KMT2A Rearrangements in Leukemias: Molecular Aspects and Therapeutic Perspectives
Abstract
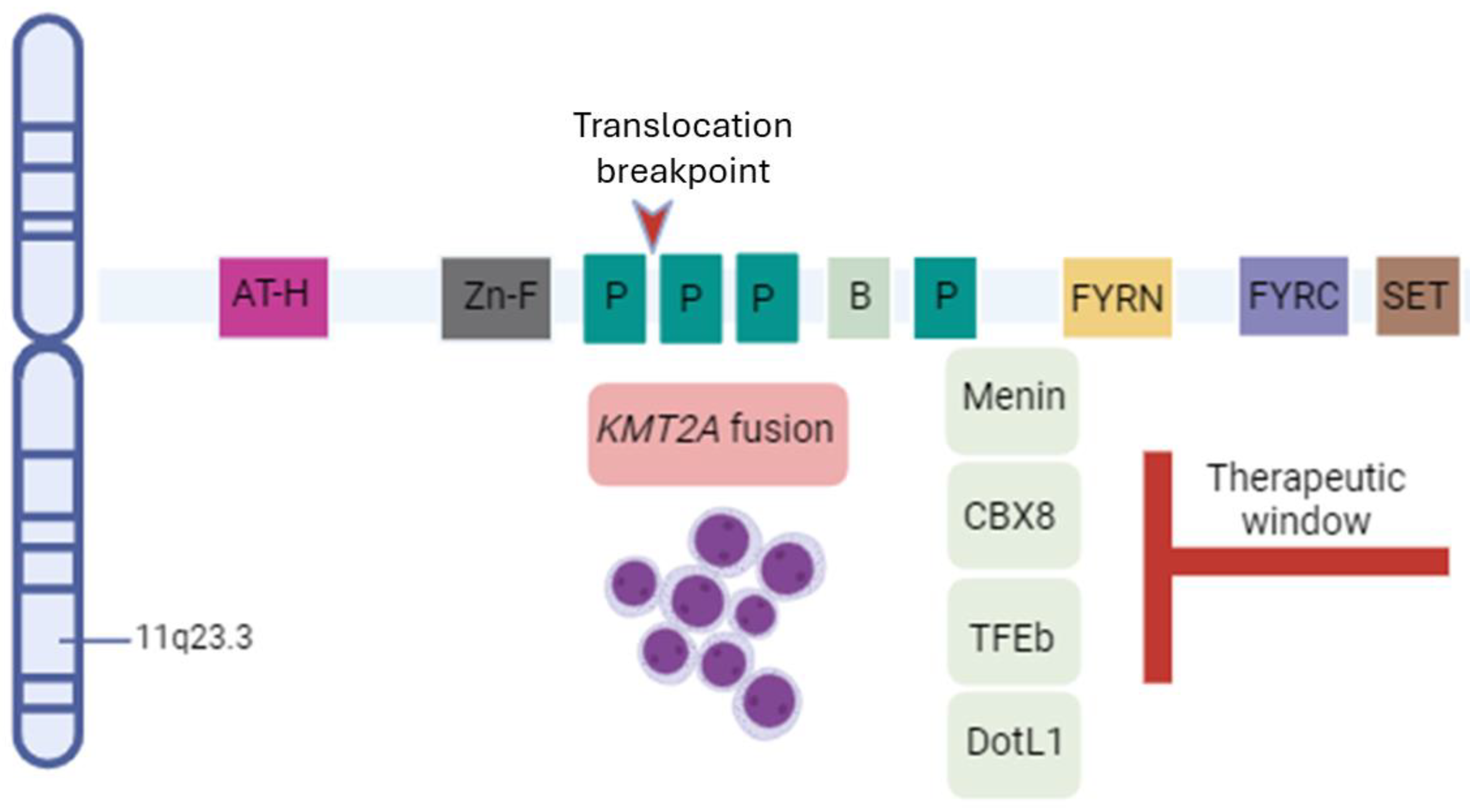
1. Introduction
2. Biology of Rearrangements
3. Clinical Aspects
3.1. Incidence, Clinical Features, and Prognosis in Infancy and Childhood
3.2. Incidence, Clinical Features, and Prognosis in Adulthood
4. Clinical Management of KMT2A-Rearranged AML and Current Clinical Practice
4.1. Chemotherapy
4.2. Less Intensive Regimens
4.3. Hematopoietic Stem Cell Transplantation
5. Menin Inhibitors
5.1. Revumenib
5.2. Ziftomenib
5.3. Other Menin Inhibitors
6. Experimental Data of Combination Therapy
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Meyer, C.; Larghero, P.; Almeida, L.B.; Burmeister, T.; Gröger, D.; Sutton, R. The KMT2A recombinome of acute leukemias in 2023. Leukemia 2023, 37, 988–1005. [Google Scholar] [CrossRef] [PubMed]
- Górecki, M.; Kozioł, I.; Kopystecka, A.; Budzyńska, J.; Zawitkowska, J.; Lejman, M. Updates in KMT2A Gene Rearrangement in Pediatric Acute Lymphoblastic Leukemia. Biomedicines 2023, 11, 821. [Google Scholar] [CrossRef]
- Larson, J.K.; Hunter-Schlichting, D.N.; Crowgey, E.L.; Mills, L.J.; Druley, T.E.; Marcotte, E.L. KMT2A-D pathogenicity, prevalence, and variation according to a population database. Cancer Med. 2023, 12, 7234–7245. [Google Scholar] [CrossRef] [PubMed]
- Lavallée, V.P.; Baccelli, I.; Krosl, J.; Wilhelm, B.; Barabé, F.; Gendron, P. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat. Genet. 2015, 47, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Steudel, C.; Wermke, M.; Schaich, M.; Schäkel, U.; Illmer, T.; Ehninger, G. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer 2003, 37, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Seto, A.; Downs, G.; King, O.; Salehi-Rad, S.; Baptista, A.; Chin, K. Genomic Characterization of Partial Tandem Duplication Involving the KMT2A Gene in Adult Acute Myeloid Leukemia. Cancers 2024, 26, 1693. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Perez, C.; Cifuentes-Riquelme, R.; Padilla, J.; de la Morena-Barrio, M.E.; Ortuño, F.J.; Garrido-Rodríguez, P. The whole is greater than the sum of its parts: Long-read sequencing for solving clinical problems in haematology. J. Cell Mol. Med. 2024, 28, e17961. [Google Scholar] [CrossRef]
- Bill, M.; Mrózek, K.; Kohlschmidt, J.; Eisfeld, A.K.; Walker, C.J.; Nicolet, D. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proc. Natl. Acad. Sci. USA 2020, 117, 26340–26346. [Google Scholar] [CrossRef]
- Hu, D.Y.; Wang, M.; Shen, K.; Pan, J.L.; Guo, Y.S.; Zhang, Z.B. A new breakpoint fusion gene involving KMT2A::EDC4 rearrangement in de novo acute myeloid leukemia. Int. J. Lab. Hematol. 2023, 45, 596–598. [Google Scholar] [CrossRef]
- Hyrenius-Wittsten, A.; Pilheden, M.; Sturesson, H.; Hansson, J.; Walsh, M.P.; Song, G. De novo activating mutations drive clonal evolution and enhance clonal fitness in KMT2A-rearranged leukemia. Nat. Commun. 2018, 9, 1770. [Google Scholar] [CrossRef]
- Dal Molin, A.; Tretti Parenzan, C.; Gaffo, E.; Borin, C.; Boldrin, E.; Meyer, L.H.; Te Kronnie, G.; Bresolin, S.; Bortoluzzi, S. Discovery of fusion circular RNAs in leukemia with KMT2A::AFF1 rearrangements by the new software CircFusion. Brief Bioinform. 2023, 24, bbac589. [Google Scholar] [CrossRef]
- Pui, C.H.; Gaynon, P.S.; Boyett, J.M.; Chessells, J.M.; Baruchel AKamps, W. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet 2002, 359, 1909–1915. [Google Scholar] [CrossRef]
- Pui, C.H.; Chessells, J.M.; Camitta, B.; Baruchel, A.; Biondi, A.; Boyett, J.M. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia 2003, 17, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Balgobind, B.V.; Raimondi, S.C.; Harbott, J.; Zimmermann, M.; Alonzo, T.A.; Auvrignon, A. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: Results of an international retrospective study. Blood 2009, 114, 2489–2496. [Google Scholar] [CrossRef]
- Szczepański, T.; Harrison, C.J.; van Dongen, J.J.M. Genetic aberrations in paediatric acute leukaemias and implications for management of patients. Lancet Oncol. 2010, 11, 880–889. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.D.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef] [PubMed]
- Teachey, D.T.; Pui, C.H. Comparative features and outcomes between paediatric T-cell and B-cell acute lymphoblastic leukaemia. Lancet Oncol. 2019, 20, e142–e154. [Google Scholar] [CrossRef] [PubMed]
- Pieters, R.; De Lorenzo, P.; Ancliffe, P.; Aversa, L.A.; Brethon, B.; Biondi, A. Outcome of Infants Younger than 1 Year with Acute Lymphoblastic Leukemia Treated with the Interfant-06 Protocol: Results from an International Phase III Randomized Study. J. Clin. Oncol. 2019, 37, 2246–2256. [Google Scholar] [CrossRef]
- Nagayama, J.; Tomizawa, D.; Koh, K.; Nagatoshi, Y.; Hotta, N.; Kishimoto, T. Infants with acute lymphoblastic leukemia and a germline MLL gene are highly curable with use of chemotherapy alone: Results from the Japan Infant Leukemia Study Group. Blood 2006, 107, 4663–4665. [Google Scholar] [CrossRef] [PubMed]
- Fazio, G.; Bardini, M.; De Lorenzo, P.; Grioni, A.; Quadri, M.; Pedace, L. Recurrent genetic fusions redefine MLL germ line acute lymphoblastic leukemia in infants. Blood 2021, 137, 1980–1984. [Google Scholar] [CrossRef]
- Boer, J.M.; Valsecchi, M.G.; Hormann, F.M.; Antić, Ž.; Zaliova, M.; Schwab, C. Favorable outcome of NUTM1-rearranged infant and pediatric B cell precursor acute lymphoblastic leukemia in a collaborative international study. Leukemia 2021, 35, 2978–2982. [Google Scholar] [CrossRef] [PubMed]
- Guest, E.M.; Kairalla, J.A.; Hilden, J.M.; Dreyer, Z.E.; Carroll, A.J.; Heerema, N.A. Outstanding outcomes in infants with KMT2A-germline acute lymphoblastic leukemia treated with chemotherapy alone: Results of the Children’s Oncology Group AALL0631 trial. Haematologica 2022, 107, 1205–1208. [Google Scholar] [CrossRef]
- Issa, G.C.; Zarka, J.; Sasaki, K.; Qiao, W.; Pak, D.; Ning, J. Predictors of outcomes in adults with acute myeloid leukemia and KMT2A rearrangements. Blood Cancer J. 2021, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Kantarjian, H.M.; Short, N.J.; Qiao, W.; Ning, J.; Cuglievan, B. Early mortality in acute myeloid leukemia with KMT2A rearrangement is associated with high risk of bleeding and disseminated intravascular coagulation. Cancer 2023, 129, 1856–1865. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Curry, C.V.; Khanlari, M.; Shi, M.; Cui, W.; Peker, D. Genetics and pathologic landscape of lineage switch of acute leukemia during therapy. Blood Cancer J. 2024, 14, 19. [Google Scholar] [CrossRef]
- Bacher, U.; Haferlach, T.; Kern, W.; Haferlach, C.; Schnittger, S. A comparative study of molecular mutations in 381 patients with myelodysplastic syndrome and in 4130 patients with acute myeloid leukemia. Haematologica 2007, 92, 744–752. [Google Scholar] [CrossRef]
- Guarnera, L.; Bravo-Perez, C.; Visconte, V. Immunotherapy in Acute Myeloid Leukemia: A Literature Review of Emerging Strategies. Bioengineering 2023, 10, 1228. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.A.; Guest, E.; Alonzo, T.A.; Gerbing, R.B.; Loken, M.R.; Brodersen, L.E. Gemtuzumab Ozogamicin Improves Event-Free Survival and Reduces Relapse in Pediatric KMT2A-Rearranged AML: Results From the Phase III Children’s Oncology Group Trial AAML0531. J. Clin. Oncol. 2021, 39, 3149–3160. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.; Hills, R.K.; Burnett, A.K.; Russell, N.H. Gemtuzumab ozogamicin in (KMT2A)-rearranged adult acute myeloid leukaemia (AML) in the UK Medical Research Council AML15 and AML16 trials. Br. J. Haematol. 2022, 196, e50–2. [Google Scholar] [CrossRef]
- Ball, B.J.; Arslan, S.; Koller, P.; Ngo, D.; Afkhami, M.; Salhotra, A. Clinical experience with venetoclax and hypomethylating agents (HMA) in patients with newly diagnosed and relapsed or refractory KMT2A-Rearranged acute myeloid leukemia (AML). Leuk Lymphoma 2022, 63, 3232–3236. [Google Scholar] [CrossRef]
- Pigneux, A.; Labopin, M.; Maertens, J.; Cordonnier, C.; Volin, L.; Socié, G. Outcome of allogeneic hematopoietic stem-cell transplantation for adult patients with AML and 11q23/MLL rearrangement (MLL-r AML). Leukemia 2015, 29, 2375–2381. [Google Scholar] [CrossRef]
- Jiang, B.; Zhao, Y.; Luo, Y.; Yu, J.; Chen, Y.; Ye, B. Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation in Adult Patients With Acute Myeloid Leukemia Harboring KMT2A Rearrangement and Its Prognostic Factors. Cell Transplant. 2024, 33, 9636897231225821. [Google Scholar] [CrossRef] [PubMed]
- Miyamura, T.; Kudo, K.; Tabuchi, K.; Ishida, H.; Tomizawa, D.; Adachi, S. Hematopoietic stem cell transplantation for pediatric acute myeloid leukemia patients with KMT2A rearrangement; A nationwide retrospective analysis in Japan. Leuk. Res. 2019, 87, 106263. [Google Scholar]
- Menghrajani, K.; Gomez-Arteaga, A.; Madero-Marroquin, R.; Zhang, M.J.; Bo-Subait, K.; Sanchez, J. Risk classification at diagnosis predicts post-HCT outcomes in intermediate-, adverse-risk, and KMT2A-rearranged AML. Blood Adv. 2022, 6, 828–847. [Google Scholar]
- Tong, J.; Zhang, L.; Liu, H.; Xu, X.; Zheng, C.; Yao, W. Umbilical cord blood transplantation can overcome the poor prognosis of KMT2A-MLLT3 acute myeloid leukemia and can lead to good GVHD-free/relapse-free survival. Ann. Hematol. 2021, 100, 1303–1309. [Google Scholar] [CrossRef]
- Antherieu, G.; Bidet, A.; Huet, S.; Hayette, S.; Migeon, M.; Boureau, L. Allogenic Stem Cell Transplantation Abrogates Negative Impact on Outcome of AML Patients with KMT2A Partial Tandem Duplication. Cancers 2021, 13, 2272. [Google Scholar] [CrossRef]
- Yokoyama, A.; Somervaille, T.C.P.; Smith, K.S.; Rozenblatt-Rosen, O.; Meyerson, M.; Cleary, M.L. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 2005, 123, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Borkin, D.; He, S.; Miao, H.; Kempinska, K.; Pollock, J.; Chase, J. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 2015, 27, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.V.; Evans, K.; Gadrey, J.Y.; Eschle, B.K.; Hatton, C.; Uckelmann, H.J. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell 2019, 36, 660–673.e11. [Google Scholar] [CrossRef]
- Klossowski, S.; Miao, H.; Kempinska, K.; Wu, T.; Purohit, T.; Kim, E. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J. Clin. Investig. 2020, 130, 981–997. [Google Scholar] [CrossRef] [PubMed]
- Issa, G.C.; Aldoss, I.; DiPersio, J.; Cuglievan, B.; Stone, R.; Arellano, M. The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature 2023, 615, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Perner, F.; Stein, E.M.; Wenge, D.V.; Singh, S.; Kim, J.; Apazidis, A. MEN1 mutations mediate clinical resistance to menin inhibition. Nature 2023, 615, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Erba, H.P.; Fathi, A.T.; Issa, G.C. Update on a Phase 1/2 First-in-Human Study of the Menin-KMT2A (MLL) Inhibitor Ziftomenib (KO-539) in Patients with Relapsed or Refractory Acute Myeloid Leukemia. Blood 2022, 140 (Suppl. S1), 153–156. [Google Scholar] [CrossRef]
- Jabbour, E.; Searle, E.; Abdul-Hay, M. A First-in-Human Phase 1 Study of the Menin-KMT2A (MLL1) Inhibitor JNJ-75276617 in Adult Patients with Relapsed/Refractory Acute Leukemia Harboring KMT2A or NPM1 Alterations. Blood 2023, 142 (Suppl. S1), 57. [Google Scholar] [CrossRef]
- Daver, N.; Zeidner, J.F.; Yuda, J. Phase 1/2 First-in-Human Study of the Menin-MLL Inhibitor DSP-5336 in Patients with Relapsed or Refractory Acute Leukemia. Blood 2023, 142 (Suppl. S1), 2911. [Google Scholar] [CrossRef]
- Fleischmann, M.; Bechwar, J.; Voigtländer, D.; Fischer, M.; Schnetzke, U.; Hochhaus, A. Synergistic Effects of the RARalpha Agonist Tamibarotene and the Menin Inhibitor Revumenib in Acute Myeloid Leukemia Cells with KMT2A Rearrangement or NPM1 Mutation. Cancers 2024, 16, 1311. [Google Scholar] [CrossRef] [PubMed]
- Ling, Q.; Zhou, Y.; Qian, Y.; Qian, J.; Zhang, Y.; Wang, J. Repressing HIF-1α-induced HDAC9 contributes to the synergistic effect of venetoclax and MENIN inhibitor in KMT2Ar AML. Biomark Res. 2023, 11, 105. [Google Scholar] [CrossRef]
- Ye, J.; Zha, J.; Shi, Y.; Li, Y.; Yuan, D.; Chen, Q. Co-inhibition of HDAC and MLL-menin interaction targets MLL-rearranged acute myeloid leukemia cells via disruption of DNA damage checkpoint and DNA repair. Clin. Epigenetics 2019, 11, 137. [Google Scholar] [CrossRef]
- Fiskus, W.; Boettcher, S.; Daver, N.; Mill, C.P.; Sasaki, K.; Birdwell, C.E. Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c). Blood Cancer J. 2022, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, B.J.; Cutler, J.A.; Bourgeois, W.; Donovan, K.A.; Gu, S.; Hatton, C. IKAROS and MENIN coordinate therapeutically actionable leukemogenic gene expression in MLL-r acute myeloid leukemia. Nat. Cancer 2022, 3, 595–613. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, W.; Cutler, J.A.; Aubrey, B.J.; Wenge, D.V.; Perner, F.; Martucci, C. Mezigdomide is effective alone and in combination with menin inhibition in preclinical models of KMT2A-r and NPM1c AML. Blood 2024, 143, 1513–1527. [Google Scholar] [CrossRef]
- Kubota, Y.; Reynold, M.; Williams, N.D. Genomic Analyses Unveil the Pathogenesis and Inform on Therapeutic Targeting in KMT2A-PTD AML. Blood 2023, 142 (Suppl. S1), 5696. [Google Scholar] [CrossRef]
- Lo-Coco, F.; Avvisati, G.; Vignetti, M.; Thiede, C.; Orlando, S.M.; Iacobelli, S. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N. Engl. J. Med. 2013, 369, 111–121. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
|
Partner Gene | Chromosomal Location | Infant AML | Pediatric AML |
Adult AML | % Total AML |
|---|---|---|---|---|---|
| MLLT3 | (9p22) | 28.4% | 36.5% | 24.4% | 30.4% |
| MLLT10 | (10p12.3) | 27.4% | 23.4% | 9.9% | 18.7% |
| KMT2A-PTD | (11q23.3) | 0 | 1.5% | 25.6% | 10.7% |
| ELL | (19p13.1) | 15.2% | 6.8% | 11.5% | 10.1% |
| AFDN | (6q27) | 1.5% | 9.03% | 10.2% | 8.1% |
| MLLT1 | (19p13.3) | 1% | 6.02% | 3.6% | 4.2% |
| MLLT11 | (1q21.3) | 6.59% | 2.15% | 0.9% | 2.4% |
| SEPTIN6 | (Xq24) | 3.55% | 2.15% | 0.45% | 1.7% |
| MLLT6 | (17q12) | 0 | 1.07% | 2.72% | 1.6% |
| EPS15 | (1p32.3) | 1.01% | 1.07% | 1.58% | 1.3% |
| SEPTIN9 | (17q25) | 1.01% | 1.29% | 1.36% | 1.3% |
| CBL | Del (11) (q23.3q23.3) | 0 | 0 | 0.22% | 0.89% |
| AFF1 | (4q21) | 1.5% | 0.2% | 1.1% | 0.8% |
| SEPTIN5 | (22q11.2) | 1.01% | 0.64% | 0.45% | 0.71% |
| ABI1 | (10p11.2) | 1.52% | 1.07% | 0 | 0.71% |
| Population | Drugs | KMT2Ar Type | Outcome | Reference # |
|---|---|---|---|---|
| Adult AML | -cytarabine/daunorubicin-based induction chemotherapy ± autologous stem cell transplantation -decitabine ± bortezomib | t(9;11) (p22;q23) t(6;11) (q27;q23) t(11;19) (q23;p13.1) t(11;19) (q23;p13.3) t/ins(10;11) (p13;q23) | % CR (42-77) % 3-year OS (41-5) | [10] |
| -venetoclax + HMA | t(9;11) (p22;q23) t(11;19) (q23;p13.1) t(6;11) (q23;q27) t(10;11) (p11.2;q23) t(4;11) (q21.3;q23) t(11;17) (q13;q23) t(11;11) (q13;q23) t/ins(10;11) (p13;q23) | % CR 83 median OS 11.05 months | [36] | |
| -allo-HSCT | t(6;11) (q27;q23) t(9;11) (p22:q23) t(11;19) (q23;p13.1) t(10;11) (p12;q23) t(1;11) (q21;q23) | % 3-year OS 59.1 | [38] | |
| -intensively treated | KMT2A-PTD | % CR 64.5 median OS 24.4 months | [42] | |
| Adult R/R AML | -venetoclax + HMA | t(9;11) (p22;q23) t(11;19) (q23;p13.1) t(6;11) (q23;q27) t(10;11) (p11.2;q23) t(4;11) (q21.3;q23) t(11;17) (q13;q23) t(11;11) (q13;q23) t/ins(10;11) (p13;q23) | % CR 17 median OS 6.05 months | [36] |
| Adult R/R AML-ALL-MPAL | -revumenib (SNDX-5613) | t(9;11) (p22:q23) t(4;11) (q21.3;q23) t(11;19) (q23;p13.1) t(6;11) (q27;q23) t(11;17) (q13;q23) Mut. NPM1 | % CR 33 | [47] |
| Pediatric AML | -intensively treated + GO | t(6;11) (q23;q27) t(10;11) (p11.2;q23) t(10;12) (p12;q23) t(4;11) (q21.3;q23) t(11;19) (q23;p13.3) | % CR 77 % 5-year OS 50 | [34] |
| -allo-HSCT | t(9;11) (p22:q23) t(11;19) (q23;p13.1) t(1;11) (q21;q23) t(6;11) (q27;q23) t(10;11) (p12;q23) t(4;11) (q21.3;q23) t(5;11) (q31;q23) t(7;11) (q22;q23) t(8;11) (q24;q23) | % 3-year OS 52.1 | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guarnera, L.; D’Addona, M.; Bravo-Perez, C.; Visconte, V. KMT2A Rearrangements in Leukemias: Molecular Aspects and Therapeutic Perspectives. Int. J. Mol. Sci. 2024, 25, 9023. https://doi.org/10.3390/ijms25169023
Guarnera L, D’Addona M, Bravo-Perez C, Visconte V. KMT2A Rearrangements in Leukemias: Molecular Aspects and Therapeutic Perspectives. International Journal of Molecular Sciences. 2024; 25(16):9023. https://doi.org/10.3390/ijms25169023
Chicago/Turabian StyleGuarnera, Luca, Matteo D’Addona, Carlos Bravo-Perez, and Valeria Visconte. 2024. "KMT2A Rearrangements in Leukemias: Molecular Aspects and Therapeutic Perspectives" International Journal of Molecular Sciences 25, no. 16: 9023. https://doi.org/10.3390/ijms25169023
APA StyleGuarnera, L., D’Addona, M., Bravo-Perez, C., & Visconte, V. (2024). KMT2A Rearrangements in Leukemias: Molecular Aspects and Therapeutic Perspectives. International Journal of Molecular Sciences, 25(16), 9023. https://doi.org/10.3390/ijms25169023








