The Impact of Growing Area on the Expression of Fruit Traits Related to Sensory Perception in Two Tomato Cultivars
Abstract
:1. Introduction
2. Results
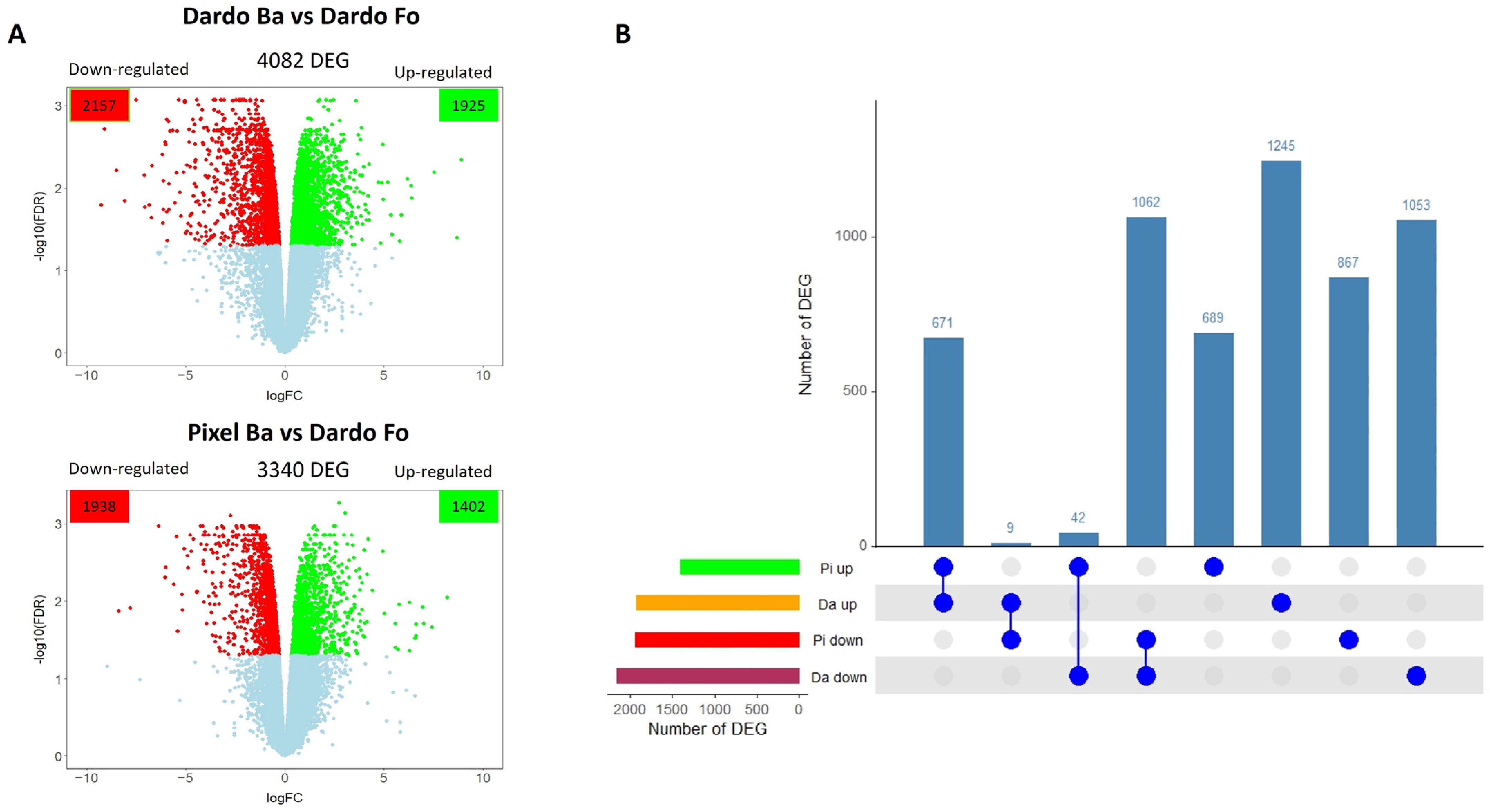
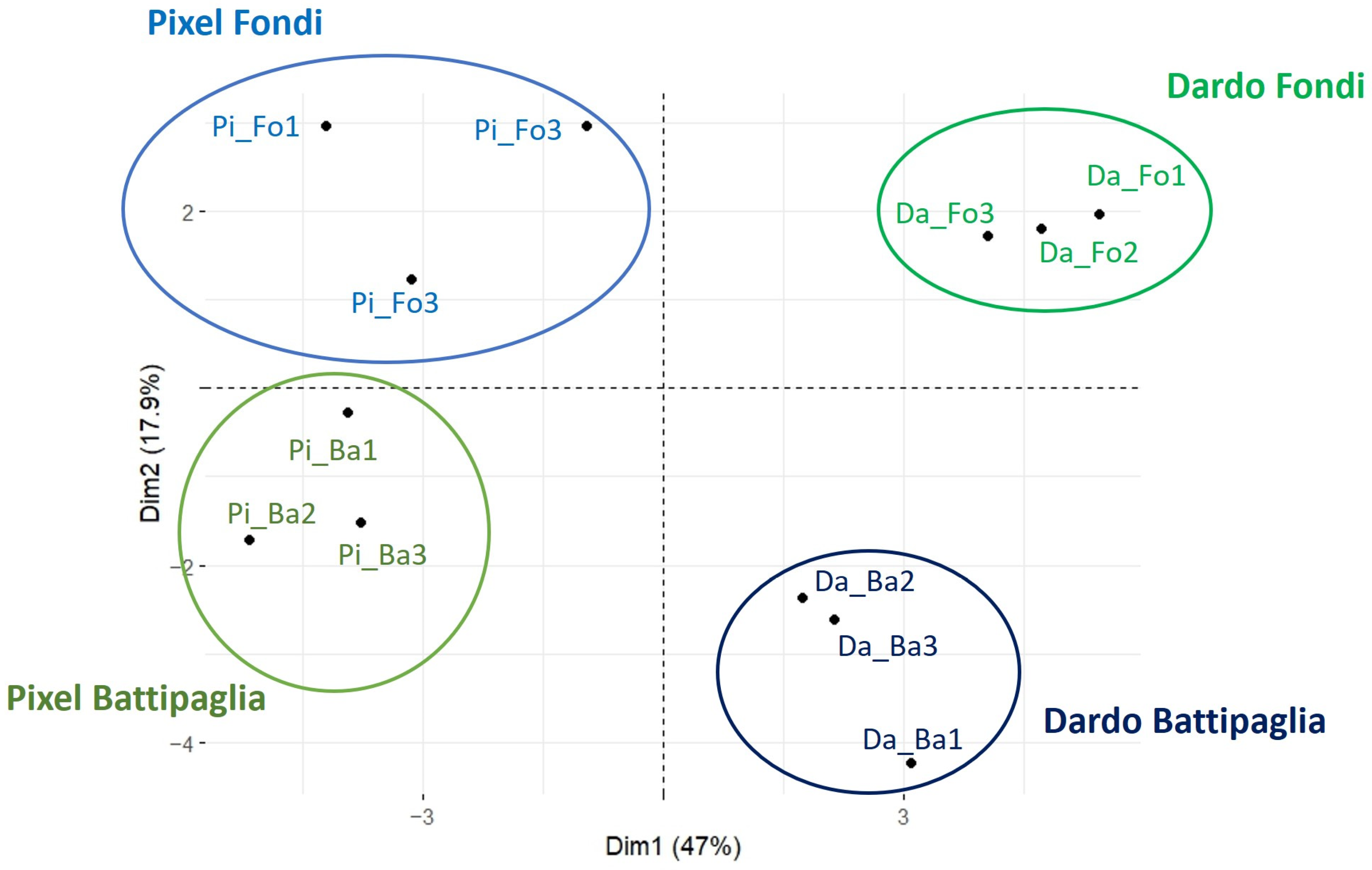
2.1. Pixel and Dardo Gene Expression Profile
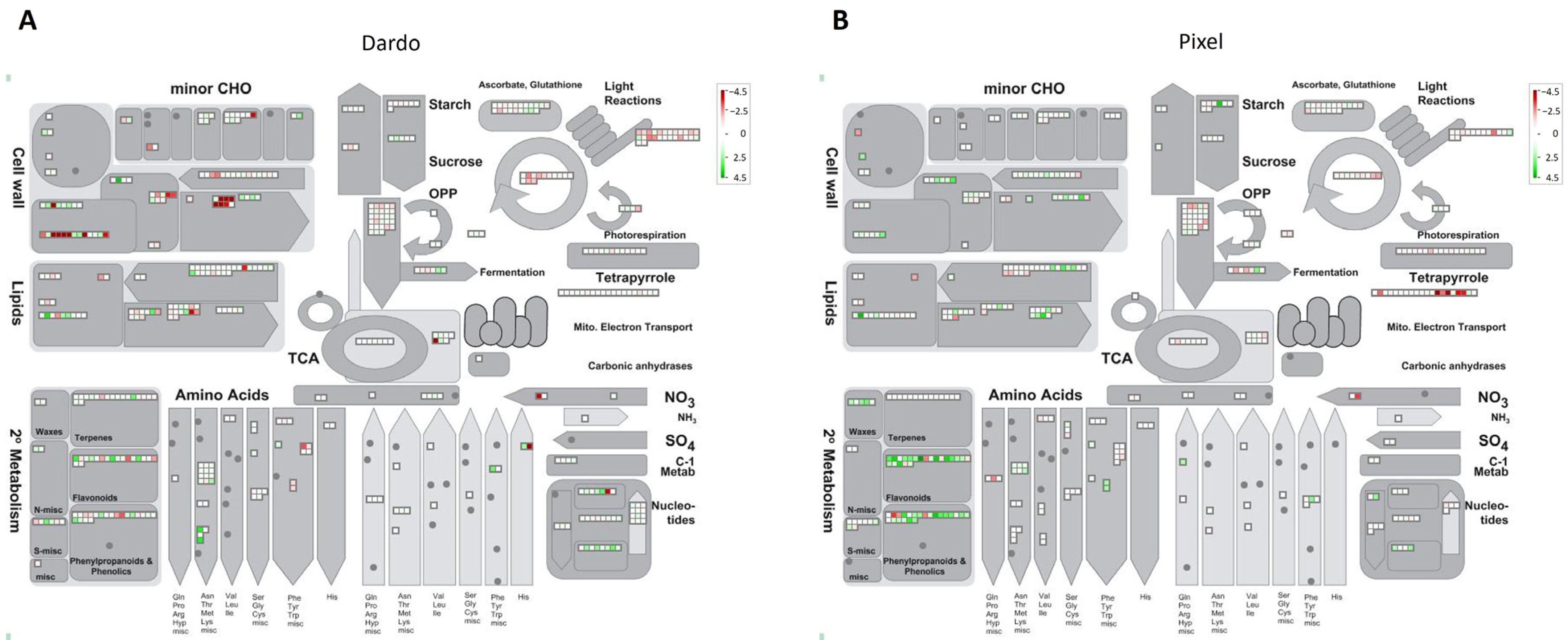
2.2. DEGs Involved in Primary Metabolism
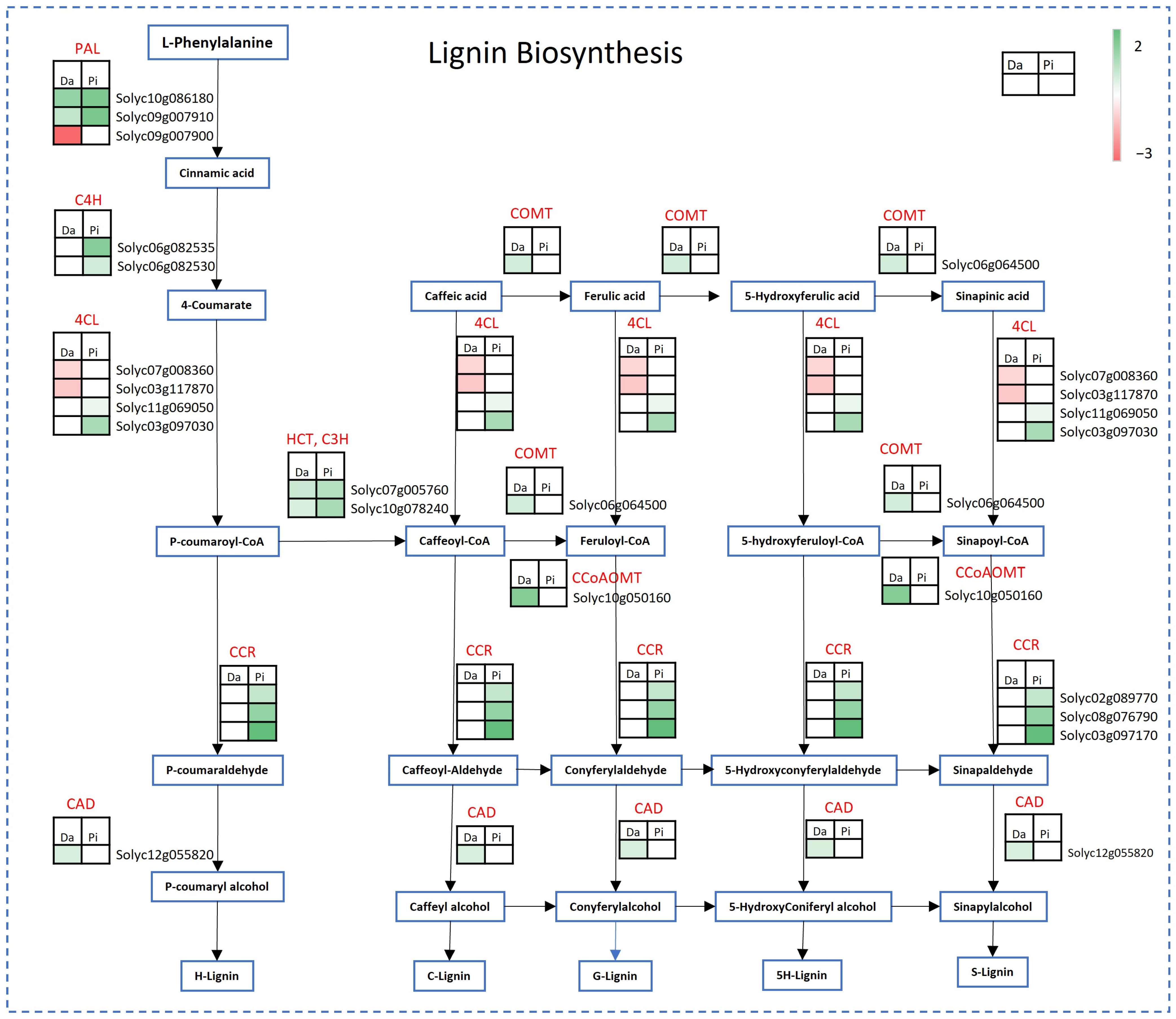
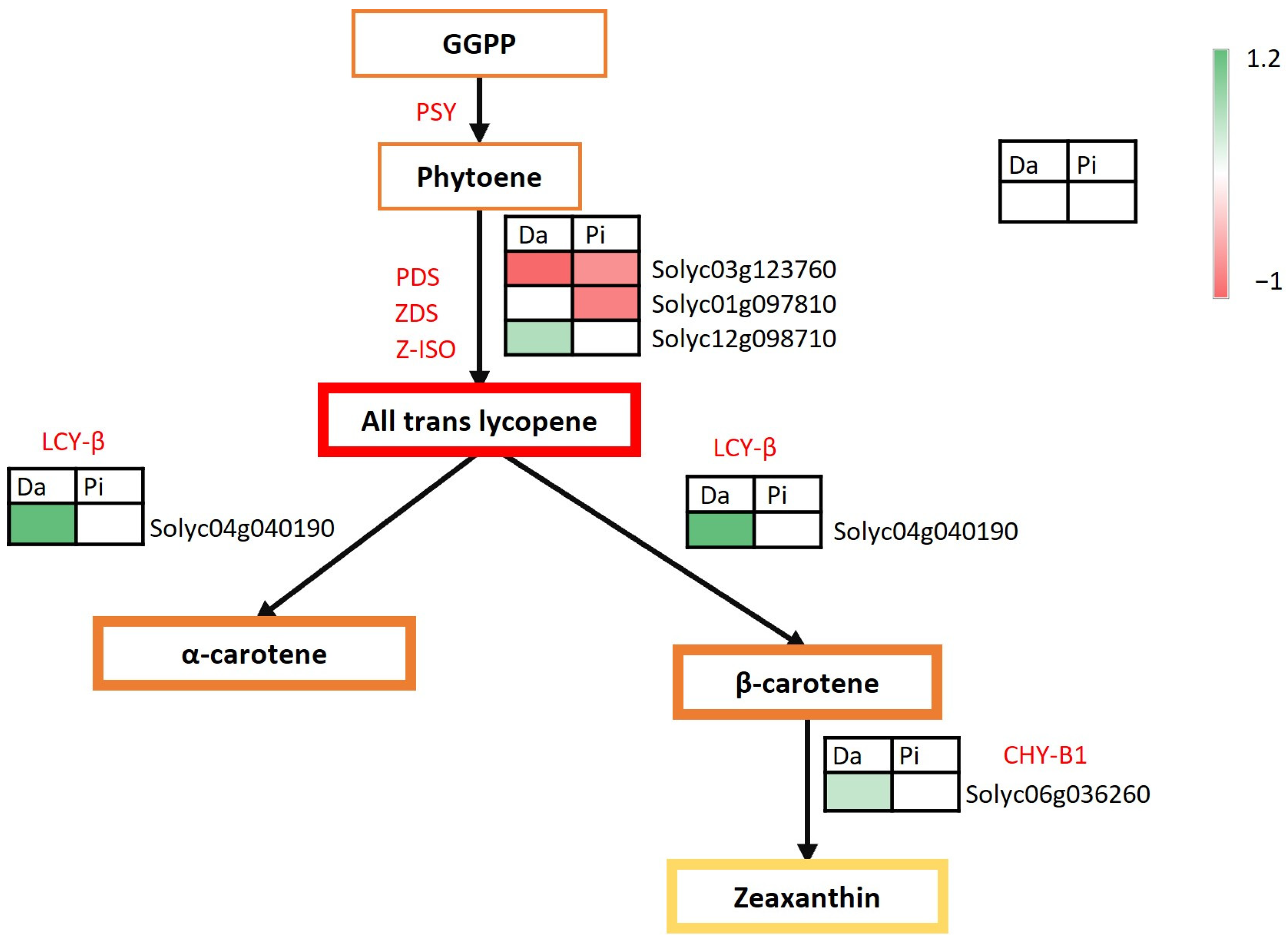
2.3. DEGs Involved in Secondary Metabolism
2.4. Sensory Evaluation
2.5. Metabolite Assessment Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Sequencing and Transcriptomic Analysis
4.3. Functional Annotation of Differentially Expressed Genes
4.4. Metabolic Analysis
4.5. Sensory Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prasanna, V.; Prabha, T.N.; Tharanathan, R.N. Fruit ripening phenomena: An overview. Crit. Rev. Food Sci. Nutr. 2007, 47, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Defilippi, B.G.; Manriquez, D.; Luengwilai, K.; Gonzalez-Aguero, M. Aroma volatiles: Biosynthesis and mechanisms of modulation during fruit ripening. Adv. Bot. Res. 2009, 50, 1–37. [Google Scholar]
- Bovy, A.; Schijlen, E.; Hall, R.D. Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): The potential for metabolomics. Metabolomics 2007, 3, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Enfissi, E.M.A.; Halket, J.M.; Truesdale, M.R.; Yu, D.; Gerrish, C.; Bramley, P. Manipulation of phytoene levels in tomato fruit: Effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 2007, 19, 3194–3211. [Google Scholar] [CrossRef] [PubMed]
- Vicente, A.R.; Saladie, M.; Rose, J.K.C.; Labavitch, J.M. The linkage between cell wall metabolism and fruit softening: Looking to the future. J. Sci. Food Agric. 2007, 87, 1435–1448. [Google Scholar] [CrossRef]
- Causse, M.; Buret, M.; Robini, K.; Verschave, P. Inheritance of nutritional and sensory quality traits in fresh market tomato and relation to consumer preferences. J. Food Sci. 2003, 68, 2342–2350. [Google Scholar] [CrossRef]
- Ercolano, M.R.; Carli, P.; Soria, A.; Cascone, A.; Fogliano, V.; Frusciante, L.; Barone, A. Biochemical, sensorial and genomic profiling of traditional Italian tomato varieties. Euphytica 2008, 164, 571–582. [Google Scholar] [CrossRef]
- Abbass, K.; Qasim, M.Z.; Song, H.; Murshed, M.; Mahmood, H.; Younis, I. A review of the global climate change impacts, adaptation, and sustainable mitigation measures. Environ. Sci. Pollut. Res. Int. 2022, 29, 42539–42559. [Google Scholar] [CrossRef] [PubMed]
- Causse, M.; Friguet, C.; Coiret, C.; Lépicier, M.; Navez, B.; Lee, M.; Holthuysen, N.; Sinesio, F.; Moneta, E.; Grandillo, S. Consumer preferences for fresh tomato at the European scale: A common segmentation on taste and firmness. J. Food Sci. 2010, 75, S531–S541. [Google Scholar] [CrossRef]
- D’Esposito, D.; Ferriello, F.; Dal Molin, A.; Diretto, G.; Sacco, A.; Minio, A.; Barone, A.; Di Monaco, R.; Cavella, S.; Tardella, L.; et al. Unraveling the complexity of transcriptomic, metabolomic and quality environmental response of tomato fruit. BMC Plant Biol. 2017, 17, 66. [Google Scholar] [CrossRef]
- Lashbrooke, J.; Adato, A.; Lotan, O.; Alkan, N.; Tsimbalist, T.; Rechav, K.; Fernandez-Moreno, J.P.; Widemann, E.; Grausem, B.; Pinot, F.; et al. The Tomato MIXTA-Like Transcription Factor Coordinates Fruit Epidermis Conical Cell Development and Cuticular Lipid Biosynthesis and Assembly. Plant Physiol. 2015, 169, 2553–2571. [Google Scholar] [CrossRef]
- Liu, M.; Gomes, B.L.; Mila, I.; Purgatto, E.; Peres, L.E.; Frasse, P.; Maza, E.; Zouine, M.; Roustan, J.P.; Bouzayen, M.; et al. Comprehensive Profiling of Ethylene Response Factor Expression Identifies Ripening-Associated ERF Genes and Their Link to Key Regulators of Fruit Ripening in Tomato. Plant Physiol. 2016, 170, 1732–1744. [Google Scholar] [CrossRef] [PubMed]
- Carli, P.; Barone, A.; Fogliano, V.; Frusciante, L.; Ercolano, M.R. Dissection of genetic and environmental factors involved in tomato organoleptic quality. BMC Plant Biol. 2011, 11, 58. [Google Scholar] [CrossRef]
- Seymour, G.B.; Østergaard, L.; Chapman, N.H.; Knapp, S.; Martin, C. Fruit development and ripening. Annu. Rev. Plant Biol. 2013, 64, 219–241. [Google Scholar] [CrossRef] [PubMed]
- Carli, P.; Arima, S.; Fogliano, V.; Tardella, L.; Frusciante, L.; Ercolano, M.R. Use of network analysis to capture key traits affecting tomato organoleptic quality. J. Exp. Bot. 2009, 60, 3379–3386. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Sakuragi, Y.; Zhu, X.; Burrell, A.J.; Mohanty, S.S.; Atwood, J.A., 3rd; Orlando, R.; Scheller, H.V.; Mohnen, D. Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan: Galacturonosyltransferase complex. Proc. Natl. Acad. Sci. USA 2011, 108, 20225–20230. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, J.; Wang, J.; Zhang, J.; Miao, H.; Jia, C.; Wang, Z.; Xu, B.; Jin, Z. MuMADS1 and MaOFP1 regulate fruit quality in a tomato ovate mutant. Plant Biotechnol. J. 2018, 16, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Nabil, S.A.N.; Azam, M.M.; Amer Eissa, A.H. Mechanical properties of tomato fruits under storage conditions. J. Appl. Sci. Res. 2012, 8, 3053–3064. [Google Scholar]
- Wang, D.; Lu, Q.; Wang, X.; Ling, H.; Huang, N. Elucidating the role of SlXTH5 in tomato fruit softening. Hortic. Plant J. 2022, 9, 777–788. [Google Scholar] [CrossRef]
- Klinghammer, M.; Tenhaken, R. Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J. Exp. Bot. 2007, 58, 3609–3621. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Bar-Peled, M. The biosynthesis of UDP-galacturonic acid in plants. Functional cloning and characterization of Arabidopsis UDP-D-glucuronic acid 4-epimerase. Plant Physiol. 2004, 136, 4256–4264. [Google Scholar] [CrossRef]
- Wills, R.; Golding, J. Postharvest: An Introduction to the Physiology and Handling of Fruit and Vegetables; NewSouth Publishing: Sydney, Australia, 2016. [Google Scholar]
- España, L.; Heredia-Guerrero, J.A.; Segado, P.; Benítez, J.J.; Heredia, A.; Domínguez, E. Biomechanical properties of the tomato (Solanum lycopersicum) fruit cuticle during development are modulated by changes in the relative amounts of its components. N. Phytol. 2014, 202, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.J.; Tieman, D.M. The genetics of fruit flavour preferences. Nat. Rev. Genet. 2018, 19, 347–356. [Google Scholar] [CrossRef]
- Baldwin, E.A.; Thompson, K.A. Effect of sugar type and concentration on perception of tomato sweetness. Proc. Fla. State Hortic. Soc. 2000, 113, 287–289. [Google Scholar]
- Goulet, C.; Kamiyoshihara, Y.; Lam, N.B.; Richard, T.; Taylor, M.G.; Tieman, D.M.; Klee, H.J. Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Mol. Plant 2015, 8, 153–162. [Google Scholar] [CrossRef]
- Kaur, G.; Abugu, M.; Tieman, D. The dissection of tomato flavor: Biochemistry, genetics, and omics. Front. Plant Sci. 2023, 14, 1144113. [Google Scholar] [CrossRef]
- Feng, L.; Wang, C.; Yang, X.; Jiao, Q.; Yin, Y. Transcriptomics and metabolomics analyses identified key genes associated with sugar and acid metabolism in sweet and sour pomegranate cultivars during the developmental period. Plant Physiol. Biochem. 2022, 181, 12–22. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, X.; Chen, S.; Meng, L.; Fu, D. Role of the tomato TAGL1 gene in regulating fruit metabolites elucidated using RNA sequence and metabolomics analyses. PLoS ONE 2018, 13, e0199083. [Google Scholar] [CrossRef]
- Min, D.; Li, Z.; Fu, X.; Wang, J.; Li, F.; Li, X.; Zhang, X. Integration of transcriptomic and metabonomic reveals molecular differences of sweetness and aroma between postharvest and vine ripened tomato fruit. Food Control 2022, 139, 109102. [Google Scholar] [CrossRef]
- Cai, B.; Li, Q.; Xu, Y.; Yang, L.; Bi, H.; Ai, X. Genome-wide analysis of the fructose 1,6-bisphosphate aldolase (FBA) gene family and functional characterization of FBA7 in tomato. Plant Physiol. Biochem. 2016, 108, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Granot, D.; David-Schwartz, R.; Kelly, G. Hexose kinases and their role in sugar-sensing and plant development. Front. Plant Sci. 2013, 4, 44. [Google Scholar] [CrossRef]
- Li, J.; Chen, G.; Zhang, J.; Shen, H.; Kang, J.; Feng, P.; Xie, Q.; Hu, Z. Suppression of a hexokinase gene, SlHXK1, leads to accelerated leaf senescence and stunted plant growth in tomato. Plant Sci. 2020, 298, 110544. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Ludewig, F.; Flügge, U.I. Role of metabolite transporters in source-sink carbon allocation. Front. Plant Sci. 2013, 4, 231. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.I.; Delhaize, E.; Frommer, W.B.; Guerinot, M.L.; Harrison, M.J.; Herrera-Estrella, L.; Horie, T.; Kochian, L.V.; Munns, R.; Nishizawa, N.K.; et al. Using membrane transporters to improve crops for sustainable food production. Nature 2013, 497, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Shammai, A.; Petreikov, M.; Yeselson, Y.; Faigenboim, A.; Moy-Komemi, M.; Cohen, S.; Cohen, D.; Besaulov, E.; Efrati, A.; Houminer, N.; et al. Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. Plant J. 2018, 96, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Tommonaro, G.; Morelli, C.F.; Rabuffetti, M.; Nicolaus, B.; De Prisco, R.; Iodice, C.; Speranza, G. Determination of flavor-potentiating compounds in different Italian tomato varieties. J. Food. Biochem. 2021, 45, e13736. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Q.; Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Erlenbach, I.; Ryba, N.J.; Zuker, C.S. The receptors for mammalian sweet and umami taste. Cell 2003, 115, 255–266. [Google Scholar] [CrossRef]
- Wu, Y.; Yuan, Y.; Jiang, W.; Zhang, X.; Ren, S.; Wang, H.; Zhang, X.; Zhang, Y. Enrichment of health-promoting lutein and zeaxanthin in tomato fruit through metabolic engineering. Synth. Syst. Biotechnol. 2022, 7, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef]
- Kou, X.H.; Zhao, Y.N.; Wu, C.; Jiang, B.L.; Zhang, Z.; Rathbun, J.R.; He, Y.; Xue, Z. SNAC4 and SNAC9 transcription factors show contrasting effects on tomato carotenoids biosynthesis and softening. Postharvest. Biol. Tec. 2018, 144, 9–19. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Tureckova, V.; Xue, G.P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Grosso, V.; Farina, A.; Giorgi, D.; Nardi, L.; Diretto, G.; Lucretti, S. A high-throughput flow cytometry system for early screening of in vitro made polyploids in Dendrobium hybrids. Plant Cell Tissue Organ Cult. 2018, 132, 57–70. [Google Scholar] [CrossRef]
- Ahrazem, O.; Argandoña, J.; Fiore, A.; Aguado, C.; Luján, R.; Rubio-Moraga, Á.; Marro, M.; Araujo-Andrade, C.; Loza-Alvarez, P.; Diretto, G.; et al. Transcriptome analysis in tissue sectors with contrasting crocins accumulation provides novel insights into apocarotenoid biosynthesis and regulation during chromoplast biogenesis. Sci. Rep. 2018, 8, 2843. [Google Scholar] [CrossRef]
- Meilgaard, M.; Vance Civille, G.; Thomas Carr, B. Sensory Evaluation Techniques, 3rd ed.; Meilgaard, M.C., Civille, G.V., Carr, B.T., Eds.; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]


| Gene ID | Functional Description | Metabolic Pathway | Pixel Ba vs. Fo (logFC) | Dardo Ba vs. Fo (logFC) |
|---|---|---|---|---|
| Solyc03g113030 | Aldose 1-epimerase | Glycolysis and gluconeogenesis | −0.50 | 0.50 |
| Solyc01g098700 | Branched-chain-amino-acid aminotransferase-like protein | L-leucine biosynthesis/L-leucine degradation I | −0.53 | 0.53 |
| Solyc03g097050 | Cellulose-synthase-like protein | Cellulose synthesis | 0.82 | −3.61 |
| Solyc03g097050 | Cellulose-synthase-like protein | Cellulose synthesis | ||
| Solyc02g089640 | Cellulose-synthase-like | Cellulose synthesis | 0.92 | −2.47 |
| Solyc01g112000 | Expansin-like protein precursor 1 | Cell wall extensibility | 1.51 | −2.84 |
| Solyc02g089440 | Glycosyltransferase family protein | Homogalacturonan biosynthesis | 0.88 | −0.68 |
| Solyc05g051850 | Putative myo-inositol-1-phosphatase | Myo-inositol biosynthesis | 0.91 | −0.68 |
| Solyc01g091840 | UDP-galactose transporter | Nucleotide sugar transport | 1.86 | −1.22 |
| Solyc12g010540 | UDP-glucuronate 4-epimerase 4 | UDP-α-D-galacturonate biosynthesis I (from UDP-D-glucuronate) | 0.63 | −1.31 |
| Pixel Battipaglia | Pixel Fondi | p-Value | Dardo Battipaglia | Dardo Fondi | p-Value | |
|---|---|---|---|---|---|---|
| Sugars/Sugar alcohol/Nucleotide sugars | ||||||
| Glucose/Fructose/Galactose/Mannose/Myo-inositol | 0.4622 ± 0.0310 | 0.6468 ± 0.0937 | 0.0104 | 0.3208 ± 0.0128 | 0.4093 ± 0.0144 | 0.0101 |
| UDP-Glucose | 0.1845 ± 0.0036 | 0.1963 ± 0.0094 | 0.3998 | 0.1678 ± 0.0049 | 0.2307 ± 0.0087 | 0.0033 |
| UDP-Galacturonic acid | 0.0038 ± 0.0009 | 0.0078 ± 0.0014 | 0.0116 | 0.0043 ± 0.0004 | 0.0089 ± 0.0022 | 0.1153 |
| Amino acids | ||||||
| Glutamine | 3.4699 ± 0.5620 | 2.6992 ± 0.5970 | 0.2490 | 6.2400 ± 0.5696 | 4.5354 ± 0.1368 | 0.0437 |
| Glutamic acid | 14.6427 ± 0.3340 | 14.9509 ± 1.0578 | 0.4793 | 9.5698 ± 0.9092 | 13.0123 ± 0.8218 | 0.0484 |
| Aspartate | 0.8360 ± 0.0373 | 0.9558 ± 0.0412 | 0.0432 | 0.7022 ± 0.0418 | 0.9397 ± 0.0396 | 0.0146 |
| 1-Aminocyclopropane-1-carboxylic acid | 0.1775 ± 0.0213 | 0.1001 ± 0.0148 | 0.0434 | 0.1591 ± 0.0055 | 0.0708 ± 0.0148 | 0.0051 |
| Organic acids | ||||||
| Ascorbic acid | 0.0629 ± 0.0071 | 0.0802 ± 0.0117 | 0.0802 | 0.0383 ± 0.0023 | 0.0503 ± 0.0021 | 0.0180 |
| Shikimic acid | 0.0071 ± 0.0002 | 0.0104 ± 0.0013 | 0.0112 | 0.0055 ± 0.0003 | 0.0078 ± 0.0004 | 0.0112 |
| Phenylpropanoids—Phenolic acids | ||||||
| p-Coumaric acid | 0.0097 ± 0.0005 | 0.0080 ± 0.0010 | 0.0194 | 0.0059 ± 0.0003 | 0.0049 ± 0.0000 | 0.0171 |
| Caffeic acid | 0.0217 ± 0.0004 | 0.0219 ± 0.0031 | 0.8907 | 0.0119 ± 0.0013 | 0.0168 ± 0.0006 | 0.0284 |
| Ferulic acid | 0.0079 ± 0.0005 | 0.0123 ± 0.0039 | 0.0810 | 0.0077 ± 0.0010 | 0.0042 ± 0.0002 | 0.0307 |
| Cinnamic acid | 14.2733 ± 0.2453 | 14.9550 ± 1.0604 | 0.1039 | 9.5698 ± 0.9092 | 13.0123 ± 0.8218 | 0.0484 |
| Phenylpropanoids—Flavonoids | ||||||
| Dihydrokaemferol | 0.0104 ± 0.0029 | 0.0082 ± 0.0006 | 0.5234 | 0.0196 ± 0.0018 | 0.0076 ± 0.0007 | 0.0032 |
| Fatty acids | ||||||
| linolenic acid | 0.0230 ± 0.0004 | 0.0413 ± 0.0095 | 0.0217 | 0.0355 ± 0.0046 | 0.0250 ± 0.0028 | 0.1202 |
| Carotenoids | ||||||
| β-carotene | 0.2434 ± 0.0176 | 0.2689 ± 0.0087 | 0.5972 | 0.4179 ± 0.0205 | 0.3504 ± 0.0023 | 0.0308 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Esposito, D.; Di Donato, A.; Puleo, S.; Nava, M.; Diretto, G.; Di Monaco, R.; Frusciante, L.; Ercolano, M.R. The Impact of Growing Area on the Expression of Fruit Traits Related to Sensory Perception in Two Tomato Cultivars. Int. J. Mol. Sci. 2024, 25, 9015. https://doi.org/10.3390/ijms25169015
D’Esposito D, Di Donato A, Puleo S, Nava M, Diretto G, Di Monaco R, Frusciante L, Ercolano MR. The Impact of Growing Area on the Expression of Fruit Traits Related to Sensory Perception in Two Tomato Cultivars. International Journal of Molecular Sciences. 2024; 25(16):9015. https://doi.org/10.3390/ijms25169015
Chicago/Turabian StyleD’Esposito, Daniela, Antimo Di Donato, Sharon Puleo, Matteo Nava, Gianfranco Diretto, Rossella Di Monaco, Luigi Frusciante, and Maria Raffaella Ercolano. 2024. "The Impact of Growing Area on the Expression of Fruit Traits Related to Sensory Perception in Two Tomato Cultivars" International Journal of Molecular Sciences 25, no. 16: 9015. https://doi.org/10.3390/ijms25169015






