Research Advancements in Salt Tolerance of Cucurbitaceae: From Salt Response to Molecular Mechanisms
Abstract
1. Introduction
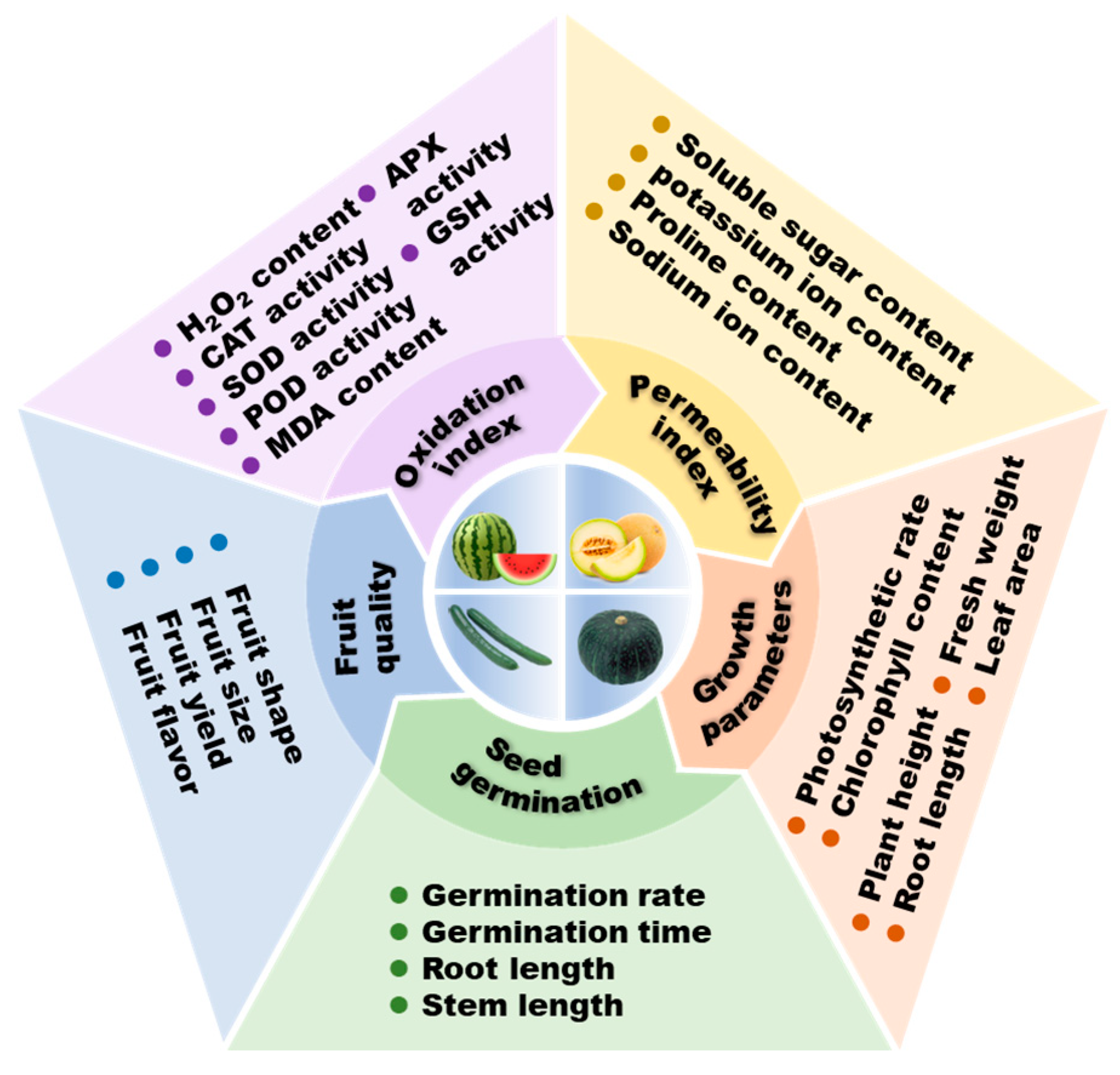
2. Salt Stress Detriment and Response
2.1. Salt Tolerance and Hazards of Different Cucurbitaceae
2.2. Salt Response of Cucurbitaceae from a Physiological Point of View
2.3. Salt Response of Cucurbitaceae from a Biochemical Point of View
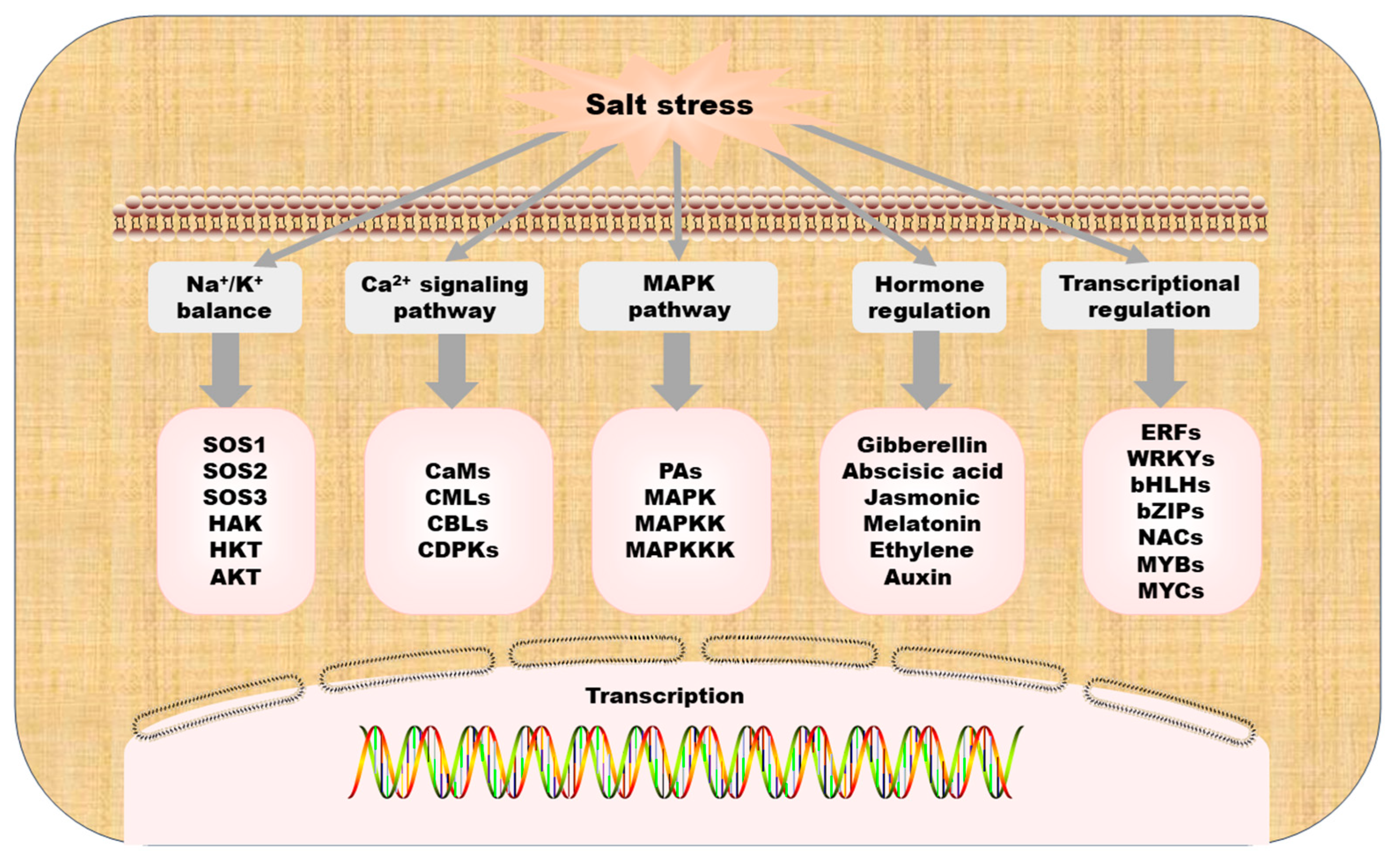
3. Research on Salt Stress at the Molecular Level
3.1. Gene Expression in Response to Stress
3.2. The Salt Overly Sensitive Pathway
3.3. Transcriptional Regulation
3.4. Hormone Regulation
3.5. Signal Transduction
4. Measures to Improve Salt Tolerance of Cucurbitaceae Plants
4.1. Nanomaterials
4.2. Graft
4.3. Polyamines
4.4. Signaling Molecules
4.5. Plant Hormones
| Measure | Species | Example | Principle | References |
|---|---|---|---|---|
| Nanoparticles | Cucumis sativus | CeO2 | Modulates the antioxidant system to improve the salt tolerance of cucumber seedlings | [131] |
| Nanoparticles | Cucumis sativus | Silicon | Silicon increased the water conductivity of the root system and improved the water balance of seedlings; silicon reduces the Na+ content and reduces ionic toxicity | [128,132] |
| Nanoparticles | Cucumis sativus | PNC | Improves K+ absorption capacity and better maintains K+/Na+ ratio | [53] |
| Nanoparticles | Cucumis sativus | Se, SeO2, Mn3O4, etc. | Regulates ion channels and transporter-related genes to maintain ion homeostasis; directly or indirectly promotes reactive oxygen species scavenging mechanisms | [147] |
| Nanoparticles | Momordica charantia | Se-CS NPs | Induces multiple defense systems to alleviate salt stress | [133] |
| Graft | Cucumis sativus | Cucumber grafted onto pumpkin | Enhances Na+ efflux and induces stomatal closure | [135,136] |
| Graft | Cucumis sativus | Cucumber grafted onto luffa | Rootstock reduces Na+ transport to the shoot, facilitating Na+/K+ balance | [3] |
| Graft | Cucumis melo | Melon grafted onto luffa | Reduces the Na+ content of leaves and reduces salt toxicity | [136] |
| Graft | Citrullus lanatus | Watermelon grafted onto bottle gourd | Accumulation of less Na+ and improved ROS scavenging capacity | [135] |
| Exogenous polyamines | Cucumis sativus | Spd | Involved in cellular autophagy, degrades damaged proteins | [21] |
| Exogenous polyamines | Cucumis sativus | Spd | Enhances the expression of SAM for the synthesis of ethylene and PA | [13] |
| Signal molecules | Cucumis sativus | H2O2 | Mediates the antioxidant enzyme system, ABA, and GA | [144] |
| Signal molecules | Cucumis melo | H2O2 | Alleviates membrane lipid peroxidation to a certain extent, activates antioxidant enzyme activity in melon under stress | [148] |
| Signal molecules | Cucumis sativus | H2S | Enhances antioxidant capacity; improves Na+/K+ balance | [149] |
| Signal molecules | Cucumis melo | NO | Improves the growth, root activity, and antioxidant enzyme activity of melon seedlings under salt stress | [130] |
| Exogenous hormones | Cucumis sativus | Melatonin | Promotes the expression of endogenous hormones; improves photosynthesis, ion homeostasis, and activates a series of downstream signals | [145] |
| Exogenous hormones | Momordica charantia | Melatonin | Regulates ionic balance, antioxidant system and secondary metabolism-related genes | [146] |
| Exogenous hormones | Cucumis sativus | EBR | Enhances antioxidant capacity, maintain ionic homeostasis, and activates salt tolerance-related signaling pathways | [65] |
| Exogenous hormones | Luffa cylindrica | MEJA | Improves photosynthetic activity and nitrogen metabolism | [61] |
| Plant growth-promoting rhizobacteria | / | PGPR | Mediates the ethylene pathway and ROS scavenging | [150] |
5. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, F.; Xie, P.; Sun, S.; Qiao, X.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; Yang, D.; et al. A Gγ Protein Regulates Alkaline Sensitivity in Crops. Science 2023, 379, eade8416. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Qin, Y.; Lv, J.; Wang, X.; Dong, H.; Dong, X.; Zhang, T.; Du, N.; Piao, F. Luffa Rootstock Enhances Salt Tolerance and Improves Yield and Quality of Grafted Cucumber Plants by Reducing Sodium Transport to the Shoot. Environ. Pollut. 2023, 316, 120521. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Mu, X.; Shao, H.; Wang, H.; Brestic, M. Global Plant-Responding Mechanisms to Salt Stress: Physiological and Molecular Levels and Implications in Biotechnology. Crit. Rev. Biotechnol. 2015, 35, 425–437. [Google Scholar] [CrossRef]
- Shelake, R.M.; Kadam, U.S.; Kumar, R.; Pramanik, D.; Singh, A.K.; Kim, J.Y. Engineering Drought and Salinity Tolerance Traits in Crops through CRISPR-Mediated Genome Editing: Targets, Tools, Challenges, and Perspectives. Plant Commun. 2022, 3, 100417. [Google Scholar] [CrossRef]
- Zhu, Y.; Yuan, G.; Gao, B.; An, G.; Li, W.; Si, W.; Sun, D.; Liu, J. Comparative Transcriptome Profiling Provides Insights into Plant Salt Tolerance in Watermelon (Citrullus lanatus). Life 2022, 12, 1033. [Google Scholar] [CrossRef]
- Niu, M.; Sun, S.; Nawaz, M.A.; Sun, J.; Cao, H.; Lu, J.; Huang, Y.; Bie, Z. Grafting Cucumber Onto Pumpkin Induced Early Stomatal Closure by Increasing ABA Sensitivity under Salinity Conditions. Front. Plant Sci. 2019, 10, 1290. [Google Scholar] [CrossRef]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y.; et al. Rice OsDOF15 Contributes to Ethylene-Inhibited Primary Root Elongation under Salt Stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Semida, W.M.; Rady, M.M.; Mohamed, G.F.; Hemida, K.A.; Alhammad, B.A.; Hassan, M.M.; Shami, A. Sequential Application of Antioxidants Rectifies Ion Imbalance and Strengthens Antioxidant Systems in Salt-Stressed Cucumber. Plants 2020, 9, 1783. [Google Scholar] [CrossRef]
- Yao, H.Y.; Xue, H.W. Phosphatidic Acid Plays Key Roles Regulating Plant Development and Stress Responses. J. Integr. Plant Biol. 2018, 60, 851–863. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Hu, L.; Zhang, S.; Zhang, M.; Jiang, W.; Wu, T.; Du, X. Rice OsWRKY50 Mediates ABA-Dependent Seed Germination and Seedling Growth, and ABA-Independent Salt Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8625. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gong, X.; Liu, W.; Kong, L.; Si, X.; Guo, S.; Sun, J. Gibberellin Mediates Spermidine-Induced Salt Tolerance and the Expression of GT-3b in Cucumber. Plant Physiol. Biochem. 2020, 152, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-Induced Reactive Oxygen Species Compartmentalization, Perception and Signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-Wide Identification of MAPK, MAPKK, and MAPKKK Gene Families and Transcriptional Profiling Analysis during Development and Stress Response in Cucumber. BMC Genom. 2015, 16, 386. [Google Scholar] [CrossRef]
- Fang, L.; Wei, X.Y.; Liu, L.Z.; Zhou, L.X.; Tian, Y.P.; Geng, C.; Li, X.D. A Tobacco Ringspot Virus-Based Vector System for Gene and microRNA Function Studies in Cucurbits. Plant Physiol. 2021, 186, 853–864. [Google Scholar] [CrossRef]
- Fu, H.; Yu, X.; Jiang, Y.; Wang, Y.; Yang, Y.; Chen, S.; Chen, Q.; Guo, Y. SALT OVERLY SENSITIVE 1 Is Inhibited by Clade D Protein Phosphatase 2C D6 and D7 in Arabidopsis thaliana. Plant Cell 2023, 35, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhou, X.; Song, H.; Zhang, M.; Jiang, C. Advances in Deciphering Salt Tolerance Mechanism in Maize. Crop J. 2023, 11, 1001–1010. [Google Scholar] [CrossRef]
- Behera, T.K.; Krishna, R.; Ansari, W.A.; Aamir, M.; Kumar, P.; Kashyap, S.P.; Pandey, S.; Kole, C. Approaches Involved in the Vegetable Crops Salt Stress Tolerance Improvement: Present Status and Way Ahead. Front. Plant Sci. 2021, 12, 787292. [Google Scholar] [CrossRef]
- Yang, Y.; Ahammed, G.J.; Wan, C.; Liu, H.; Chen, R.; Zhou, Y. Comprehensive Analysis of TIFY Transcription Factors and Their Expression Profiles under Jasmonic Acid and Abiotic Stresses in Watermelon. Int. J. Genom. 2019, 2019, 6813086. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wen, W.; Shi, Z.; Gu, Q.; Ahammed, G.J.; Cao, K.; Shah Jahan, M.; Shu, S.; Wang, J.; et al. Hydrogen Peroxide Mediates Spermidine-Induced Autophagy to Alleviate Salt Stress in Cucumber. Autophagy 2021, 17, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Mishra, J.; Arora, N.K. Plant Growth Promoting Bacteria for Combating Salinity Stress in Plants-Recent Developments and Prospects: A Review. Microbiol. Res. 2021, 252, 126861. [Google Scholar] [CrossRef]
- Yang, L.I.; Kai, L.; Jipeng, W.; Lan, Z.; Xin, L.I.; Wenyan, H.; Qingyun, L.I.; University, H.A. Effects of Various Concentrations of EGCG on Seed Germination and Resistance in Cucumber under NaCl Stress. Acta Agric. Zhejiangensis 2018, 30, 1160–1167. [Google Scholar]
- Li, L.; Du, L.; Cao, Q.; Yang, Z.; Liu, Y.; Yang, H.; Duan, X.; Meng, Z. Salt Tolerance Evaluation of Cucumber Germplasm under Sodium Chloride Stress. Plants 2023, 12, 2927. [Google Scholar] [CrossRef] [PubMed]
- Chevilly, S.; Dolz-Edo, L.; Martínez-Sánchez, G.; Morcillo, L.; Vilagrosa, A.; López-Nicolás, J.M.; Blanca, J.; Yenush, L.; Mulet, J.M. Distinctive Traits for Drought and Salt Stress Tolerance in Melon (Cucumis melo L.). Front. Plant Sci. 2021, 12, 777060. [Google Scholar] [CrossRef]
- Marium, A.; Kausar, A.; Ali Shah, S.M.; Ashraf, M.Y.; Akhtar, N.; Akram, M.; Riaz, M. Assessment of Cucumber Genotypes for Salt Tolerance Based on Germination and Physiological Indices. Dose Response 2019, 17, 1559325819889809. [Google Scholar] [CrossRef]
- Yasar, F.; Kusvuran, S.; Ellialtioglu, S. Determination of Anti-Oxidant Activities in Some Melon (Cucumis melo L.) Varieties and Cultivars under Salt Stress. J. Hortic. Sci. Biotechnol. 2006, 81, 627–630. [Google Scholar] [CrossRef]
- Liu, D.; Dong, S.; Miao, H.; Liu, X.; Li, C.; Han, J.; Zhang, S.; Gu, X. A Large-Scale Genomic Association Analysis Identifies the Candidate Genes Regulating Salt Tolerance in Cucumber (Cucumis sativus L.) Seedlings. Int. J. Mol. Sci. 2022, 23, 8260. [Google Scholar] [CrossRef]
- Liu, D.; Dong, S.; Bo, K.; Miao, H.; Li, C.; Zhang, Y.; Zhang, S.; Gu, X. Identification of QTLs Controlling Salt Tolerance in Cucumber (Cucumis sativus L.) Seedlings. Plants 2021, 10, 85. [Google Scholar] [CrossRef]
- Sarabi, B.; Bolandnazar, S.; Ghaderi, N.; Ghashghaie, J. Genotypic Differences in Physiological and Biochemical Responses to Salinity Stress in Melon (Cucumis melo L.) Plants: Prospects for Selection of Salt Tolerant Landraces. Plant Physiol. Biochem. 2017, 119, 294–311. [Google Scholar] [CrossRef] [PubMed]
- Tarchoun, N.; Saadaoui, W.; Mezghani, N.; Pavli, O.I.; Falleh, H.; Petropoulos, S.A. The Effects of Salt Stress on Germination, Seedling Growth and Biochemical Responses of Tunisian Squash (Cucurbita maxima Duchesne) Germplasm. Plants 2022, 11, 800. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Farid, I.B.; Marghany, M.R.; Rowezek, M.M.; Sheded, M.G. Effect of Salinity Stress on Growth and Metabolomic Profiling of Cucumis sativus and Solanum lycopersicum. Plants 2020, 9, 1626. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Elango, D.; Liu, H.; Jin, D.; Wang, X.; Wu, Y. Metabolome and Transcriptome Analysis Reveals Molecular Mechanisms of Watermelon under Salt Stress. Environ. Exp. Bot. 2023, 206, 105200. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Tian, J.; Li, J.; Sun, J.; He, L.; Guo, S.; Tezuka, T. Proteomic Study Participating the Enhancement of Growth and Salt Tolerance of Bottle Gourd Rootstock-Grafted Watermelon Seedlings. Plant Physiol. Biochem. 2012, 58, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Ondrasek, G.; Romic, D.; Rengel, Z.; Romic, M.; Zovko, M. Cadmium Accumulation by Muskmelon under Salt Stress in Contaminated Organic Soil. Sci. Total Environ. 2009, 407, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Akrami, M.; Arzani, A. Inheritance of Fruit Yield and Quality in Melon (Cucumis melo L.) Grown under Field Salinity Stress. Sci. Rep. 2019, 9, 7249. [Google Scholar] [CrossRef]
- Sanoubar, R.; Orsini, F.; Gianquinto, G. Ionic Partitioning and Stomatal Regulation: Dissecting Functional Elements of the Genotypic Basis of Salt Stress Adaptation in Grafted Melon. Plant Signal. Behav. 2013, 8, e27334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Akrami, M.; Arzani, A.; Majnoun, Z. Leaf Ion Content, Yield and Fruit Quality of Field-Grown Melon under Saline Conditions. Ex. Agric. 2019, 55, 707–722. [Google Scholar] [CrossRef]
- Wei, S.; Wang, L.; Zhang, Y.; Huang, D. Identification of Early Response Genes to Salt Stress in Roots of Melon (Cucumis melo L.) Seedlings. Mol. Biol. Rep. 2013, 40, 2915–2926. [Google Scholar] [CrossRef] [PubMed]
- Sohail, H.; Noor, I.; Nawaz, M.; Ma, M.; Shireen, F.; Huang, Y.; Yang, L.; Bie, Z. Genome-Wide Identification of Plasma-Membrane Intrinsic Proteins in Pumpkin and Functional Characterization of CmoPIP1-4 under Salinity Stress. Environ. Exp. Bot. 2022, 202, 104995. [Google Scholar] [CrossRef]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root Respiratory Burst Oxidase Homologue-Dependent H2O2 Production Confers Salt Tolerance on a Grafted Cucumber by Controlling Na+ Exclusion and Stomatal Closure. J. Exp. Bot. 2018, 69, 3465–3476. [Google Scholar] [CrossRef]
- Rouphael, Y.; Cardarelli, M.; Rea, E.; Colla, G. Improving Melon and Cucumber Photosynthetic Activity, Mineral Composition, and Growth Performance under Salinity Stress by Grafting onto Cucurbita Hybrid Rootstocks. Photosynthetica 2012, 50, 180–188. [Google Scholar] [CrossRef]
- Huang, Y.; Cao, H.; Yang, L.; Chen, C.; Shabala, L.; Xiong, M.; Niu, M.; Liu, J.; Zheng, Z.; Zhou, L.; et al. Tissue-Specific Respiratory Burst Oxidase Homolog-Dependent H2O2 Signaling to the Plasma Membrane H+-ATPase Confers Potassium Uptake and Salinity Tolerance in Cucurbitaceae. J. Exp. Bot. 2019, 70, 5879–5893. [Google Scholar] [CrossRef]
- Fu, A.; Zheng, Y.; Guo, J.; Grierson, D.; Zhao, X.; Wen, C.; Liu, Y.; Li, J.; Zhang, X.; Yu, Y.; et al. Telomere-to-Telomere Genome Assembly of Bitter Melon (Momordica charantia L. Var. Abbreviata Ser.) Reveals Fruit Development, Composition and Ripening Genetic Characteristics. Hortic. Res. 2022, 10, uhac228. [Google Scholar] [CrossRef]
- Cebrián, G.; Iglesias-Moya, J.; García, A.; Martínez, J.; Romero, J.; Regalado, J.J.; Martínez, C.; Valenzuela, J.L.; Jamilena, M. Involvement of Ethylene Receptors in the Salt Tolerance Response of Cucurbita pepo. Hortic. Res. 2021, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709.e5. [Google Scholar] [CrossRef]
- Steinhorst, L.; He, G.; Moore, L.K.; Schültke, S.; Schmitz-Thom, I.; Cao, Y.; Hashimoto, K.; Andrés, Z.; Piepenburg, K.; Ragel, P.; et al. A Ca2+-Sensor Switch for Tolerance to Elevated Salt Stress in Arabidopsis. Dev. Cell 2022, 57, 2081–2094.e7. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; He, G.; Li, J.; Perez-Hormaeche, J.; Becker, T.; Luo, M.; Wallrad, L.; Gao, J.; Li, J.; Pardo, J.M.; et al. A Salt Stress-Activated GSO1-SOS2-SOS1 Module Protects the Arabidopsis Root Stem Cell Niche by Enhancing Sodium Ion Extrusion. EMBO J. 2023, 42, e113004. [Google Scholar] [CrossRef]
- Flores-León, A.; García-Martínez, S.; González, V.; Garcés-Claver, A.; Martí, R.; Julián, C.; Sifres, A.; Pérez-de-Castro, A.; Díez, M.J.; López, C.; et al. Grafting Snake Melon [Cucumis melo L. Subsp. Melo Var. Flexuosus (L.) Naudin] in Organic Farming: Effects on Agronomic Performance; Resistance to Pathogens; Sugar, Acid, and VOC Profiles; and Consumer Acceptance. Front. Plant Sci. 2021, 12, 613845. [Google Scholar] [CrossRef] [PubMed]
- Akrami, M.; Arzani, A. Physiological Alterations Due to Field Salinity Stress in Melon (Cucumis melo L.). Acta Physiol. Plant. 2018, 40, 91. [Google Scholar] [CrossRef]
- Qian, Z.J.; Song, J.J.; Chaumont, F.; Ye, Q. Differential Responses of Plasma Membrane Aquaporins in Mediating Water Transport of Cucumber Seedlings under Osmotic and Salt Stresses. Plant Cell Environ. 2015, 38, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.L.; Tian, Y.; Li, L.; Yu, M.; Hou, R.-P.; Ren, X.M. H2S Alleviates Salinity Stress in Cucumber by Maintaining the Na+/K+ Balance and Regulating H2S Metabolism and Oxidative Stress Response. Front. Plant Sci. 2019, 10, 678. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, L.; Zhu, L.; Cui, L.; Yang, L.; Wu, H.; Bie, Z. CsAKT1 Is a Key Gene for the CeO2 Nanoparticle’s Improved Cucumber Salt Tolerance: A Validation from CRISPR-Cas9 Lines. Environ. Sci. Nano 2022, 9, 4367–4381. [Google Scholar] [CrossRef]
- Peng, Y.; Cao, H.; Peng, Z.; Zhou, L.; Sohail, H.; Cui, L.; Yang, L.; Huang, Y.; Bie, Z. Transcriptomic and Functional Characterization Reveals CsHAK5;3 as a Key Player in K+ Homeostasis in Grafted Cucumbers under Saline Conditions. Plant Sci. 2023, 326, 111509. [Google Scholar] [CrossRef]
- Gao, L.W.; Yang, S.L.; Wei, S.W.; Huang, D.F.; Zhang, Y.D. Supportive Role of the Na+ Transporter CmHKT1;1 from Cucumis melo in Transgenic Arabidopsis Salt Tolerance through Improved K+/Na+ Balance. Plant Mol. Biol. 2020, 103, 561–580. [Google Scholar] [CrossRef]
- Wei, L.; Liu, L.; Chen, Z.; Huang, Y.; Yang, L.; Wang, P.; Xue, S.; Bie, Z. CmCNIH1 Improves Salt Tolerance by Influencing the Trafficking of CmHKT1;1 in Pumpkin. Plant J. 2023, 114, 1353–1368. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Véry, A.A.; Wang, L.M.; Deng, Y.W.; Sentenac, H.; Huang, D.F. A K+ Channel from Salt-Tolerant Melon Inhibited by Na+. New Phytol. 2011, 189, 856–868. [Google Scholar] [CrossRef]
- Yu, J.; Yu, J.; Liao, W.; Xie, J.; Wu, Y. Ethylene Was Involved in Ca2+-Regulated Na+ Homeostasis, Na+ Transport and Cell Ultrastructure During Adventitious Rooting in Cucumber Explants under Salt Stress. J. Plant Biol. 2020, 63, 311–320. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, L.; Feng, L.; Zhang, M.; Hu, D.; Tie, J.; Liao, W. Hydrogen Sulfide Promotes Adventitious Root Development in Cucumber under Salt Stress by Enhancing Antioxidant Ability. Plants 2022, 11, 935. [Google Scholar] [CrossRef]
- Zhang, G.; Ding, Q.; Wei, B. Genome-Wide Identification of Superoxide Dismutase Gene Families and Their Expression Patterns under Low-Temperature, Salt and Osmotic Stresses in Watermelon and Melon. 3 Biotech 2021, 11, 194. [Google Scholar] [CrossRef]
- Parihar, P.; Singh, R.; Singh, A.; Prasad, S.M. Role of Oxylipin on Luffa Seedlings Exposed to NaCl and UV-B Stresses: An Insight into Mechanism. Plant Physiol. Biochem. 2021, 167, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, J.; Chen, H.; Wang, Z.; Gu, X.; Wei, C.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Exogenous Melatonin Confers Salt Stress Tolerance to Watermelon by Improving Photosynthesis and Redox Homeostasis. Front. Plant Sci. 2017, 8, 295. [Google Scholar] [CrossRef]
- Shu, S.; Guo, S.-R.; Sun, J.; Yuan, L.Y. Effects of Salt Stress on the Structure and Function of the Photosynthetic Apparatus in Cucumis Sativus and Its Protection by Exogenous Putrescine. Physiol. Plant 2012, 146, 285–296. [Google Scholar] [CrossRef]
- Gurmani, A.R.; Khan, S.U.; Ali, A.; Rubab, T.; Schwinghamer, T.; Jilani, G.; Farid, A.; Zhang, J. Salicylic Acid and Kinetin Mediated Stimulation of Salt Tolerance in Cucumber (Cucumis sativus L.) Genotypes Varying in Salinity Tolerance. Hortic. Environ. Biotechnol. 2018, 59, 461–471. [Google Scholar] [CrossRef]
- He, X.; Wan, Z.; Jin, N.; Jin, L.; Zhang, G.; Lyu, J.; Liu, Z.; Luo, S.; Yu, J. Enhancement of Cucumber Resistance under Salt Stress by 2, 4-Epibrassinolide Lactones. Front. Plant Sci. 2022, 13, 1023178. [Google Scholar] [CrossRef]
- Chen, J.B.; Zhang, F.R.; Huang, D.F.; Zhang, L.D.; Zhang, Y.D. Transcriptome Analysis of Transcription Factors in Two Melon (Cucumis melo L.) Cultivars under Salt Stress. Plant Physiol. J. 2014, 50, 150–158. [Google Scholar]
- Yan, Y.; Sun, M.; Li, Y.; Wang, J.; He, C.; Yu, X. The CsGPA1-CsAQPs Module Is Essential for Salt Tolerance of Cucumber Seedlings. Plant Cell Rep. 2020, 39, 1301–1316. [Google Scholar] [CrossRef]
- Li, J.; Yang, C.; Xu, J.; Lu, H.; Liu, J. The Hot Science in Rice Research: How Rice Plants Cope with Heat Stress. Plant Cell Environ. 2023, 46, 1087–1103. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Liu, Y.; Lu, L.; Zhang, J.; Chen, S.; Wang, B. Comparison of Tolerant and Susceptible Cultivars Revealed the Roles of Circular RNAs in Rice Responding to Salt Stress. Plant Growth Regul. 2022, 96, 243–254. [Google Scholar] [CrossRef]
- Wang, Z.; Li, N.; Yu, Q.; Wang, H. Genome-Wide Characterization of Salt-Responsive miRNAs, circRNAs and Associated ceRNA Networks in Tomatoes. Int. J. Mol. Sci. 2021, 22, 12238. [Google Scholar] [CrossRef]
- Liu, P.; Zhu, Y.; Liu, H.; Liang, Z.; Zhang, M.; Zou, C.; Yuan, G.; Gao, S.; Pan, G.; Shen, Y.; et al. A Combination of a Genome-Wide Association Study and a Transcriptome Analysis Reveals circRNAs as New Regulators Involved in the Response to Salt Stress in Maize. Int. J. Mol. Sci. 2022, 23, 9755. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Jia, J.H.; Yang, L.; Xia, Y.C.; Zhang, H.L.; Jia, J.B.; Zhou, R.; Nie, P.Y.; Yin, J.L.; Ma, D.F.; et al. Identification of Cucumber Circular RNAs Responsive to Salt Stress. BMC Plant Biol. 2019, 19, 164. [Google Scholar] [CrossRef]
- Bin, L.I.; Guo, S.R.; Sun, J.; Juan, L.I. Effects of Exogenous Spermidine on Free Polyamine Content and Polyamine Biosynthesis Gene Expression in Cucumber Seedlings under Salt Stress. Chin. J. Eco-Agric. 2011, 19, 93–97. [Google Scholar] [CrossRef]
- Reda, M.; Golicka, A.; Kabała, K.; Janicka, M. Involvement of NR and PM-NR in NO Biosynthesis in Cucumber Plants Subjected to Salt Stress. Plant Sci. 2018, 267, 55–64. [Google Scholar] [CrossRef]
- Yan, S.; Che, G.; Ding, L.; Chen, Z.; Liu, X.; Wang, H.; Zhao, W.; Ning, K.; Zhao, J.; Tesfamichael, K.; et al. Different Cucumber CsYUC Genes Regulate Response to Abiotic Stresses and Flower Development. Sci. Rep. 2016, 6, 20760. [Google Scholar] [CrossRef]
- Zijian, Y.; Yong, Z.; Lingli, G.; Guanghua, L.; Qiang, L.; Yaping, X.; Lunwei, J.; Yingui, Y.; Agriculture, S.O.; University, J.A. Expression of Cucumber CsCAT3 Gene under Stress and Its Salt Tolerance in Transgenic Arabidopsis Thaliana. Mol. Plant Breed. 2018, 16, 807–812. [Google Scholar]
- Shu, S.; Tang, Y.; Zhou, X.; Jahan, M.S.; Sun, J.; Wang, Y.; Guo, S. Physiological Mechanism of Transglutaminase-Mediated Improvement in Salt Tolerance of Cucumber Seedlings. Plant Physiol. Biochem. 2020, 156, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Fan, H.; Lu, H.; Li, Y.; Guo, Y.; Liu, C.; Chai, L.; Du, C. Cucumis sativus PHLOEM PROTEIN 2-A1 like Gene Positively Regulates Salt Stress Tolerance in Cucumber Seedlings. Plant Mol. Biol. 2023, 111, 493–504. [Google Scholar] [CrossRef]
- Hou, K.; Wang, Y.; Tao, M.Q.; Jahan, M.S.; Shu, S.; Sun, J.; Guo, S.R. Characterization of the CsPNG1 Gene from Cucumber and Its Function in Response to Salinity Stress. Plant Physiol. Biochem. 2020, 150, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Jang, H.A.; Lee, S.S.; Cho, H.S.; Lee, D.H.; Choi, D.; Kwon, S.Y. Cucumber Pti1-L Is a Cytoplasmic Protein Kinase Involved in Defense Responses and Salt Tolerance. J. Plant Physiol. 2014, 171, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, T.; Han, J.; Ren, Z. Genome-Wide Identification and Characterization of Cucumber bHLH Family Genes and the Functional Characterization of CsbHLH041 in NaCl and ABA Tolerance in Arabidopsis and Cucumber. BMC Plant Biol. 2020, 20, 272. [Google Scholar] [CrossRef]
- Zhang, X.M.; Yu, H.J.; Sun, C.; Deng, J.; Zhang, X.; Liu, P.; Li, Y.Y.; Li, Q.; Jiang, W.J. Genome-Wide Characterization and Expression Profiling of the NAC Genes under Abiotic Stresses in Cucumis sativus. Plant Physiol. Biochem. 2017, 113, 98–109. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-Wide Analysis of the WRKY Gene Family in the Cucumber Genome and Transcriptome-Wide Identification of WRKY Transcription Factors That Respond to Biotic and Abiotic Stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef]
- Zhu, H.; He, M.; Jahan, M.; Wu, J.; Gu, Q.; Shu, S.; Sun, J.; Guo, S. CsCDPK6, a CsSAMS1-Interacting Protein, Affects Polyamine/Ethylene Biosynthesis in Cucumber and Enhances Salt Tolerance by Overexpression in Tobacco. Int. J. Mol. Sci. 2021, 22, 11133. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, M.; Liu, W.; Jahan, M.S.; Gu, Q.; Shu, S.; Sun, J.; Guo, S. CsPAO2 Improves Salt Tolerance of Cucumber through the Interaction with CsPSA3 by Affecting Photosynthesis and Polyamine Conversion. Int. J. Mol. Sci. 2022, 23, 12413. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, X.; Gao, R.; Li, Y.; Ding, Z.; Huang, Y.; Yao, K.; Yao, Y.; Liang, C.; Liao, W. CsSHMT3 Gene Enhances the Growth and Development in Cucumber Seedlings under Salt Stress. Plant Mol. Biol. 2024, 114, 52. [Google Scholar] [CrossRef]
- Kabała, K.; Reda, M.; Wdowikowska, A.; Janicka, M. Role of Plasma Membrane NADPH Oxidase in Response to Salt Stress in Cucumber Seedlings. Antioxidants 2022, 11, 1534. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wang, L.; Zhao, J.; Li, Y.; Huang, R.; Jiang, X.; Zhou, X.; Zhu, X.; He, Y.; He, Y.; et al. Genome-Wide Characterization of the C2H2 Zinc-Finger Genes in Cucumis sativus and Functional Analyses of Four CsZFPs in Response to Stresses. BMC Plant Biol. 2020, 20, 359. [Google Scholar] [CrossRef]
- Zhang, R.; Dong, Y.; Li, Y.; Ren, G.; Chen, C.; Jin, X. SLs Signal Transduction Gene CsMAX2 of Cucumber Positively Regulated to Salt, Drought and ABA Stress in Arabidopsis Thaliana L. Gene 2023, 864, 147282. [Google Scholar] [CrossRef]
- Li, S.; Huang, M.; Di, Q.; Ji, T.; Wang, X.; Wei, M.; Shi, Q.; Li, Y.; Gong, B.; Yang, F. The Functions of a Cucumber Phospholipase D Alpha Gene (CsPLDα) in Growth and Tolerance to Hyperosmotic Stress. Plant Physiol. Biochem. 2015, 97, 175–186. [Google Scholar] [CrossRef]
- Li, J.; Song, C.; Li, H.; Wang, S.; Hu, L.; Yin, Y.; Wang, Z.; He, W. Comprehensive Analysis of Cucumber RAV Family Genes and Functional Characterization of CsRAV1 in Salt and ABA Tolerance in Cucumber. Front. Plant Sci. 2023, 14, 1115874. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Wu, J.; Wang, J.; Wang, Y.; Guo, S.; Shu, S. Identification of Cucumber S-Adenosylmethionine Decarboxylase Genes and Functional Analysis of CsSAMDC3 in Salt Tolerance. Front. Plant Sci. 2023, 14, 1076153. [Google Scholar] [CrossRef]
- Li, S.; Sun, M.; Miao, L.; Di, Q.; Lv, L.; Yu, X.; Yan, Y.; He, C.; Wang, J.; Shi, A.; et al. Multifaceted Regulatory Functions of CsBPC2 in Cucumber under Salt Stress Conditions. Hortic. Res. 2023, 10, uhad051. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, Z.; Wang, F.; Lv, H.; Cao, M.; Zhang, N.; Li, F.; Wang, H.; Li, X.; Yuan, X.; et al. A Tubby-like Protein CsTLP8 Acts in the ABA Signaling Pathway and Negatively Regulates Osmotic Stresses Tolerance during Seed Germination. BMC Plant Biol. 2021, 21, 340. [Google Scholar] [CrossRef]
- Rus, A.; Lee, B.; Muñoz-Mayor, A.; Sharkhuu, A.; Miura, K.; Zhu, J.K.; Bressan, R.A.; Hasegawa, P.M. AtHKT1 Facilitates Na+ Homeostasis and K+ Nutrition in Planta. Plant Physiol. 2004, 136, 2500–2511. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shen, L.; Han, X.; He, G.; Fan, W.; Li, Y.; Yang, S.; Zhang, Z.; Yang, Y.; Jin, W.; et al. Phosphatidic Acid–Regulated SOS2 Controls Sodium and Potassium Homeostasis in Arabidopsis under Salt Stress. EMBO J. 2023, 42, e112401. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.K.; Song, R.F.; Guo, J.X.; Zhang, Y.; Zuo, J.X.; Chen, H.H.; Liao, C.Y.; Hu, X.Y.; Ren, F.; Lu, Y.T.; et al. CycC1;1–WRKY75 Complex-Mediated Transcriptional Regulation of SOS1 Controls Salt Stress Tolerance in Arabidopsis. Plant Cell 2023, 35, 2570–2591. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.; Zhou, W.; Li, J.; Luo, M.; Scheres, B.; Guo, Y. On Salt Stress, PLETHORA Signaling Maintains Root Meristems. Dev. Cell 2023, 58, 1657–1669.e5. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a Putative Calcium Sensor, Interacts with the Protein Kinase SOS2 to Protect Arabidopsis Shoots from Salt Stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Xia, Y.; Xie, Q.; Cao, Y.; Wang, Z.; Hao, G.; Song, J.; Zhou, Y.; Jiang, X. The Protein Kinase Complex CBL10-CIPK8-SOS1 Functions in Arabidopsis to Regulate Salt Tolerance. J. Exp. Bot. 2020, 71, 1801–1814. [Google Scholar] [CrossRef]
- Li, B.; Wang, D.; Han, L.; Zhang, Y.; Zhang, L.; Liu, D. Effect of Polyamines on SOS2 Family Gene Expression in Cucumber under Salt Stress. Acta Bot. Boreal. Occident. Sin. 2020, 40, 1855–1865. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic Acid Dynamics, Signaling, and Functions in Plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Yan, X.; Yue, Z.; Pan, X.; Si, F.; Li, J.; Chen, X.; Li, X.; Luan, F.; Yang, J.; Zhang, X.; et al. The HD-ZIP Gene Family in Watermelon: Genome-Wide Identification and Expression Analysis under Abiotic Stresses. Genes 2022, 13, 2242. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, Y.; Wan, C.; Li, J.; Yang, Y.; Chen, J. Genome-Wide Characterization and Expression Analysis of the Dof Gene Family Related to Abiotic Stress in Watermelon. PeerJ 2020, 8, e8358. [Google Scholar] [CrossRef]
- Song, Q.; Joshi, M.; Joshi, V. Transcriptomic Analysis of Short-Term Salt Stress Response in Watermelon Seedlings. Int. J. Mol. Sci. 2020, 21, 6036. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Wang, Y.; Wang, J.; Li, Y.; Zhang, Y.; Zhao, X.; Xu, J.; Li, Z.; Zhao, T.; Wang, W.; et al. The bHLH Transcription Factor Gene, OsbHLH38, Plays a Key Role in Controlling Rice Salt Tolerance. J. Integr. Plant Biol. 2023, 65, 1859–1873. [Google Scholar] [CrossRef]
- Peng, Y.; Cao, H.; Cui, L.; Wang, Y.; Wei, L.; Geng, S.; Yang, L.; Huang, Y.; Bie, Z. CmoNAC1 in Pumpkin Rootstocks Improves Salt Tolerance of Grafted Cucumbers by Binding to the Promoters of CmoRBOHD1, CmoNCED6, CmoAKT1;2 and CmoHKT1;1 to Regulate H2O2, ABA Signaling and K+/Na+ Homeostasis. Hortic. Res. 2023, 10, uhad157. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Wei, S.; Gao, L.; Zhang, Y.; Zhang, F.; Yang, X.; Huang, D. Genome-Wide Investigation of the NAC Transcription Factor Family in Melon (Cucumis melo L.) and Their Expression Analysis under Salt Stress. Plant Cell Rep. 2016, 35, 1827–1839. [Google Scholar] [CrossRef]
- Boonyaves, K.; Ang, M.C.-Y.; Park, M.; Cui, J.; Khong, D.T.; Singh, G.P.; Koman, V.B.; Gong, X.; Porter, T.K.; Choi, S.W.; et al. Near-Infrared Fluorescent Carbon Nanotube Sensors for the Plant Hormone Family Gibberellins. Nano Lett. 2023, 23, 916–924. [Google Scholar] [CrossRef]
- Ribba, T.; Garrido-Vargas, F.; O’Brien, J.A. Auxin-Mediated Responses under Salt Stress: From Developmental Regulation to Biotechnological Applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The Antagonistic Action of Abscisic Acid and Cytokinin Signaling Mediates Drought Stress Response in Arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef]
- Habibpourmehraban, F.; Wu, Y.; Masoomi-Aladizgeh, F.; Amirkhani, A.; Atwell, B.J.; Haynes, P.A. Pre-Treatment of Rice Plants with ABA Makes Them More Tolerant to Multiple Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 9628. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, H.J.; Zhao, B.; Sun, Q.Q.; Cao, Y.Y.; Li, R.; Wu, X.X.; Weeda, S.; Li, L.; Ren, S.; et al. The RNA-Seq Approach to Discriminate Gene Expression Profiles in Response to Melatonin on Cucumber Lateral Root Formation. J. Pineal Res. 2014, 56, 39–50. [Google Scholar] [CrossRef]
- Chang, J.; Guo, Y.; Yan, J.; Zhang, Z.; Yuan, L.; Wei, C.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X.; et al. The Role of Watermelon Caffeic Acid O-Methyltransferase (ClCOMT1) in Melatonin Biosynthesis and Abiotic Stress Tolerance. Hortic. Res. 2021, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Nie, X.; Zhang, T.; Li, S.; Wang, X.; Du, X.; Tong, W.; Song, W. Melatonin: A Small Molecule but Important for Salt Stress Tolerance in Plants. Int. J. Mol. Sci. 2019, 20, 709. [Google Scholar] [CrossRef]
- Monihan, S.M.; Magness, C.A.; Yadegari, R.; Smith, S.E.; Schumaker, K.S. Arabidopsis CALCINEURIN B-LIKE10 Functions Independently of the SOS Pathway during Reproductive Development in Saline Conditions. Plant Physiol. 2016, 171, 369–379. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, C.; Tang, R.J.; Xu, H.X.; Lan, W.Z.; Zhao, F.; Luan, S. Calcineurin B-Like Proteins CBL4 and CBL10 Mediate Two Independent Salt Tolerance Pathways in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2421. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Cell Signaling under Salt, Water and Cold Stresses. Curr. Opin. Plant Biol. 2001, 4, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xiong, X.; Arif, S.; Gao, L.; Zhao, L.; Shah, I.H.; Zhang, Y. A Calmodulin-like CmCML13 from Cucumis melo Improved Transgenic Arabidopsis Salt Tolerance through Reduced Shoot’s Na+, and Also Improved Drought Resistance. Plant Physiol. Biochem. 2020, 155, 271–283. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, C.; Kang, X.; Zhang, L.; Wang, J.; Zheng, S.; Zhang, T. Hydrogen Sulfide and Nitric Oxide Are Involved in Melatonin-Induced Salt Tolerance in Cucumber. Plant Physiol. Biochem. 2021, 167, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhuang, B.; Wu, Q.; Zhang, H.; Nie, J.; Jing, W.; Yang, L.; Zhang, W. Phosphatidic Acid Promotes the Activation and Plasma Membrane Localization of MKK7 and MKK9 in Response to Salt Stress. Plant Sci. 2019, 287, 110190. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Jing, W.; Zhang, W. The Mitogen-Activated Protein Kinase Cascade MKK1–MPK4 Mediates Salt Signaling in Rice. Plant Sci. 2014, 227, 181–189. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, C.; Wang, H.; Li, L.; Wang, C. The Function of MAPK Cascades in Response to Various Stresses in Horticultural Plants. Front. Plant Sci. 2020, 11, 952. [Google Scholar] [CrossRef]
- Song, Q.; Li, D.; Dai, Y.; Liu, S.; Huang, L.; Hong, Y.; Zhang, H.; Song, F. Characterization, Expression Patterns and Functional Analysis of the MAPK and MAPKK Genes in Watermelon (Citrullus lanatus). BMC Plant Biol. 2015, 15, 298. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Xing, Q.; Yue, L.; Qi, H. Genome-Wide Identification of Mitogen-Activated Protein Kinase (MAPK) Cascade and Expression Profiling of CmMAPKs in Melon (Cucumis melo L.). PLoS ONE 2020, 15, e0232756. [Google Scholar] [CrossRef]
- Mastrochirico, M.; Spanò, R.; Mascia, T. Grafting to Manage Infections of the Emerging Tomato Leaf Curl New Delhi Virus in Cucurbits. Plants 2022, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Jia, J.; Lian, Z.; Hu, Y.; Guo, J.; Huo, H.; Zhu, Y.; Gong, H. Silicon Enhances the Salt Tolerance of Cucumber through Increasing Polyamine Accumulation and Decreasing Oxidative Damage. Ecotoxicol. Environ. Saf. 2019, 169, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Yuan, L.Y.; Guo, S.R.; Sun, J.; Yuan, Y.H. Effects of Exogenous Spermine on Chlorophyll Fluorescence, Antioxidant System and Ultrastructure of Chloroplasts in Cucumis sativus L. under Salt Stress. Plant Physiol. Biochem. 2013, 63, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Ding, F.; Wang, X.; Wei, M. Exogenous Nitric Oxide Protect Cucumber Roots against Oxidative Stress Induced by Salt Stress. Plant Physiol. Biochem. 2007, 45, 542–550. [Google Scholar] [CrossRef]
- Chen, L.; Peng, Y.; Zhu, L.; Huang, Y.; Bie, Z.; Wu, H. CeO2 Nanoparticles Improved Cucumber Salt Tolerance Is Associated with Its Induced Early Stimulation on Antioxidant System. Chemosphere 2022, 299, 134474. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, P.; Chen, D.; Yin, L.; Li, H.; Deng, X. Silicon Enhanced Salt Tolerance by Improving the Root Water Uptake and Decreasing the Ion Toxicity in Cucumber. Front. Plant Sci. 2015, 6, 759. [Google Scholar] [CrossRef]
- Sheikhalipour, M.; Mohammadi, S.A.; Esmaielpour, B.; Spanos, A.; Mahmoudi, R.; Mahdavinia, G.R.; Milani, M.H.; Kahnamoei, A.; Nouraein, M.; Antoniou, C.; et al. Seedling Nanopriming with Selenium-Chitosan Nanoparticles Mitigates the Adverse Effects of Salt Stress by Inducing Multiple Defence Pathways in Bitter Melon Plants. Int. J. Biol. Macromol. 2023, 242, 124923. [Google Scholar] [CrossRef]
- Lang, X.; Zhao, X.; Zhao, J.; Ren, T.; Nie, L.; Zhao, W. MicroRNA Profiling Revealed the Mechanism of Enhanced Cold Resistance by Grafting in Melon (Cucumis melo L.). Plants 2024, 13, 1016. [Google Scholar] [CrossRef] [PubMed]
- Bőhm, V.; Fekete, D.; Balázs, G.; Gáspár, L.; Kappel, N. Salinity Tolerance of Grafted Watermelon Seedlings. Acta Biol. Hung. 2017, 68, 412–427. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, S.; Li, H.; Sun, H.; Lu, N.; Shu, S.; Sun, J. Resistance of Cucumber Grafting Rootstock Pumpkin Cultivars to Chilling and Salinity Stresses. Hortic. Sci. Technol. 2017, 35, 220–231. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, J.; Wen, W.; Sun, S.; Shu, S.; Guo, S. Transcriptome and Proteome Analysis Identifies Salt Stress Response Genes in Bottle Gourd Rootstock-Grafted Watermelon Seedlings. Agronomy 2023, 13, 618. [Google Scholar] [CrossRef]
- Li, B.; He, L.; Guo, S.; Li, J.; Yang, Y.; Yan, B.; Sun, J.; Li, J. Proteomics Reveal Cucumber Spd-Responses under Normal Condition and Salt Stress. Plant Physiol. Biochem. 2013, 67, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shu, S.; Li, C.; Sun, J.; Guo, S. Spermidine-Mediated Hydrogen Peroxide Signaling Enhances the Antioxidant Capacity of Salt-Stressed Cucumber Roots. Plant Physiol. Biochem. 2018, 128, 152–162. [Google Scholar] [CrossRef]
- Hongna, C.; Junmei, S.; Leyuan, T.; Xiaori, H.; Guolin, L.; Xianguo, C. Exogenous Spermidine Priming Mitigates the Osmotic Damage in Germinating Seeds of Leymus chinensis under Salt-Alkali Stress. Front. Plant Sci. 2021, 12, 701538. [Google Scholar] [CrossRef]
- He, M.W.; Wang, Y.; Wu, J.Q.; Shu, S.; Sun, J.; Guo, S.R. Isolation and Characterization of S-Adenosylmethionine Synthase Gene from Cucumber and Responsive to Abiotic Stress. Plant Physiol. Biochem. 2019, 141, 431–445. [Google Scholar] [CrossRef]
- Li, H.; Liu, S.S.; Yi, C.Y.; Wang, F.; Zhou, J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Hydrogen Peroxide Mediates Abscisic Acid-Induced HSP70 Accumulation and Heat Tolerance in Grafted Cucumber Plants. Plant Cell Environ. 2014, 37, 2768–2780. [Google Scholar] [CrossRef]
- Luo, S.; Liu, Z.; Wan, Z.; He, X.; Lv, J.; Yu, J.; Zhang, G. Foliar Spraying of NaHS Alleviates Cucumber Salt Stress by Maintaining Na+/K+ Balance and Activating Salt Tolerance Signaling Pathways. Plants 2023, 12, 2450. [Google Scholar] [CrossRef]
- Santos, A.S.; Almeida, J.F.; Silva, M.S.D.; Nóbrega, J.S.; Queiroga, T.B.D.; Pereira, J.A.R.; Linné, J.A.; Gomes, F.A.L. The Influence of H2O2 Application Methods on Melon Plants Submitted to Saline Stress. J. Agric. Sci. 2019, 11, 245. [Google Scholar] [CrossRef]
- Zhang, H.J.; Zhang, N.; Yang, R.C.; Wang, L.; Sun, Q.Q.; Li, D.B.; Cao, Y.Y.; Weeda, S.; Zhao, B.; Ren, S. Melatonin Promotes Seed Germination under High Salinity by Regulating Antioxidant Systems, ABA and GA(4) Interaction in Cucumber (Cucumis sativus L.). J. Pineal Res. 2014, 57, 269–279. [Google Scholar]
- Sheikhalipour, M.; Mohammadi, S.A.; Esmaielpour, B.; Zareei, E.; Kulak, M.; Ali, S.; Nouraein, M.; Bahrami, M.K.; Gohari, G.; Fotopoulos, V. Exogenous Melatonin Increases Salt Tolerance in Bitter Melon by Regulating Ionic Balance, Antioxidant System and Secondary Metabolism-Related Genes. BMC Plant Biol. 2022, 22, 380. [Google Scholar] [CrossRef]
- Amerian, M.; Palangi, A.; Gohari, G.; Ntatsi, G. Enhancing Salinity Tolerance in Cucumber through Selenium Biofortification and Grafting. BMC Plant Biol. 2024, 24, 24. [Google Scholar] [CrossRef]
- Zhu, L.J.; Yan, Q.J.; Chen, G.S.; Hu, J.; Luo, M.; Yang, Y. Exogenous H2O2 Promotes Seed Germination under High Salinity by Regulating Antioxidant Enzymes, ABA and GA Interaction in Cucumber (Cucumis sativus). Plant Physiol. J. 2019, 55, 342–348. [Google Scholar]
- Singh, V.P.; Srivastava, P.K.; Prasad, S.M. Nitric Oxide Alleviates Arsenic-Induced Toxic Effects in Ridged Luffa Seedlings. Plant Physiol. Biochem. 2013, 71, 155–163. [Google Scholar] [CrossRef]
- Feng, L.; Li, Q.; Zhou, D.; Jia, M.; Liu, Z.; Hou, Z.; Ren, Q.; Ji, S.; Sang, S.; Lu, S.; et al. B. subtilis CNBG-PGPR-1 Induces Methionine to Regulate Ethylene Pathway and ROS Scavenging for Improving Salt Tolerance of Tomato. Plant J. 2024, 117, 193–211. [Google Scholar] [CrossRef]
- Liang, L.; Tang, W.; Lian, H.; Sun, B.; Huang, Z.; Sun, G.; Li, X.; Tu, L.; Li, H.; Tang, Y. Grafting Promoted Antioxidant Capacity and Carbon and Nitrogen Metabolism of Bitter Gourd Seedlings under Heat Stress. Front. Plant Sci. 2022, 13, 1074889. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, T.; Ni, L.; Chen, C.; Jiang, J.; Cui, Z.; Wang, S.; Xu, F.; Yan, R.; Jiang, M. Phosphorylation of OsRbohB by the CaM-Dependent Protein Kinase OsDMI3 Promotes H2O2 Production to Potentiate ABA Responses in Rice. Mol. Plant 2023, 16, S1674205223001016. [Google Scholar] [CrossRef]
- Qin, R.; Hu, Y.; Chen, H.; Du, Q.; Yang, J.; Li, W.X. MicroRNA408 Negatively Regulates Salt Tolerance by Affecting Secondary Cell Wall Development in Maize. Plant Physiol. 2023, 192, kiad135. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, S.; Zhang, Y.; Tan, J.; Li, X.; Chu, X.; Xu, B.; Tian, Y.; Sun, Y.; Li, B.; et al. A Telomere-to-Telomere Gap-Free Reference Genome of Watermelon and Its Mutation Library Provide Important Resources for Gene Discovery and Breeding. Mol. Plant 2022, 15, 1268–1284. [Google Scholar] [CrossRef]
- Wei, M.; Huang, Y.; Mo, C.; Wang, H.; Zeng, Q.; Yang, W.; Chen, J.; Zhang, X.; Kong, Q. Telomere-to-Telomere Genome Assembly of Melon (Cucumis melo L. Var. Inodorus) Provides a High-Quality Reference for Meta-QTL Analysis of Important Traits. Hortic. Res. 2023, 10, uhad189. [Google Scholar] [CrossRef]

| Gene Name | Specific Mechanism | Key Regulatory Classification | References |
|---|---|---|---|
| CsAKT1 | Involved in K+ uptake under salt stress | Ionic homeostasis | [53] |
| CsHAK5;3 | Involved in K+ uptake under salt stress | Ionic homeostasis | [54] |
| CsSOS1/2/3 | Facilitates Na+ extrusion, regulates intracellular Na+/H+ homeostasis | Ionic homeostasis | [73] |
| CsAQPs | Involved in water molecule transport | Ionic homeostasis | [67] |
| CsABF | ABA-responsive element, involved in salt stress | Hormone regulation | [7] |
| CsERF | Ethylene response element, regulates salt stress through ethylene | Hormone regulation | [74] |
| CsYUC | Upregulated to elevate the auxin level | Hormone regulation | [75] |
| CsCAT3 | Degrades H2O2 into H2O and O2, scavenges reactive oxygen species | ROS regulation | [76] |
| CsTGase | Increases endogenous PA content and ROS scavenger capacity | ROS regulation | [77] |
| CsMAPKs | Involved in the signaling pathway of salt stress response, as well as the response to plant hormones | Signal transduction | [15] |
| CsPP2-A1 | Osmoregulation and reactive oxygen species (ROS) homeostasis | Multiple regulation | [78] |
| CsATG | Autophagy gene, degrades proteins or organelles damaged by salt stress | Autophagy | [21] |
| CsSAM | Activates PA metabolic pathway, increases PA quantity | Signal transduction | [13] |
| CsPNG1 | Involved in the ER-associated degradation pathway (ERAD) | Unknown | [79] |
| CscircRNAs | Potentially mediates transcription, signal transduction, cell cycle, metabolic adaptation, and ion homeostasis in salt stress response | Multiple regulation | [72] |
| CsPti1-L | A gene for a cytoplasmic protein kinase that may be involved in ABA signaling | Signal transduction | [80] |
| CsbHLH041 | Enhances salt and ABA tolerance in cucumber seedlings | Transcriptional regulation | [81] |
| CsNAC032 | The upregulation of salt stress may play a defensive role | Transcriptional regulation | [82] |
| CsMYBs | Involved in hormone signaling | Transcriptional regulation | [81] |
| CsWRKY27, 41, 50 | It may be involved in ABA signaling pathway and Ros scavenging pathway | Transcriptional regulation | [83] |
| CsCDPK6 | A gene for a membrane protein that is highly expressed under salt stress | Signal transduction | [84] |
| CsPAO2 | Genes for key enzymes in polyamine metabolism | Signal transduction | [85] |
| CsPAS3 | A gene that interacts with the PAO2 protein and is involved in polyamine transformation | ROS regulation | [85] |
| CsSHMT3 | Serine hydroxymethyl transferase gene | Multiple regulation | [86] |
| CsRBOHs | Genes for the enzyme NADPH oxidase | ROS regulation | [87] |
| CsZFPs | May be involved in the regulation of plant hormones and/or abiotic stress responses | Transcriptional regulation | [88] |
| CsMAX2 | A key gene of the strigolactones signal transduction pathway to improve stress tolerance | Hormone regulation | [89] |
| CsPLDα | A second messenger PA is produced to participate in the salt reaction | Signal transduction | [90] |
| CsRAV1 | RAV transcription factor gene, an important regulator of salt response | Transcriptional regulation | [91] |
| CsGPA1 | Inhibited the expression of CsAQPs in roots and leaves and reduced water content | Osmotic regulation | [67] |
| CsSAMDC3 | Transcription regulates antioxidant enzyme activity | ROS regulation | [92] |
| CsBPC2 | Transcription regulates ABA biosynthesis and expression of genes associated with ABA signaling | Hormone regulation | [93] |
| CsTLP8 | Affects antioxidant enzyme activity and negatively regulates it, which may be related to ABA | ROS regulation | [94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Yu, W.; Xu, X.; Wang, Y.; Wang, B.; Xu, S.; Lan, Q.; Wang, Y. Research Advancements in Salt Tolerance of Cucurbitaceae: From Salt Response to Molecular Mechanisms. Int. J. Mol. Sci. 2024, 25, 9051. https://doi.org/10.3390/ijms25169051
Chen C, Yu W, Xu X, Wang Y, Wang B, Xu S, Lan Q, Wang Y. Research Advancements in Salt Tolerance of Cucurbitaceae: From Salt Response to Molecular Mechanisms. International Journal of Molecular Sciences. 2024; 25(16):9051. https://doi.org/10.3390/ijms25169051
Chicago/Turabian StyleChen, Cuiyun, Wancong Yu, Xinrui Xu, Yiheng Wang, Bo Wang, Shiyong Xu, Qingkuo Lan, and Yong Wang. 2024. "Research Advancements in Salt Tolerance of Cucurbitaceae: From Salt Response to Molecular Mechanisms" International Journal of Molecular Sciences 25, no. 16: 9051. https://doi.org/10.3390/ijms25169051
APA StyleChen, C., Yu, W., Xu, X., Wang, Y., Wang, B., Xu, S., Lan, Q., & Wang, Y. (2024). Research Advancements in Salt Tolerance of Cucurbitaceae: From Salt Response to Molecular Mechanisms. International Journal of Molecular Sciences, 25(16), 9051. https://doi.org/10.3390/ijms25169051






