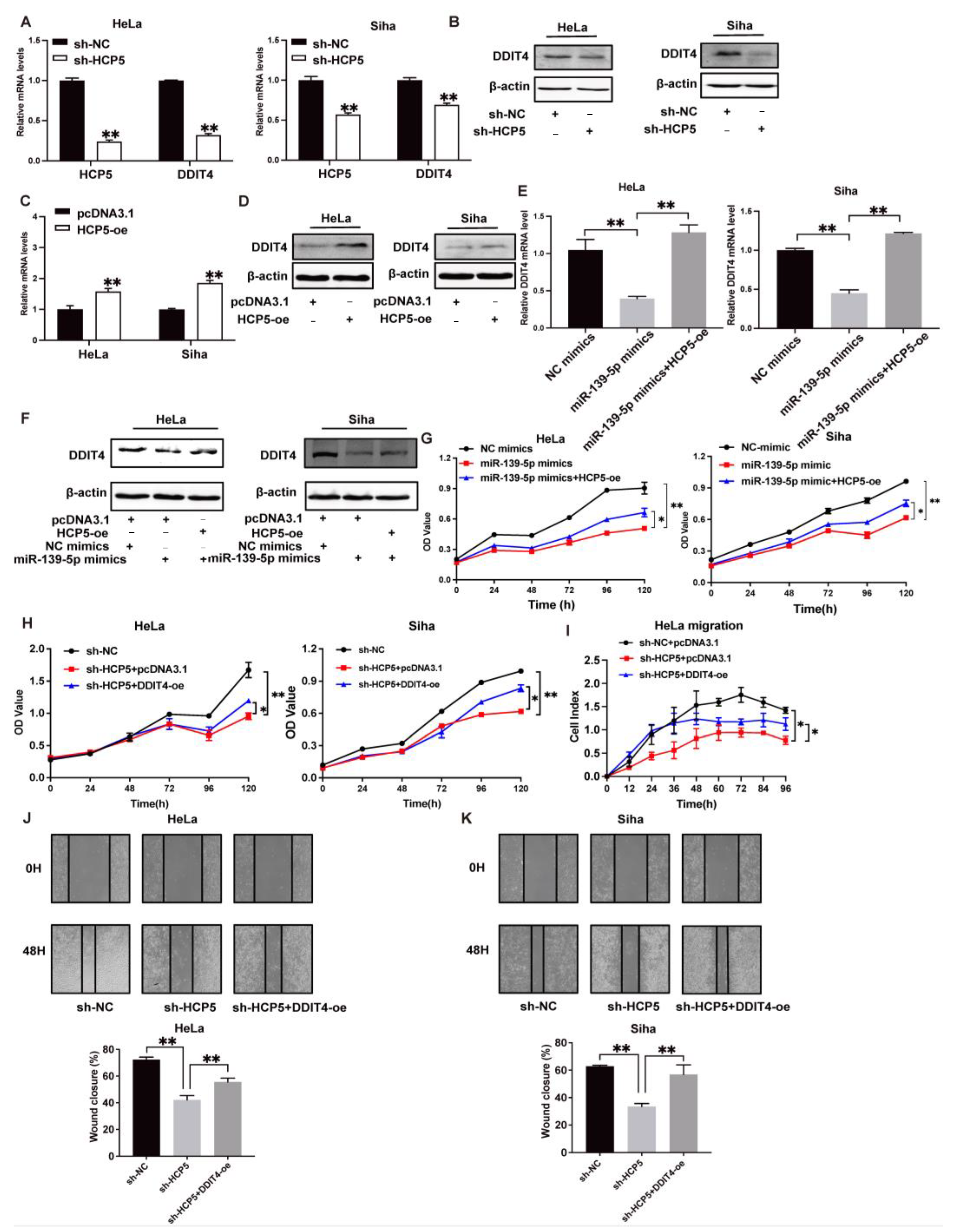
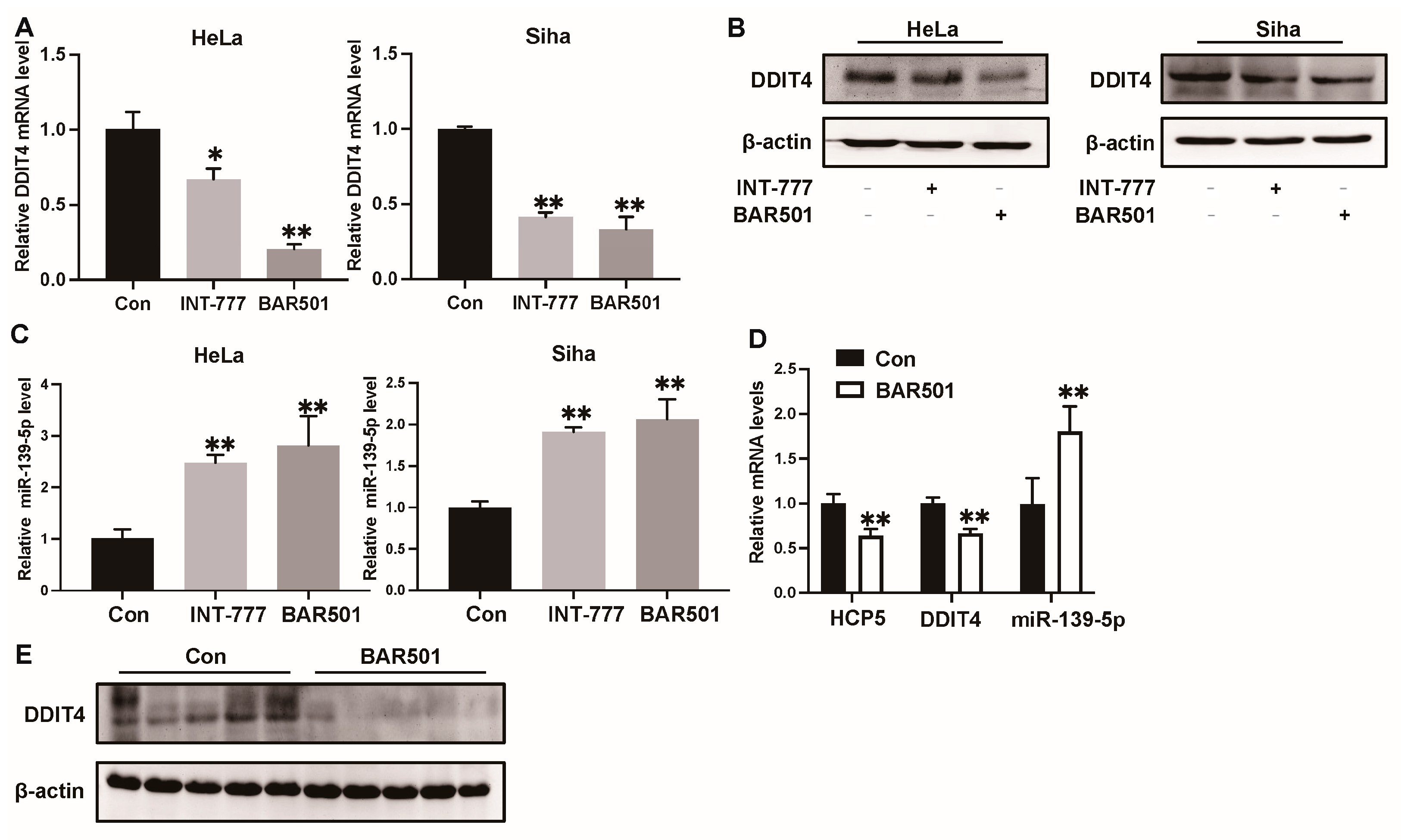
2.4. HCP5 Functions as a Sponge to miR-139-5p to Target DDIT4 in CC Cells
2.5. Activation of TGR5 Inhibits CC Progression by HCP5, Which Promotes CC Progression by Modulating the miR-139-5p/DDIT4 Axis In Vivo and In Vitro
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Plasmid Construction and Cell Transfection
4.3. RNA Isolation, Reverse Transcription and qRT-PCR
4.4. Subcellular Fractionation
4.5. Cell Proliferation Assay
4.6. Cell Colony Formation Assay
4.7. Wound-Healing Assays
4.8. Real-Time Cellular Analysis (RTCA)
4.9. Cell Apoptosis Assays
4.10. Western Blot Assays
4.11. Luciferase Reporter Assay
4.12. RIP Assay
4.13. ChIP Assay
4.14. Animal Experiments
4.15. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Small, W., Jr.; Bacon, M.A.; Bajaj, A.; Chuang, L.T.; Fisher, B.J.; Harkenrider, M.M.; Jhingran, A.; Kitchener, H.C.; Mileshkin, L.R.; Viswanathan, A.N.; et al. Cervical cancer: A global health crisis. Cancer 2017, 123, 2404–2412. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Ladurner, A.; Zehl, M.; Grienke, U.; Hofstadler, C.; Faur, N.; Pereira, F.C.; Berry, D.; Dirsch, V.M.; Rollinger, J.M. Allspice and Clove as Source of Triterpene Acids Activating the G Protein-Coupled Bile Acid Receptor TGR5. Front. Pharmacol. 2017, 8, 468. [Google Scholar] [CrossRef]
- Guo, C.; Chen, W.D.; Wang, Y.D. TGR5, Not Only a Metabolic Regulator. Front. Physiol. 2016, 7, 646. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 802–816. [Google Scholar] [CrossRef]
- Wang, Y.D.; Chen, W.D.; Yu, D.; Forman, B.M.; Huang, W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef]
- Guo, C.; Qi, H.; Yu, Y.; Zhang, Q.; Su, J.; Yu, D.; Huang, W.; Chen, W.D.; Wang, Y.D. The G-Protein-Coupled Bile Acid Receptor Gpbar1 (TGR5) Inhibits Gastric Inflammation Through Antagonizing NF-κB Signaling Pathway. Front. Pharmacol. 2015, 6, 287. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Duan, G.; Wei, D.; Zhao, C.; Ma, Y. The Bile Acid Membrane Receptor TGR5 in Cancer: Friend or Foe? Molecules 2022, 27, 5292. [Google Scholar] [CrossRef]
- Su, J.; Zhang, Q.; Qi, H.; Wu, L.; Li, Y.; Yu, D.; Huang, W.; Chen, W.D.; Wang, Y.D. The G-protein-coupled bile acid receptor Gpbar1 (TGR5) protects against renal inflammation and renal cancer cell proliferation and migration through antagonizing NF-κB and STAT3 signaling pathways. Oncotarget 2017, 8, 54378–54387. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, X.; Chen, B.; You, W.; Xue, S.; Qin, H.; Jiang, H. The membrane bile acid receptor TGR5 drives cell growth and migration via activation of the JAK2/STAT3 signaling pathway in non-small cell lung cancer. Cancer Lett. 2018, 412, 194–207. [Google Scholar] [CrossRef]
- Sabeena, S. Role of noncoding RNAs with emphasis on long noncoding RNAs as cervical cancer biomarkers. J. Med. Virol. 2023, 95, e28525. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, L.; Wang, J.; Liu, W. The role of long noncoding RNAs in therapeutic resistance in cervical cancer. Front. Cell Dev. Biol. 2022, 10, 1060909. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.; Lan, X.; Qiu, Q.; Zhou, Q.; Li, J.; Wu, M.; Liu, P.; Zhang, H.; Lu, B.; Lu, Y.; et al. LncRNA SFTA1P promotes cervical cancer progression by interaction with PTBP1 to facilitate TPM4 mRNA degradation. Cell Death Dis. 2022, 13, 936. [Google Scholar] [CrossRef]
- Lu, M.; Gao, Q.; Wang, Y.; Ren, J.; Zhang, T. LINC00511 promotes cervical cancer progression by regulating the miR-497-5p/MAPK1 axis. Apoptosis 2022, 27, 800–811. [Google Scholar] [CrossRef]
- Liao, Y.; Huang, J.; Liu, P.; Zhang, C.; Liu, J.; Xia, M.; Shang, C.; Ooi, S.; Chen, Y.; Qin, S.; et al. Downregulation of LNMAS orchestrates partial EMT and immune escape from macrophage phagocytosis to promote lymph node metastasis of cervical cancer. Oncogene 2022, 41, 1931–1943. [Google Scholar] [CrossRef] [PubMed]
- Walch-Rückheim, B.; Pahne-Zeppenfeld, J.; Fischbach, J.; Wickenhauser, C.; Horn, L.C.; Tharun, L.; Büttner, R.; Mallmann, P.; Stern, P.; Kim, Y.J.; et al. STAT3/IRF1 Pathway Activation Sensitizes Cervical Cancer Cells to Chemotherapeutic Drugs. Cancer Res. 2016, 76, 3872–3883. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Hashimoto, A.; Muromoto, R.; Kitai, Y.; Oritani, K.; Matsuda, T. Central Roles of STAT3-Mediated Signals in Onset and Development of Cancers: Tumorigenesis and Immunosurveillance. Cells 2022, 11, 2618. [Google Scholar] [CrossRef]
- Pu, F.F.; Shi, D.Y.; Chen, T.; Liu, Y.X.; Zhong, B.L.; Zhang, Z.C.; Liu, W.J.; Wu, Q.; Wang, B.C.; Shao, Z.W.; et al. SP1-induced long non-coding RNA SNHG6 facilitates the carcinogenesis of chondrosarcoma through inhibiting KLF6 by recruiting EZH2. Cell Death Dis. 2021, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, Y.; Chen, C.H.; Wen, Y.; Zhou, Z.; Yang, C.; Sun, J.; Du, G.; Wu, J.; Mao, X.; et al. KLF5-induced lncRNA IGFL2-AS1 promotes basal-like breast cancer cell growth and survival by upregulating the expression of IGFL1. Cancer Lett. 2021, 515, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Tang, T.; Chen, M.; Dong, B.; Sun, W.; Wu, N.; Chen, H.; Feng, Q.; Yang, X.; Jin, R.; et al. C-Myc-activated long non-coding RNA LINC01050 promotes gastric cancer growth and metastasis by sponging miR-7161-3p to regulate SPZ1 expression. J. Exp. Clin. Cancer Res. 2021, 40, 351. [Google Scholar] [CrossRef]
- Sun, Y.; Tian, Y.; He, J.; Tian, Y.; Zhang, G.; Zhao, R.; Zhu, W.J.; Gao, P. Linc01133 contributes to gastric cancer growth by enhancing YES1-dependent YAP1 nuclear translocation via sponging miR-145-5p. Cell Death Dis. 2022, 13, 51. [Google Scholar] [CrossRef] [PubMed]
- Wahlström, A.; Aydin, Ö.; Olsson, L.M.; Sjöland, W.; Henricsson, M.; Lundqvist, A.; Marschall, H.U.; Franken, R.; van de Laar, A.; Gerdes, V.; et al. Alterations in bile acid kinetics after bariatric surgery in patients with obesity with or without type 2 diabetes. EBioMedicine 2024, 2, 105265. [Google Scholar] [CrossRef]
- Hosseini, S.A.; Haddadi, M.H.; Fathizadeh, H.; Nemati, F.; Aznaveh, H.M.; Taraj, F.; Aghabozorgizadeh, A.; Gandomkar, G.; Bazazzadeh, E. Long non-coding RNAs and gastric cancer: An update of potential biomarkers and therapeutic applications. Biomed. Pharmacother. 2023, 163, 114407. [Google Scholar] [CrossRef] [PubMed]
- Ranga, S.; Yadav, R.; Chhabra, R.; Chauhan, M.B.; Tanwar, M.; Yadav, C.; Kadian, L.; Ahuja, P. Long non-coding RNAs as critical regulators and novel targets in cervical cancer: Current status and future perspectives. Apoptosis 2023, 28, 925–942. [Google Scholar] [CrossRef]
- Su, J.; Deng, L.; Wang, Y.D. Roles and Mechanisms of Long Non-Coding RNAs in Breast Cancer. Int. J. Mol. Sci. 2022, 24, 89. [Google Scholar] [CrossRef]
- Song, P.; Yang, F.; Jin, H.; Wang, X. The regulation of protein translation and its implications for cancer. Signal Transduct. Target. Ther. 2021, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Zhao, A.; Kong, F.; Jiang, L.; Wang, J. Long Non-Coding RNA LINC00511 Accelerates Proliferation and Invasion in Cervical Cancer Through Targeting miR-324-5p/DRAM1 Axis. Onco Targets Ther. 2020, 13, 10245–10256. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Wang, Q.; Ji, M.; Guo, X.; Li, L.; Su, X. Exosomal lncRNA UCA1 modulates cervical cancer stem cell self-renewal and differentiation through microRNA-122-5p/SOX2 axis. J. Transl. Med. 2021, 19, 229. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, B.; Huang, A.; Ren, C.; Wang, L.; Zhu, T.; Xiong, J.; Ding, W.; Wang, H. LncRNA HCP5 enhances the proliferation and migration of cervical cancer via miR-216a-5p/CDC42 axis. J. Cancer 2022, 13, 1882–1894. [Google Scholar] [CrossRef]
- Yu, Y.; Shen, H.M.; Fang, D.M.; Meng, Q.J.; Xin, Y.H. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4812–4819. [Google Scholar] [PubMed]
- Srinivas, T.; Mathias, C.; Oliveira-Mateos, C.; Guil, S. Roles of lncRNAs in brain development and pathogenesis: Emerging therapeutic opportunities. Mol. Ther. 2023, 31, 1550–1561. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, R.; Fang, L.; Ge, X.; Chen, L.; Zhou, M.; Zhou, Y.; Xiong, W.; Hu, Y.; Tang, X.; et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics 2019, 9, 2460–2474. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Xu, J.; Wang, M.; Xu, G.; Zhang, N.; Wang, G.; Zhao, Y. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018, 9, 372. [Google Scholar] [CrossRef]
- Tirado-Hurtado, I.; Fajardo, W.; Pinto, J.A. DNA Damage Inducible Transcript 4 Gene: The Switch of the Metabolism as Potential Target in Cancer. Front. Oncol. 2018, 8, 106. [Google Scholar] [CrossRef]
- Ho, K.H.; Chen, P.H.; Chou, C.M.; Shih, C.M.; Lee, Y.T.; Cheng, C.H.; Chen, K.C. A Key Role of DNA Damage-Inducible Transcript 4 (DDIT4) Connects Autophagy and GLUT3-Mediated Stemness to Desensitize Temozolomide Efficacy in Glioblastomas. Neurotherapeutics 2020, 17, 1212–1227. [Google Scholar] [CrossRef]
- Du, F.; Sun, L.; Chu, Y.; Li, T.; Lei, C.; Wang, X.; Jiang, M.; Min, Y.; Lu, Y.; Zhao, X.; et al. DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways. Cancer Commun. 2018, 38, 45. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Chen, Z.; Zhang, M.; Zhang, M.; Lu, X.; Li, C.; Miao, L. DDIT4 overexpression associates with poor prognosis in lung adenocarcinoma. J. Cancer 2021, 12, 6422–6428. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.T.; Zheng, B.; Cao, S.Q.; Ding, J.C.; Hu, G.S.; Liu, W.; Chen, C. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol. Cancer 2022, 21, 69. [Google Scholar] [CrossRef]
- Guan, Y.F.; Huang, Q.L.; Ai, Y.L.; Chen, Q.T.; Zhao, W.X.; Wang, X.M.; Wu, Q.; Chen, H.Z. Nur77-activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene 2020, 39, 2408–2423. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, J.; Zhao, Y.; Chen, W.-D.; Wang, Y.-D. Activation of the G Protein-Coupled Bile Acid Receptor TGR5 Modulates the HCP5/miR-139-5p/DDIT4 Axis to Antagonize Cervical Cancer Progression. Int. J. Mol. Sci. 2024, 25, 8932. https://doi.org/10.3390/ijms25168932
Su J, Zhao Y, Chen W-D, Wang Y-D. Activation of the G Protein-Coupled Bile Acid Receptor TGR5 Modulates the HCP5/miR-139-5p/DDIT4 Axis to Antagonize Cervical Cancer Progression. International Journal of Molecular Sciences. 2024; 25(16):8932. https://doi.org/10.3390/ijms25168932
Chicago/Turabian StyleSu, Jia, Yiqi Zhao, Wei-Dong Chen, and Yan-Dong Wang. 2024. "Activation of the G Protein-Coupled Bile Acid Receptor TGR5 Modulates the HCP5/miR-139-5p/DDIT4 Axis to Antagonize Cervical Cancer Progression" International Journal of Molecular Sciences 25, no. 16: 8932. https://doi.org/10.3390/ijms25168932
APA StyleSu, J., Zhao, Y., Chen, W.-D., & Wang, Y.-D. (2024). Activation of the G Protein-Coupled Bile Acid Receptor TGR5 Modulates the HCP5/miR-139-5p/DDIT4 Axis to Antagonize Cervical Cancer Progression. International Journal of Molecular Sciences, 25(16), 8932. https://doi.org/10.3390/ijms25168932





