Density Gradient Centrifugation Is an Effective Tool to Isolate Cancer Stem-like Cells from Hypoxic and Normoxia Triple-Negative Breast Cancer Models
Abstract
:1. Introduction
2. Results
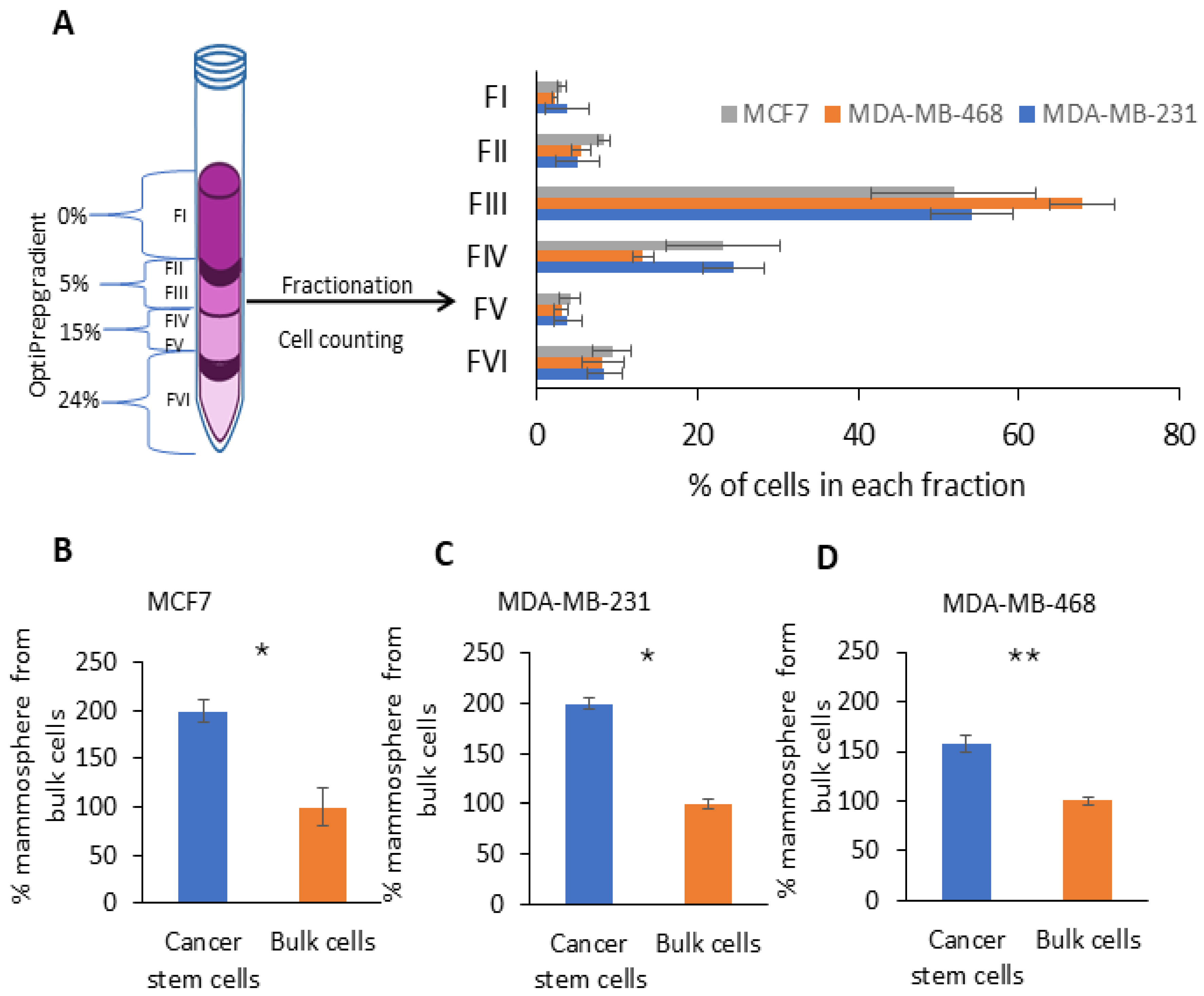
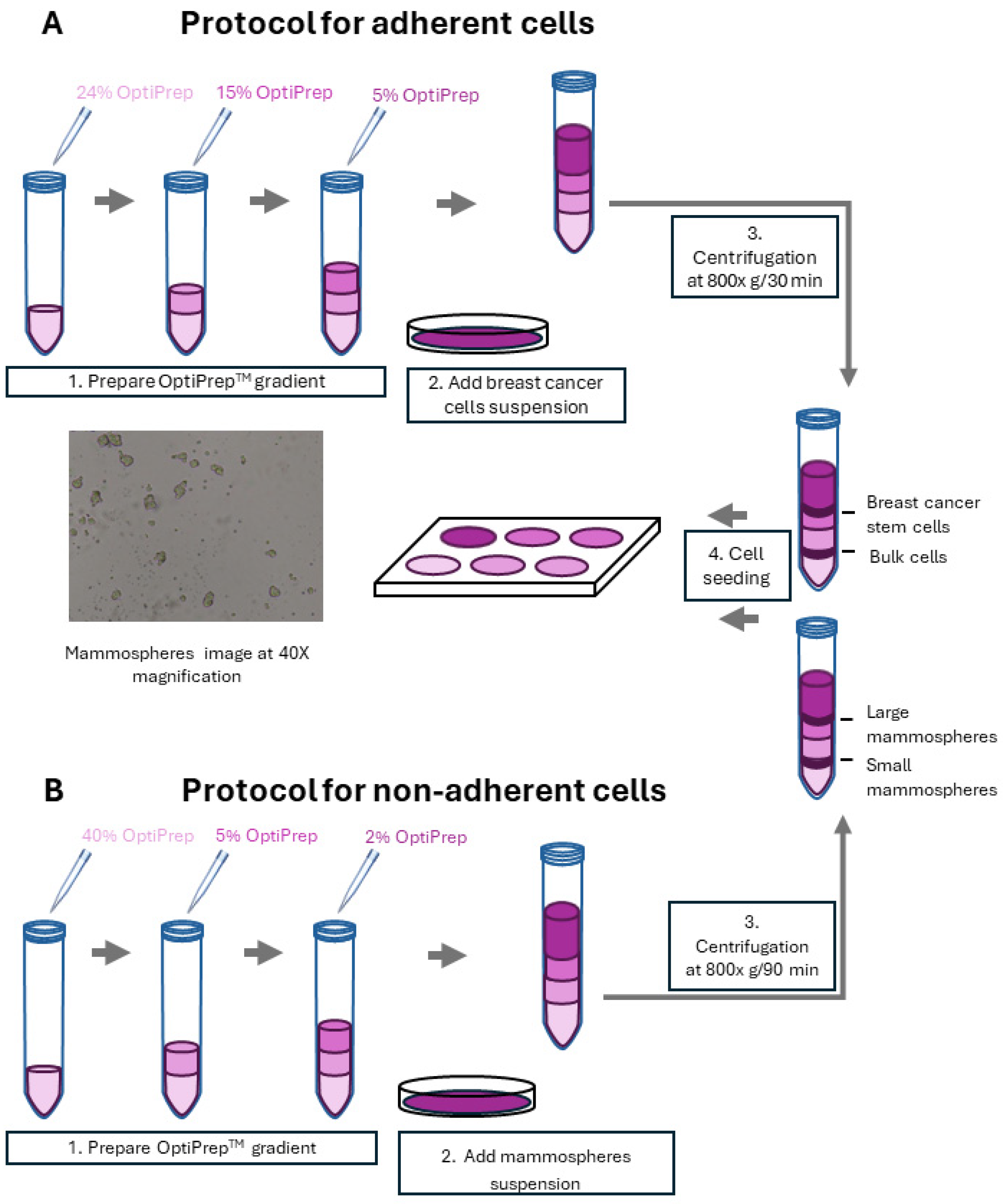
2.1. Fractionation of Breast Cancer Cells by Density Gradient Centrifugation and CSCs’ Enriched Fraction Determination via Non-Adherent 3D Mammosphere Assay
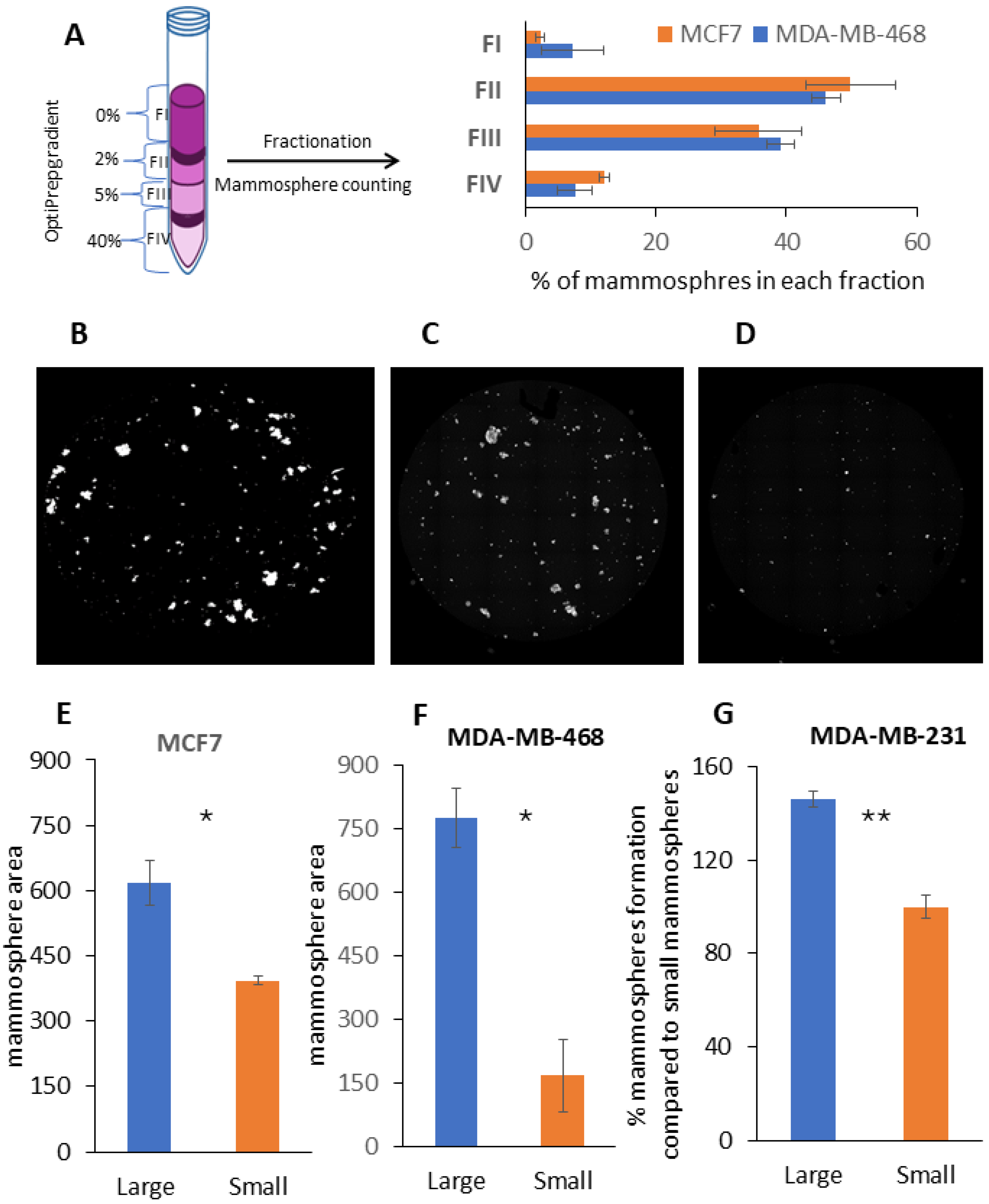
2.2. Fractionation of Mammospheres by Size Using Density Gradient Centrifugation
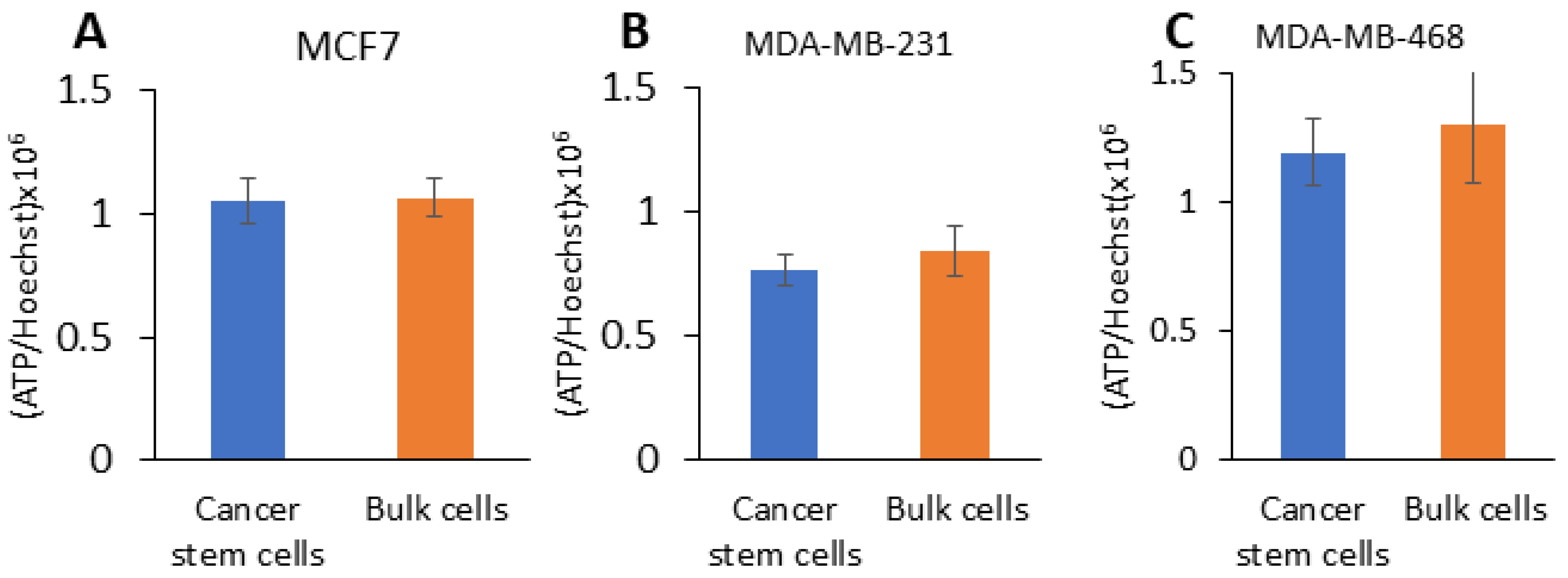
2.3. CSCs from TNBC Have Hybrid Metabolism and CSCs from Large Mammospheres Have Increased Glycolytic Capacity
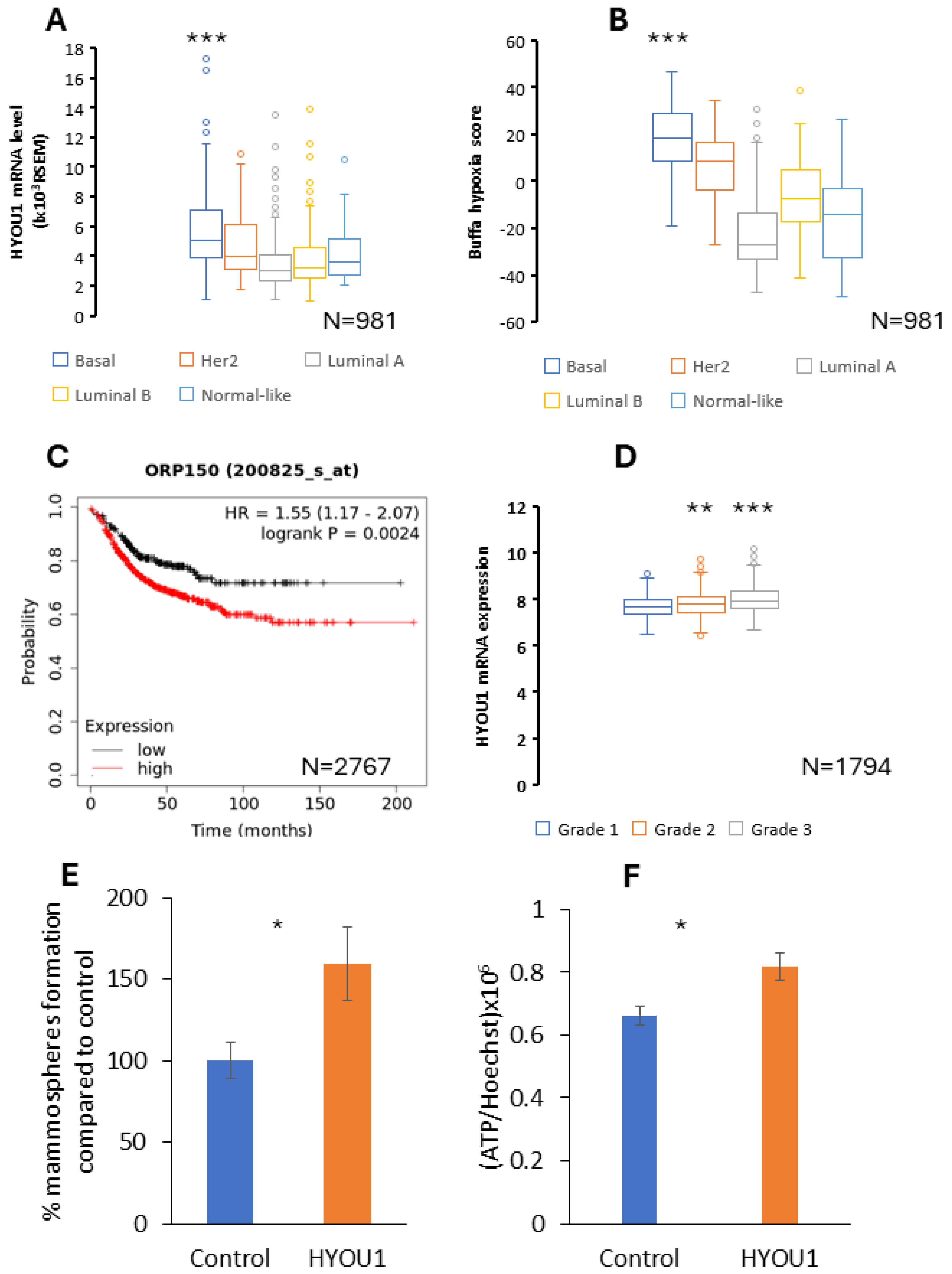
2.4. HYOU1 Is Highly Expressed in Hyoxic TNBC and HYOU1 Is Related with Aggressiveness, Stemness, Hyper-Metabolic Activity, Metastasis and Poor Prognosis of Breast Caner
3. Discussion
4. Materials and Methods
4.1. Experimental Model Cell Lines
4.2. Gradient-Based Breast CSCs and Hypoxic Mammosphere Separation
4.3. Three-Dimensional Mammosphere Growth and Three-Dimensional Mammosphere Formation Assay and Colony Formation Assay
4.4. Fluorescent Microscopy Analysis
4.5. ATP Assay Using Cell-Titer-Glo
4.6. Metabolic Flux Analysis for Density Gradient Separated Breast Cancer Cells
4.7. Viral Transduction of HYOU1 Gene
4.8. Bioinformatics Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 66. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Tasdogan, A.; Faubert, B.; Ramesh, V.; Ubellacker, J.M.; Shen, B.; Solmonson, A.; Murphy, M.M.; Gu, Z.; Gu, W.; Martin, M.; et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 2020, 577, 115–120. [Google Scholar] [CrossRef]
- Yu, L.; Lu, M.; Jia, D.; Ma, J.; Ben-Jacob, E.; Levine, H.; Kaipparettu, B.A.; Onuchic, J.N. Modeling the genetic regulation of cancer metabolism: Interplay between glycolysis and oxidative phosphorylation. Cancer Res. 2017, 77, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Lehuédé, C.; Dupuy, F.; Rabinovitch, R.; Jones, R.G.; Siegel, P.M. Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 2016, 76, 5201–5208. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, M.; Scatena, C.; Naccarato, A.G.; Sotgia, F.; Lisanti, M.P. Bedaquiline, an FDA-approved drug, inhibits mitochondrial ATP production and metastasis in vivo, by targeting the gamma subunit (ATP5F1C) of the ATP synthase. Cell Death Differ. 2021, 28, 2797–2817. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Lu, M.; Jung, K.H.; Park, J.H.; Yu, L.; Onuchic, J.N.; Kaipparettu, B.A.; Levine, H. Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. USA 2019, 116, 3909–3918. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Palmgren, V.A.; Danen, E.H.; Le Dévédec, S.E. Acute vs. chronic vs. intermittent hypoxia in breast Cancer: A review on its application in in vitro research. Mol. Biol. Rep. 2022, 49, 10961–10973. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lin, Q.; Glazer, P.M.; Yun, Z. Hypoxic Tumor Microenvironment and Cancer Cell Differentiation. Curr. Mol. Med. 2009, 9, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Tan, Y.; Gao, G.; Wu, S.; Zhang, J.; Tang, H.; Zhu, H.; Yang, A. Development and Verification of a Prognostic Stemness-Related Gene Signature in Triple-Negative Breast Cancer. J. Oncol. 2023, 2023, 6242355. [Google Scholar] [CrossRef]
- De Francesco, E.M.; Maggiolini, M.; Tanowitz, H.B.; Sotgia, F.; Lisanti, M.P. Targeting hypoxic cancer stem cells (CSCs) with Doxycycline: Implications for optimizing anti-angiogenic therapy. Oncotarget 2017, 8, 56126–56142. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K.; Kharazinejad, E.; Majidpoor, J.; Ahadi, R. Hypoxia in solid tumors: A key promoter of cancer stem cell (CSC) resistance. J. Cancer Res. Clin. Oncol. 2020, 146, 19–31. [Google Scholar] [CrossRef]
- Rao, S.; Oyang, L.; Liang, J.; Yi, P.; Han, Y.; Luo, X.; Xia, L.; Lin, J.; Tan, S.; Hu, J.; et al. Biological function of HYOU1 in tumors and other diseases. OncoTargets Ther. 2021, 14, 1727–1735. [Google Scholar] [CrossRef]
- Lee, M.; Song, Y.; Choi, I.; Lee, S.-Y.; Kim, S.; Kim, S.-H.; Kim, J.; Seo, H.R. Expression of hyou1 via reciprocal crosstalk between nsclc cells and huvecs control cancer progression and chemoresistance in tumor spheroids. Mol. Cells 2021, 44, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Franco, S.S.; Szczesna, K.; Iliou, M.S.; Al-Qahtani, M.; Mobasheri, A.; Kobolák, J.; Dinnyés, A. In vitro models of cancer stem cells and clinical applications. BMC Cancer 2016, 16, 23–49. [Google Scholar] [CrossRef]
- Sin, W.C.; Lim, C.L. Breast cancer stem cells—From origins to targeted therapy. Stem Cell Investig. 2017, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Fu, N.Y.; Visvader, J.E. Halting triple negative breast cancer by targeting PROCR. Cell Res. 2019, 29, 875–876. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.Y.; Zhang, H.; Yang, J.; Chen, Y.; Lu, H. Asymmetric Cell Division and Tumor Heterogeneity. Front. Cell Dev. Biol. 2022, 10, 938685. [Google Scholar] [CrossRef]
- Liu, W.-H.; Wang, X.; You, N.; Tao, K.-S.; Wang, T.; Tang, L.-J.; Dou, K.-F. Efficient enrichment of hepatic cancer stem-like cells from a primary Rat HCC model via a density gradient centrifugation-centered method. PLoS ONE 2012, 7, e35720. [Google Scholar] [CrossRef]
- Sabelström, H.; Quigley, D.A.; Fenster, T.; Foster, D.J.; Fuchshuber, C.A.; Saxena, S.; Yuan, E.; Li, N.; Paterno, F.; Phillips, J.J.; et al. High density is a property of slow-cycling and treatment-resistant human glioblastoma cells. Exp. Cell Res. 2019, 378, 76–86. [Google Scholar] [CrossRef]
- Kovacovicova, K.; Vinciguerra, M. Isolation of senescent cells by iodixanol (OptiPrep) density gradient-based separation. Cell Prolif. 2019, 52, e12674. [Google Scholar] [CrossRef]
- Mandujano-Tinoco, E.A.; Gallardo-Pérez, J.C.; Marín-Hernández, A.; Moreno-Sánchez, R.; Rodríguez-Enríquez, S. Anti-mitochondrial therapy in human breast cancer multi-cellular spheroids. Biochim. Biophys. Acta BBA Mol. Cell Res. 2013, 1833, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.-G.; Lee, S.-H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Choi, J.-H.; Nam, J.-S. Targeting cancer stem cells in triple-negative breast cancer. Cancers 2019, 11, 965. [Google Scholar] [CrossRef]
- Zhang, X.; Powell, K.; Li, L. Breast cancer stem cells: Biomarkers, identification and isolation methods, regulating mechanisms, cellular origin, and beyond. Cancers 2020, 12, 3765. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qi, D.; Hsieh, T.-C.; Huang, J.H.; Wu, J.M.; Wu, E. Trailblazing perspectives on targeting breast cancer stem cells. Pharmacol. Ther. 2021, 223, 107800. [Google Scholar] [CrossRef]
- Ponti, D.; Costa, A.; Zaffaroni, N.; Pratesi, G.; Petrangolini, G.; Coradini, D.; Pilotti, S.; Pierotti, M.A.; Daidone, M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005, 65, 5506–5511. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Wicinski, J.; Cervera, N.; Finetti, P.; Hur, M.-H.; Diebel, M.E.; Monville, F.; Dutcher, J.; et al. Breast cancer cell lines contain functional cancer stem sells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009, 69, 1302–1313. [Google Scholar] [CrossRef]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ma, S.; Tong, M. Metabolic Plasticity of Cancer Stem Cells in Response to Microenvironmental Cues. Cancers 2022, 14, 5345. [Google Scholar] [CrossRef] [PubMed]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Reue, K.; Frohnen, P.; Chan, M.; Alhiyari, Y.; Dratver, M.B.; Pajonk, F. Metabolic differences in breast cancer stem cells and differentiated progeny. Breast Cancer Res. Treat. 2014, 146, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Koit, A.; Shevchuk, I.; Ounpuu, L.; Klepinin, A.; Chekulayev, V.; Timohhina, N.; Tepp, K.; Puurand, M.; Truu, L.; Heck, K.; et al. Mitochondrial Respiration in Human Colorectal and Breast Cancer Clinical Material Is Regulated Differently. Oxidative Med. Cell. Longev. 2017, 2017, 1372640. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, F.; Fiorillo, M.; Lisanti, M.P. Mitochondrial markers predict recurrence, metastasis and tamoxifen-resistance in breast cancer patients: Early detection of treatment failure with companion diagnostics. Oncotarget 2017, 8, 68730–68745. [Google Scholar] [CrossRef] [PubMed]
- Fovez, Q.; Quesnel, B.; Laine, W.; Khamari, R.; Berthon, C.; Goursaud, L.; Germain, N.; Preudhomme, C.; Marchetti, P.; Kluza, J. Mitochondrial Spare Reserve Capacity: A New Predictive Metabolic Biomarker for Aggressiveness of Acute Myeloid Leukemia. Blood 2020, 136, 7. [Google Scholar] [CrossRef]
- Teh, J.T.; Zhu, W.L.; Newgard, C.B.; Casey, P.J.; Wang, M. Respiratory capacity and reserve predict cell sensitivity to mitochondria inhibitors: Mechanism-based markers to identify metformin-responsive cancers. Mol. Cancer Ther. 2019, 18, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Isono, T.; Chano, T.; Yonese, J.; Yuasa, T. Therapeutic inhibition of mitochondrial function induces cell death in starvation-resistant renal cell carcinomas. Sci. Rep. 2016, 6, 25669. [Google Scholar] [CrossRef]
- Park, M.; Sunwoo, K.; Kim, Y.-J.; Won, M.; Xu, Y.; Kim, J.; Pu, Z.; Li, M.; Kim, J.Y.; Seo, J.H.; et al. Cutting Off H+ Leaks on the Inner Mitochondrial Membrane: A Proton Modulation Approach to Selectively Eradicate Cancer Stem Cells. J. Am. Chem. Soc. 2023, 145, 4647–4658. [Google Scholar] [CrossRef]
- Ayyasamy, V.; Owens, K.M.; Desouki, M.M.; Liang, P.; Bakin, A.; Thangaraj, K.; Buchsbaum, D.J.; LoBuglio, A.F.; Singh, K.K. Cellular model of Warburg Effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PLoS ONE 2011, 6, e24792. [Google Scholar] [CrossRef]
- Marchetti, P.; Fovez, Q.; Germain, N.; Khamari, R.; Kluza, J. Mitochondrial spare respiratory capacity: Mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. 2020, 34, 13106–13124. [Google Scholar] [CrossRef]
- Cordani, M.; Butera, G.; Dando, I.; Torrens-Mas, M.; Butturini, E.; Pacchiana, R.; Oppici, E.; Cavallini, C.; Gasperini, S.; Tamassia, N.; et al. Mutant p53 blocks SESN1/AMPK/PGC-1α/UCP2 axis increasing mitochondrial O2ˉ· production in cancer cells. Br. J. Cancer 2018, 119, 994–1008. [Google Scholar] [CrossRef]
- Andugulapati, S.B.; Sundararaman, A.; Lahiry, M.; Rangarajan, A. AMP-activated protein kinase promotes breast cancer stemness and drug resistance. Dis. Model. Mech. 2022, 15, dmm049203. [Google Scholar] [CrossRef] [PubMed]
- Elia, I.; Schmieder, R.; Christen, S.; Fendt, S.M. Organ-Specific Cancer Metabolism and Its Potential for Therapy Ilaria: Adipokines and the Endocrine Role of Adipose Tissues. Handb. Exp. Pharmacol. 2015. [Google Scholar] [CrossRef]
- Dai, S.; Peng, Y.; Zhu, Y.; Xu, D.; Zhu, F.; Xu, W.; Chen, Q.; Zhu, X.; Liu, T.; Hou, C.; et al. Glycolysis promotes the progression of pancreatic cancer and reduces cancer cell sensitivity to gemcitabine. Biomed. Pharmacother. 2020, 121, 109521. [Google Scholar] [CrossRef]
- Yang, T.; Shu, X.; Zhang, H.-W.; Sun, L.-X.; Yu, L.; Liu, J.; Sun, L.-C.; Yang, Z.-H.; Ran, Y.-L. Enolase 1 regulates stem cell-like properties in gastric cancer cells by stimulating glycolysis. Cell Death Dis. 2020, 11, 870. [Google Scholar] [CrossRef] [PubMed]
- Ciavardelli, D.; Rossi, C.; Barcaroli, D.; Volpe, S.; Consalvo, A.; Zucchelli, M.; De Cola, A.; Scavo, E.; Carollo, R.; D’Agostino, D.; et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 2014, 5, e1336. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L.; Jiang, W.; Lu, W.; Yang, J.; Yang, W. B cell lymphoma with different metabolic characteristics show distinct sensitivities to metabolic inhibitors. J. Cancer 2018, 9, 1582–1591. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Cui, Y.; Xu, S.; Wu, X.; Huang, Y.; Zhou, W.; Wang, S.; Fu, Z.; Xie, H. Altered glycolysis results in drug-resistant in clinical tumor therapy (Review). Oncol. Lett. 2021, 21, 369. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Miao, W.; Huang, M.; Li, L.; Dai, X.; Wang, Y. Elevated hexokinase II expression confers acquired resistance to 4-hydroxytamoxifen in breast cancer cells. Mol. Cell. Proteom. 2019, 18, 2273–2284. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, H.; Yu, W.; Qiao, F.; Su, X.; Xu, H. Downregulation of hexokinase 2 improves radiosensitivity of breast cancer. Transl. Cancer Res. 2019, 8, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Wintzell, M.; Löfstedt, L.; Johansson, J.; Pedersen, A.B.; Fuxe, J.; Shoshan, M. Repeated cisplatin treatment can lead to a multiresistant tumor cell population with stem cell features and sensitivity to 3-bromopyruvate. Cancer Biol. Ther. 2012, 13, 1454–1462. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, H.; Liu, Y.; Wu, J.; Wang, C.; Hou, X.; Chen, X.; Yang, G.; Zhao, L.; Che, H.; et al. miR-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells. Cancer Lett. 2013, 333, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Yeerken, D.; Hong, R.; Wang, Y.; Gong, Y.; Liu, R.; Yang, D.; Li, J.; Fan, J.; Chen, J.; Zhang, W.; et al. PFKP is transcriptionally repressed by BRCA1/ZBRK1 and predicts prognosis in breast cancer. PLoS ONE 2020, 15, e0233750. [Google Scholar] [CrossRef] [PubMed]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Li, X.; Luo, Z.; Liu, J.; Fan, Z. Cetuximab Reverses the Warburg Effect by Inhibiting HIF-1–Regulated LDH-A. Mol. Cancer Ther. 2013, 12, 2187–2199. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Dai, W.; Zhang, Q.; Feng, J.; Wu, L.; Liu, T.; Yu, Q.; Xu, S.; Wang, W.; et al. Genistein suppresses aerobic glycolysis and induces hepatocellular carcinoma cell death. Br. J. Cancer 2017, 117, 1518–1528. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef]
- Sebestyén, A.; Kopper, L.; Dankó, T.; Tímár, J. Hypoxia Signaling in Cancer: From Basics to Clinical Practice. Pathol. Oncol. Res. 2021, 27, 1609802. [Google Scholar] [CrossRef]
- Nejad, A.E.; Najafgholian, S.; Rostami, A.; Sistani, A.; Shojaeifar, S.; Esparvarinha, M.; Nedaeinia, R.; Javanmard, S.H.; Taherian, M.; Ahmadlou, M.; et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: A novel approach to developing treatment. Cancer Cell Int. 2021, 21, 62. [Google Scholar] [CrossRef]
- Klepinin, A.; Miller, S.; Reile, I.; Puurand, M.; Rebane-Klemm, E.; Klepinina, L.; Vija, H.; Zhang, S.; Terzic, A.; Dzeja, P.; et al. Stable Isotope Tracing Uncovers Reduced γ/β-ATP Turnover and Metabolic Flux Through Mitochondrial-Linked Phosphotransfer Circuits in Aggressive Breast Cancer Cells. Front. Oncol. 2022, 12, 892195. [Google Scholar] [CrossRef] [PubMed]
- Hao, A.; Wang, Y.; Zhang, X.; Li, J.; Li, Y.; Li, D.; Kulik, G.; Sui, G. Long non-coding antisense RNA HYOU1-AS is essential to human breast cancer development through competitive binding hnRNPA1 to promote HYOU1 expression. Biochim. Biophys. Acta BBA Mol. Cell Res. 2021, 1868, 118951. [Google Scholar] [CrossRef] [PubMed]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Sargiacomo, C.; Stonehouse, S.; Moftakhar, Z.; Sotgia, F.; Lisanti, M.P. MitoTracker Deep Red (MTDR) Is a Metabolic Inhibitor for Targeting Mitochondria and Eradicating Cancer Stem Cells (CSCs), With Anti-Tumor and Anti-Metastatic Activity In Vivo. Front. Oncol. 2021, 11, 678343. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2009, 123, 725–731. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sargiacomo, C.; Klepinin, A. Density Gradient Centrifugation Is an Effective Tool to Isolate Cancer Stem-like Cells from Hypoxic and Normoxia Triple-Negative Breast Cancer Models. Int. J. Mol. Sci. 2024, 25, 8958. https://doi.org/10.3390/ijms25168958
Sargiacomo C, Klepinin A. Density Gradient Centrifugation Is an Effective Tool to Isolate Cancer Stem-like Cells from Hypoxic and Normoxia Triple-Negative Breast Cancer Models. International Journal of Molecular Sciences. 2024; 25(16):8958. https://doi.org/10.3390/ijms25168958
Chicago/Turabian StyleSargiacomo, Camillo, and Aleksandr Klepinin. 2024. "Density Gradient Centrifugation Is an Effective Tool to Isolate Cancer Stem-like Cells from Hypoxic and Normoxia Triple-Negative Breast Cancer Models" International Journal of Molecular Sciences 25, no. 16: 8958. https://doi.org/10.3390/ijms25168958
APA StyleSargiacomo, C., & Klepinin, A. (2024). Density Gradient Centrifugation Is an Effective Tool to Isolate Cancer Stem-like Cells from Hypoxic and Normoxia Triple-Negative Breast Cancer Models. International Journal of Molecular Sciences, 25(16), 8958. https://doi.org/10.3390/ijms25168958




