EPA and DHA Enhance CACT Promoter Activity by GABP/NRF2
Abstract
:1. Introduction
2. Results
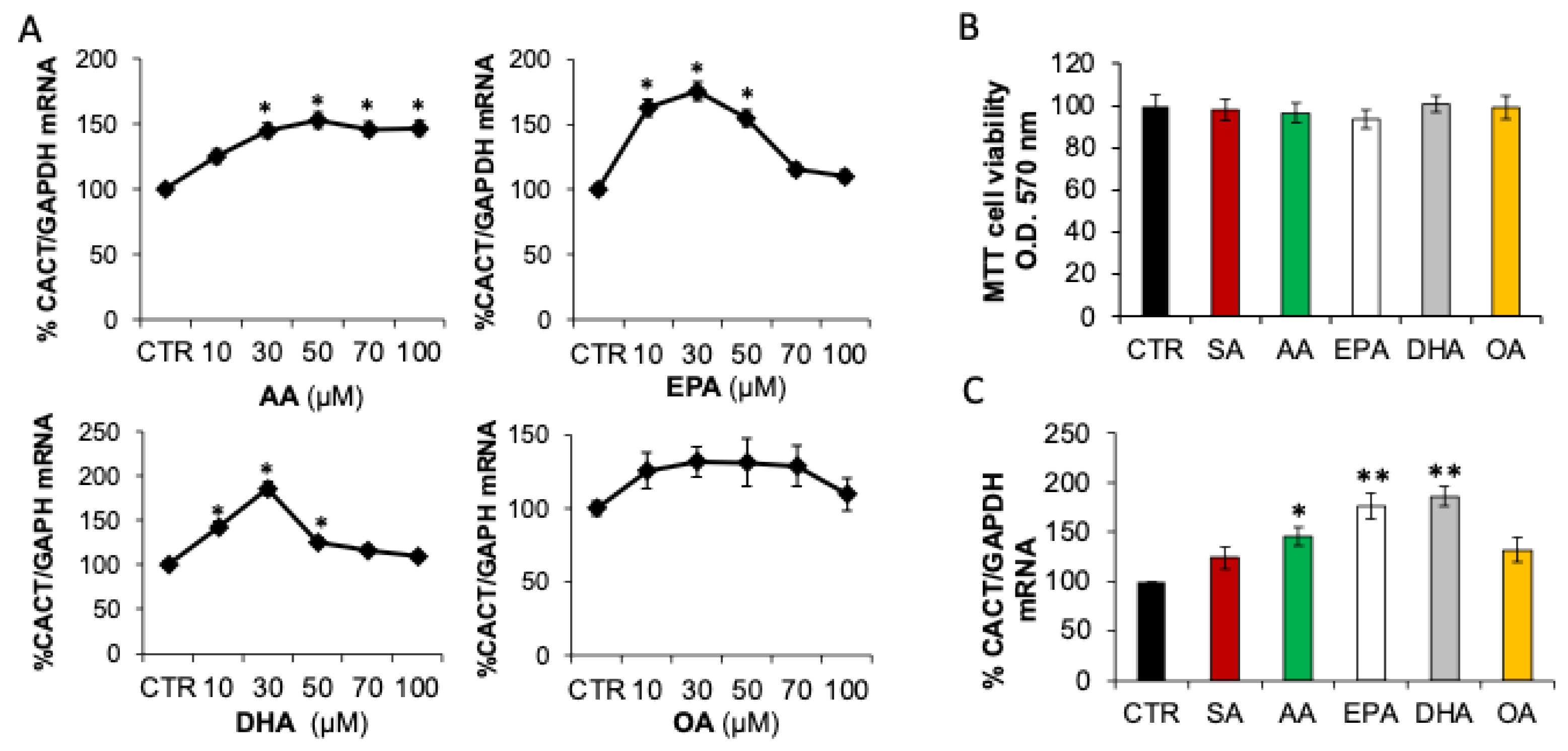
2.1. Fatty Acids Differently Affect CACT Expression
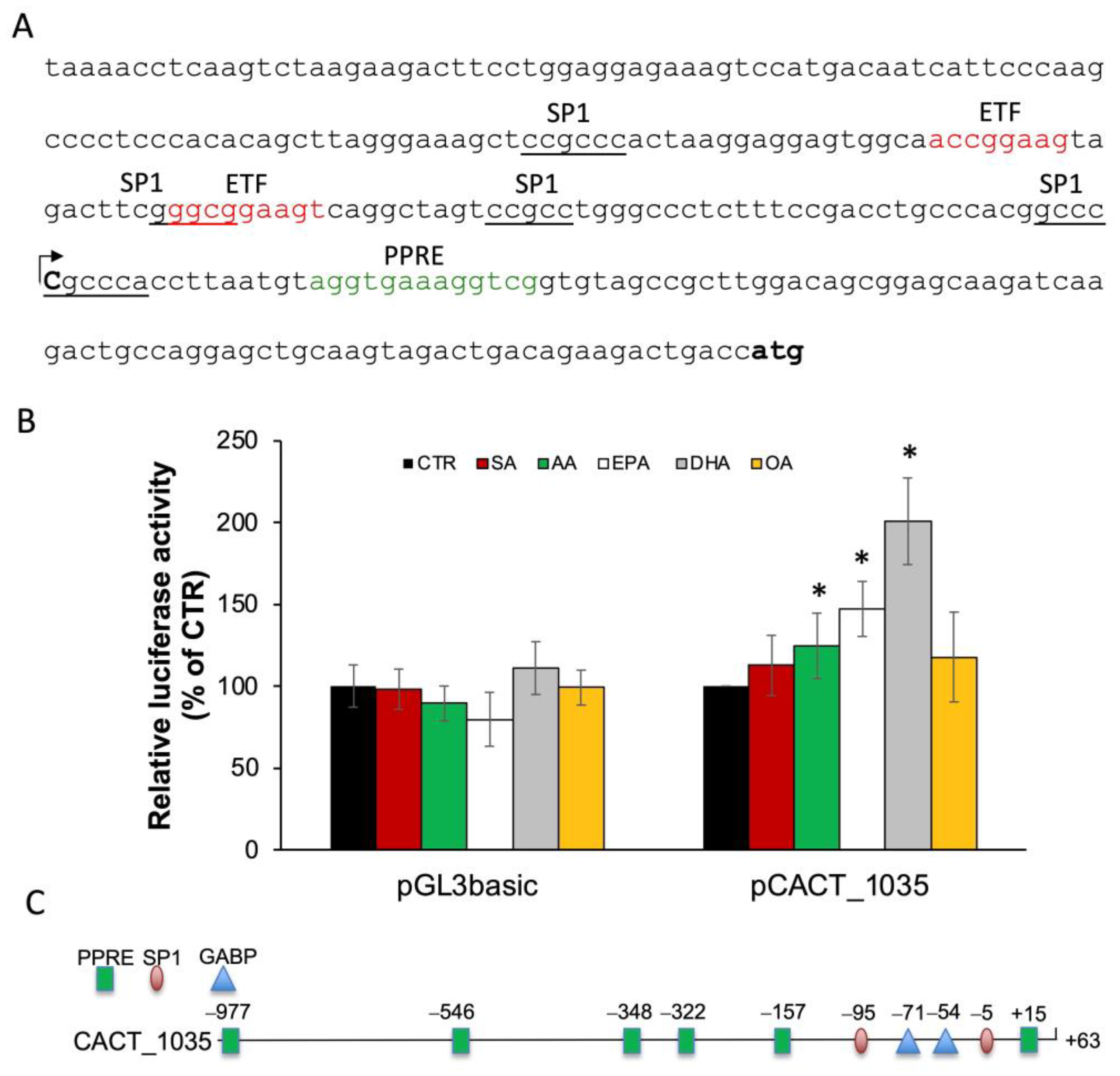
2.2. Structural Organization of CACT Promoter and 5′-UTR
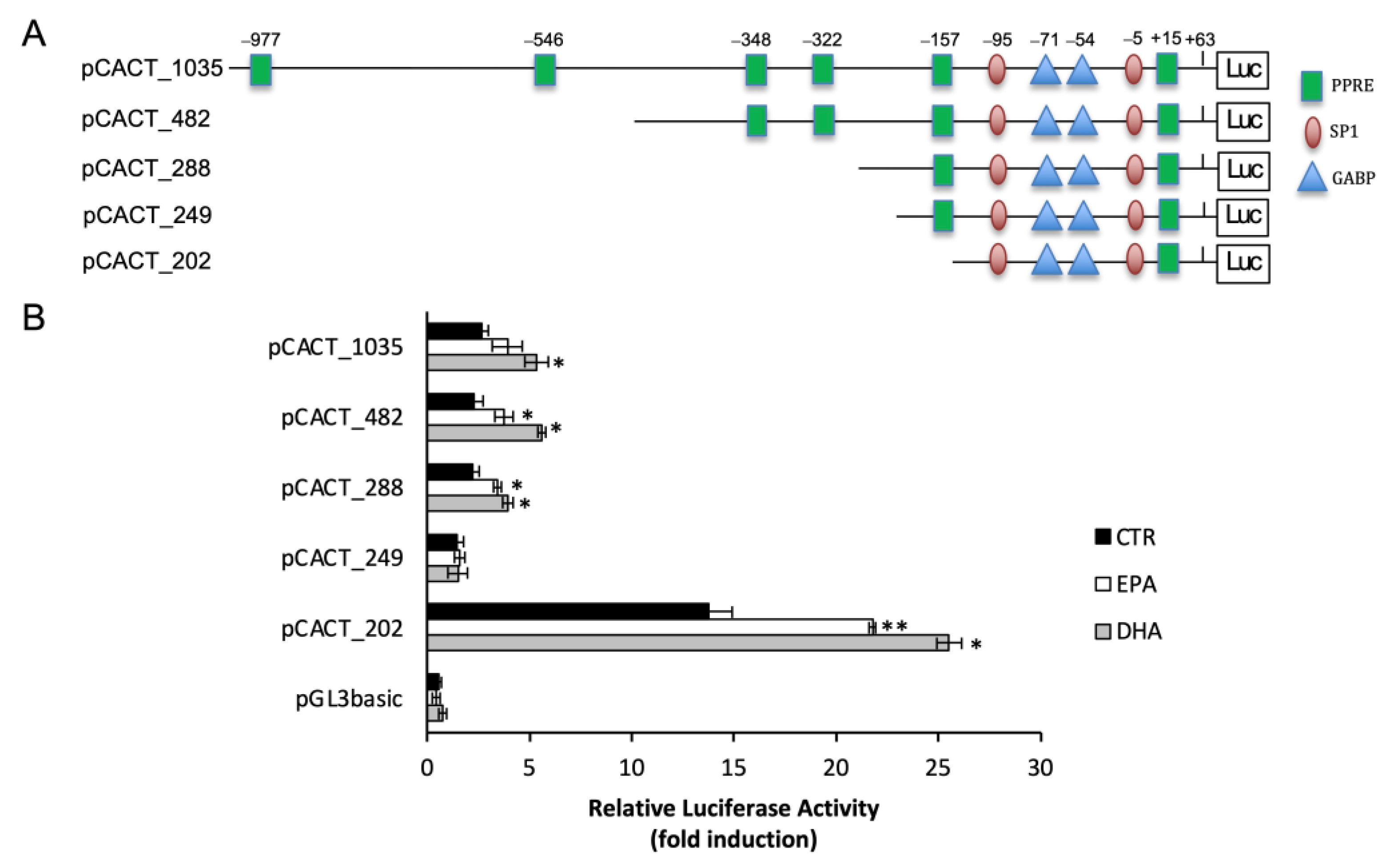
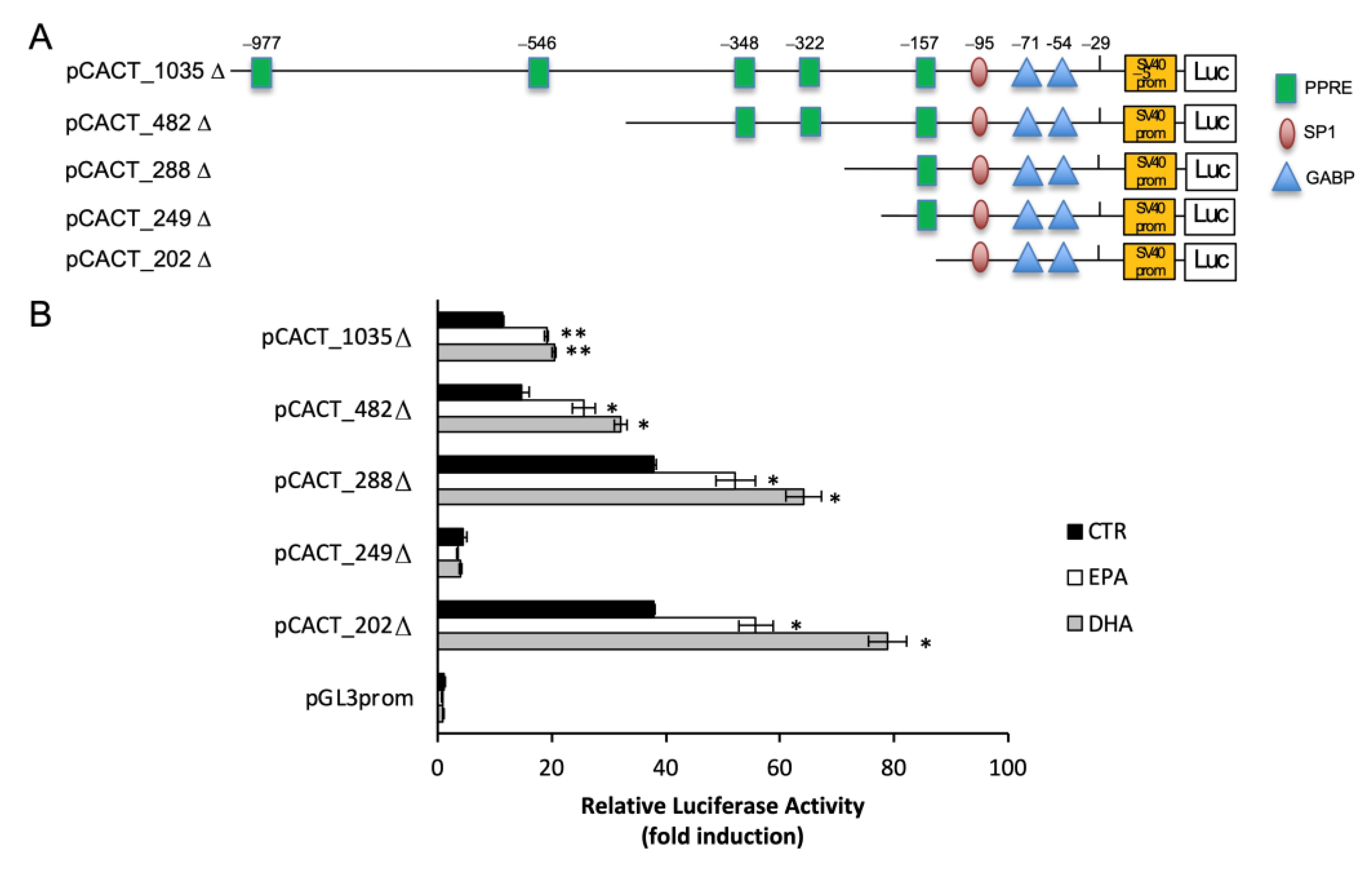
2.3. n-3 PUFA Increased CACT Promoter Activity
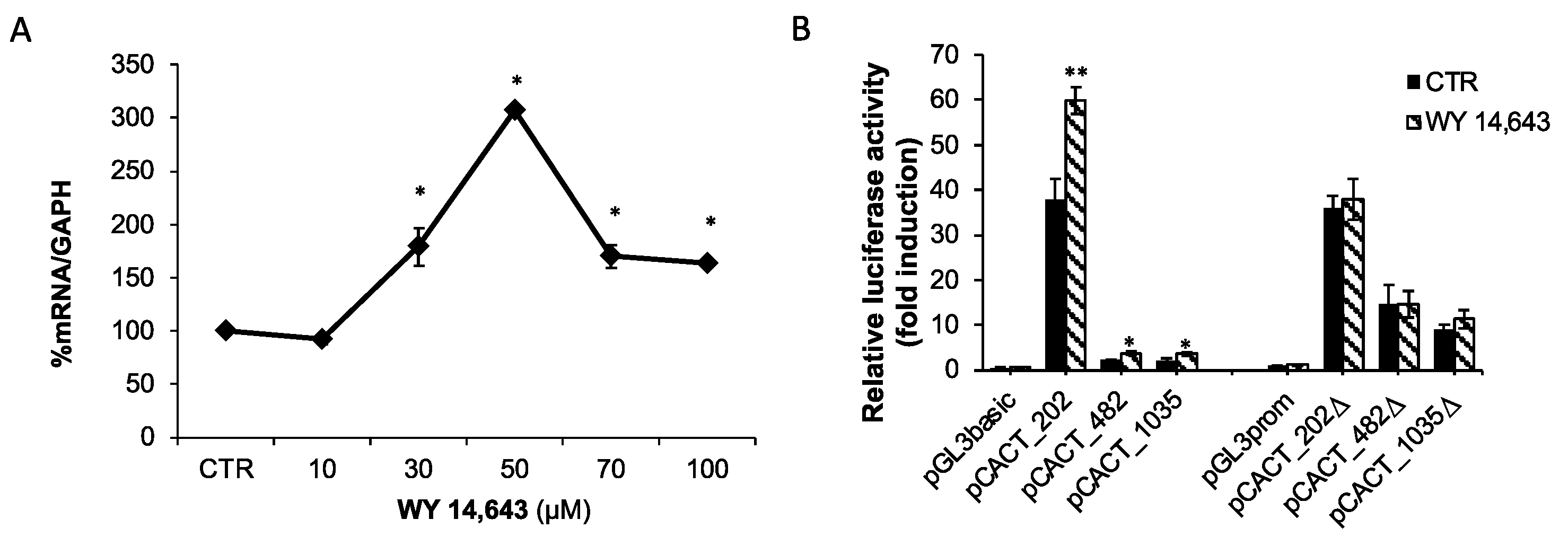
2.4. Effect of the PPARα Agonist WY-14,643 on CACT Expression and Promoter Activity
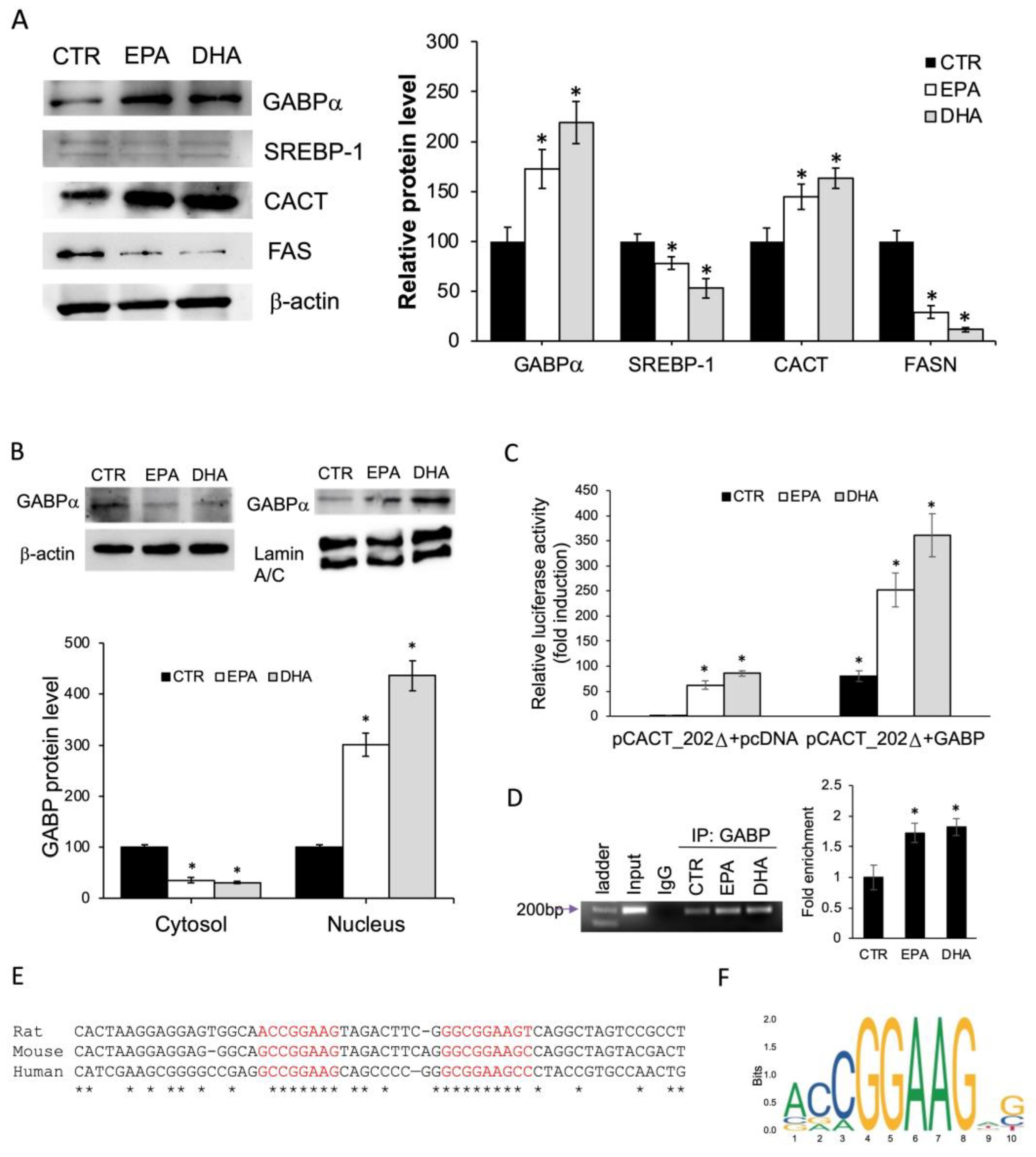
2.5. n-3 PUFA Enhanced CACT Expression via GABP/NRF2
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatments
4.2. Isolation of RNA and Real-Time PCR Analysis
4.3. 5′ Rapid Amplification of cDNA Ends (RACE)
4.4. Bioinformatics Analysis
4.5. Plasmid Vector Constructs and Transient Transfection Assay
4.6. Preparation of Nuclear and Cytosolic Protein Extracts
4.7. Western Blotting Analysis
4.8. Chromatin Immunoprecipitation (ChIP) Assay
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pégorier, J.P.; Le May, C.; Girard, J. Control of gene expression by fatty acids. J. Nutr. 2004, 134, 2444S–2449S. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, F. N-3 Polyunsaturated Fatty Acids and Inflammation in Obesity: Local Effect and Systemic Benefit. Biomed. Res. Int. 2015, 2015, 581469. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, S.J.; Feskens, E.J.; Bos, M.B.; Hoelen, D.W.; Heijligenberg, R.; Bromhaar, M.G.; de Groot, L.C.; de Vries, J.H.; Müller, M.; Afman, L.A. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am. J. Clin. Nutr. 2009, 90, 1656–1664. [Google Scholar] [CrossRef]
- Joffe, Y.T.; Collins, M.; Goedecke, J.H. The relationship between dietary fatty acids and inflammatory genes on the obese phenotype and serum lipids. Nutrients 2013, 5, 1672–1705. [Google Scholar] [CrossRef] [PubMed]
- Enos, R.T.; Davis, J.M.; Velázquez, K.T.; McClellan, J.L.; Day, S.D.; Carnevale, K.A.; Murphy, E.A. Influence of dietary saturated fat content on adiposity, macrophage behavior, inflammation, and metabolism: Composition matters. J. Lipid. Res. 2013, 54, 152–163. [Google Scholar] [CrossRef]
- Jump, D.B.; Tripathy, S.; Depner, C.M. Fatty acid-regulated transcription factors in the liver. Annu. Rev. Nutr. 2013, 33, 249–269. [Google Scholar] [CrossRef]
- Giudetti, A.M.; Stanca, E.; Siculella, L.; Gnoni, G.V.; Damiano, F. Nutritional and Hormonal Regulation of Citrate and Carnitine/Acylcarnitine Transporters: Two Mitochondrial Carriers Involved in Fatty Acid Metabolism. Int. J. Mol. Sci. 2016, 17, 817. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Yahagi, N.; Izumida, Y.; Nishi, M.; Kubota, M.; Teraoka, Y.; Yamamoto, T.; Matsuzaka, T.; Nakagawa, Y.; Sekiya, M.; et al. Polyunsaturated fatty acids selectively suppress sterol regulatory element-binding protein-1 through proteolytic processing and autoloop regulatory circuit. J. Biol. Chem. 2010, 285, 11681–11691. [Google Scholar] [CrossRef]
- Liput, K.P.; Lepczyński, A.; Ogłuszka, M.; Nawrocka, A.; Poławska, E.; Grzesiak, A.; Ślaska, B.; Pareek, C.S.; Czarnik, U.; Pierzchała, M. Effects of Dietary n-3 and n-6 Polyunsaturated Fatty Acids in Inflammation and Cancerogenesis. Int. J. Mol. Sci. 2021, 22, 6965. [Google Scholar] [CrossRef]
- Minihane, A.M. Nutrient gene interactions in lipid metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Siculella, L.; Sabetta, S.; Damiano, F.; Giudetti, A.M.; Gnoni, G.V. Different dietary fatty acids have dissimilar effects on activity and gene expression of mitochondrial tricarboxylate carrier in rat liver. FEBS Lett. 2004, 578, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Damiano, F.; Gnoni, G.V.; Siculella, L. Functional analysis of rat liver citrate carrier promoter: Differential responsiveness to polyunsaturated fatty acids. Biochem. J. 2009, 417, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Salati, L.M.; Szeszel-Fedorowicz, W.; Tao, H.; Gibson, M.A.; Amir-Ahmady, B.; Stabile, L.P.; Hodge, D.L. Nutritional regulation of mRNA processing. J. Nutr. 2004, 134, 2437–2443. [Google Scholar] [CrossRef]
- Jump, D.B. Fatty acid regulation of hepatic lipid metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 115–120. [Google Scholar] [CrossRef]
- Duplus, E.; Glorian, M.; Forest, C. Fatty acid regulation of gene transcription. J. Biol. Chem. 2000, 275, 30749–30752. [Google Scholar] [CrossRef]
- Worsch, S.; Heikenwalder, M.; Hauner, H.; Bader, B.L. Dietary n-3 long-chain polyunsaturated fatty acids upregulate energy dissipating metabolic pathways conveying anti-obesogenic effects in mice. Nutr. Metab. 2018, 15, 65. [Google Scholar] [CrossRef]
- El-Gharba, A.; Vockley, J. Inborn Errors of Metabolism with Myopathy: Defects of Fatty Acid Oxidation and the Carnitine Shuttle System. Pediatr. Clin. N. Am. 2018, 65, 317–335. [Google Scholar] [CrossRef]
- Rubio-Gozalbo, M.E.; Bakker, J.A.; Waterham, H.R.; Wanders, R.J. Carnitine-acylcarnitine translocase deficiency, clinical, biochemical and genetic aspects. Mol. Asp. Med. 2004, 25, 521–532. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Pierre, G.; Macdonald, A.; Gray, G.; Hendriksz, C.; Preece, M.A.; Chakrapani, A. Prospective treatment in carnitine-acylcarnitine translocase deficiency. J. Inherit. Metab. Dis. 2007, 30, 815. [Google Scholar] [CrossRef] [PubMed]
- Priore, P.; Stanca, E.; Gnoni, G.V.; Siculella, L. Dietary fat types differently modulate the activity and expression of mitochondrial carnitine/acylcarnitine translocase in rat liver. Biochim. Biophys. Acta 2012, 1821, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Goto, T. A review of the studies on food-derived factors which regulate energy metabolism via the modulation of lipid-sensing nuclear receptors. Biosci. Biotechnol. Biochem. 2019, 83, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Deckelbaum, R.J.; Torrejon, C. The omega-3 fatty acid nutritional landscape: Health benefits and sources. J. Nutr. 2012, 142, 587S–591S. [Google Scholar] [CrossRef] [PubMed]
- Gutgesell, A.; Wen, G.; König, B.; Koch, A.; Spielmann, J.; Stangl, G.I.; Eder, K.; Ringseis, R. Mouse carnitine-acylcarnitine translocase (CACT) is transcriptionally regulated by PPARalpha and PPARdelta in liver cells. Biochim. Biophys. Acta 2009, 1790, 1206–1216. [Google Scholar] [CrossRef]
- Mangiullo, R.; Gnoni, A.; Damiano, F.; Siculella, L.; Zanotti, F.; Papa, S.; Gnoni, G.V. 3,5-diiodo-L-thyronine upregulates rat-liver mitochondrial F(o)F(1)-ATP synthase by GA-binding protein/nuclear respiratory factor-2. Biochim. Biophys. Acta 2010, 1797, 233–240. [Google Scholar] [CrossRef]
- Rossmeisl, M.; Medrikova, D.; van Schothorst, E.M.; Pavlisova, J.; Kuda, O.; Hensler, M.; Bardova, K.; Flachs, P.; Stankova, B.; Vecka, M.; et al. Omega-3 phospholipids from fish suppress hepatic steatosis by integrated inhibition of biosynthetic pathways in dietary obese mice. Biochim. Biophys. Acta 2014, 1841, 267–278. [Google Scholar] [CrossRef]
- Kroupova, P.; van Schothorst, E.M.; Keijer, J.; Bunschoten, A.; Vodicka, M.; Irodenko, I.; Oseeva, M.; Zacek, P.; Kopecky, J.; Rossmeisl, M.; et al. Omega-3 phospholipids from krill oil enhance intestinal fatty acid oxidation more effectively than omega-3 triacylglycerols in high-fat diet-fed obese mice. Nutrients 2020, 12, 2037. [Google Scholar] [CrossRef]
- Shibabaw, T. Omega-3 polyunsaturated fatty acids: Anti-inflammatory and anti-hypertriglyceridemia mechanisms in cardiovascular disease. Mol. Cell. Biochem. 2021, 476, 993–1003. [Google Scholar] [CrossRef]
- Katsnelson, G.; Ceddia, R.B. Docosahexaenoic and eicosapentaenoic fatty acids differentially regulate glucose and fatty acid metabolism in L6 rat skeletal muscle cells. Am. J. Physiol. Cell. Physiol. 2020, 319, C1120–C1129. [Google Scholar] [CrossRef]
- Clarke, S.D.; Gasperikova, D.; Nelson, C.; Lapillonne, A.; Heird, W.C. Fatty acid regulation of gene expression: A genomic explanation for the benefits of the mediterranean diet. Ann. N. Y. Acad. Sci. 2002, 967, 283–298. [Google Scholar] [CrossRef]
- Lee, J.; Choi, Y.R.; Kim, M.; Park, J.M.; Kang, M.; Oh, J.; Lee, C.J.; Park, S.; Kang, S.M.; Manabe, I.; et al. Common and differential effects of docosahexaenoic acid and eicosapentaenoic acid on helper T-cell responses and associated pathways. BMB Rep. 2021, 54, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Dias, B.V.; Gomes, S.V.; Castro, M.L.D.C.; Carvalho, L.C.F.; Breguez, G.S.; de Souza, D.M.S.; Ramos, C.O.; Sant’Ana, M.R.; Nakandakari, S.C.B.R.; Araujo, C.M.; et al. EPA/DHA and linseed oil have different effects on liver and adipose tissue in rats fed with a high-fat diet. Prostaglandins Other Lipid Mediat. 2022, 159, 106622. [Google Scholar] [CrossRef]
- Emami, K.H.; Burke, T.W.; Smale, S.T. Sp1 activation of a TATA-less promoter requires a species-specific interaction involving transcription factor IID. Nucleic Acids Res. 1998, 26, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Tonazzi, A.; Giangregorio, N.; Console, L.; Palmieri, F.; Indiveri, C. The Mitochondrial Carnitine Acyl-carnitine Carrier (SLC25A20): Molecular Mechanisms of Transport, Role in Redox Sensing and Interaction with Drugs. Biomolecules 2021, 11, 521. [Google Scholar] [CrossRef]
- Vitoria, I.; Martín-Hernández, E.; Peña-Quintana, L.; Bueno, M.; Quijada-Fraile, P.; Dalmau, J.; Molina-Marrero, S.; Pérez, B.; Merinero, B. Carnitine-acylcarnitine translocase deficiency: Experience with four cases in Spain and review of the literature. JIMD Rep. 2015, 20, 11–20. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Convertini, P.; Infantino, V.; Scarcia, P.; Todisco, S.; Palmieri, F. Statins, fibrates and retinoic acid upregulate mitochondrial acylcarnitine carrier gene expression. Biochem. Biophys. Res. Commun. 2009, 388, 643–647. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, K.; Takeuchi, K.; Inada, H.; Yamasaki, D.; Ishimoto, K.; Tanaka, T.; Hamakubo, T.; Sakai, J.; Kodama, T.; Doi, T. Regulation of the human SLC25A20 expression by peroxisome proliferator-activated receptor alpha in human hepatoblastoma cells. Biochem. Biophys. Res. Commun. 2009, 389, 501–505. [Google Scholar] [CrossRef]
- Gacias, M.; Pérez-Martí, A.; Pujol-Vidal, M.; Marrero, P.F.; Haro, D.; Relat, J. PGC-1β regulates mouse carnitine-acylcarnitine translocase through estrogen-related receptor α. Biochem. Biophys. Res. Commun. 2012, 423, 838–843. [Google Scholar] [CrossRef]
- Convertini, P.; Infantino, V.; Bisaccia, F.; Palmieri, F.; Iacobazzi, V. Role of FOXA and Sp1 in mitochondrial acylcarnitine carrier gene expression in different cell lines. Biochem. Biophys. Res. Commun. 2011, 404, 376–381. [Google Scholar] [CrossRef]
- Louet, J.F.; Chatelain, F.; Decaux, J.F.; Park, E.A.; Kohl, C.; Pineau, T.; Girard, J.; Pegorier, J.P. Long-chain fatty acids regulate liver carnitine palmitoyltransferase I gene (L-CPT I) expression through a peroxisome-proliferator-activated receptor alpha (PPARalpha)-independent pathway. Biochem. J. 2001, 354, 189–197. [Google Scholar] [CrossRef]
- Bruni, F.; Polosa, P.L.; Gadaleta, M.N.; Cantatore, P.; Roberti, M. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J. Biol. Chem. 2010, 285, 3939–3948. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.F.; Drumea, K.; Mott, S.; Wang, J.; Rosmarin, A.G. GABP transcription factor (nuclear respiratory factor 2) is required for mitochondrial biogenesis. Mol. Cell. Biol. 2014, 34, 3194–3201. [Google Scholar] [CrossRef] [PubMed]
- Scarpulla, R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta 2002, 1576, 1–14. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, S.H.; Jeong, C.; Han, Y.H.; Lee, M.O. RORα-GABP-TFAM axis alleviates myosteatosis with fatty atrophy through reinforcement of mitochondrial capacity. J. Cachexia Sarcopenia Muscle 2024, 15, 615–630. [Google Scholar] [CrossRef]
- Ewaschuk, J.B.; Almasud, A.; Mazurak, V.C. Role of n-3 fatty acids in muscle loss and myosteatosis. Appl. Physiol. Nutr. Metab. 2014, 39, 654–662. [Google Scholar] [CrossRef]
- Ebadi, M.; Tsien, C.; Bhanji, R.A.; Dunichand-Hoedl, A.R.; Rider, E.; Motamedrad, M.; Mazurak, V.C.; Baracos, V.; Montano-Loza, A.J. Myosteatosis in Cirrhosis: A Review of Diagnosis, Pathophysiological Mechanisms and Potential Interventions. Cells 2022, 11, 1216. [Google Scholar] [CrossRef] [PubMed]
- Niopek, K.; Üstünel, B.E.; Seitz, S.; Sakurai, M.; Zota, A.; Mattijssen, F.; Wang, X.; Sijmonsma, T.; Feuchter, Y.; Gail, A.M.; et al. A Hepatic GAbp-AMPK Axis Links Inflammatory Signaling to Systemic Vascular Damage. Cell. Rep. 2017, 20, 1422–1434. [Google Scholar] [CrossRef]
- Nammo, T.; Udagawa, H.; Funahashi, N.; Kawaguchi, M.; Uebanso, T.; Hiramoto, M.; Nishimura, W.; Yasuda, K. Genome-wide profiling of histone H3K27 acetylation featured fatty acid signalling in pancreatic beta cells in diet-induced obesity in mice. Diabetologia 2018, 61, 2608–2620. [Google Scholar] [CrossRef]
| Gene | Accession Number | Sequences 5′-3′ |
|---|---|---|
| Slc25a20 | NW_047802 | promFOR1: GAATTCGGTACCAAATTCCTCCACGGTACCTC |
| promFOR2: GAATTCGGTACCCCCAAAGAGGCCAAAAGAGG | ||
| promFOR3: GAATTCGGTACCTTCACCTCTTTACCATGCCC | ||
| promFOR4: GAATTCGGTACCCTATGCATCTTTTCACTTTGTC | ||
| promFOR5: AATTCGGTACCCCACAGAGGACCAGAGAAAG | ||
| promREV: CTCGAGAAGCTTGTCTTGATCTTGCTCCGCT | ||
| promREV1: GGTACCCTCGAGAGGCGGACTAGCCTGACTTC | ||
| NM_053965 | FOR: GTGTGCTTCTTTGGGTTTGG REV: TTCTCCAGGGGTCATGATTC | |
| Gapdh | NM_017008 | REV: GCCGCCTGCTTCACCACCTTCT FOR: GCATGGCCTTCCGTGTTCCTACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanca, E.; Spedicato, F.; Giudetti, A.M.; Giannotti, L.; Di Chiara Stanca, B.; Damiano, F.; Siculella, L. EPA and DHA Enhance CACT Promoter Activity by GABP/NRF2. Int. J. Mol. Sci. 2024, 25, 9095. https://doi.org/10.3390/ijms25169095
Stanca E, Spedicato F, Giudetti AM, Giannotti L, Di Chiara Stanca B, Damiano F, Siculella L. EPA and DHA Enhance CACT Promoter Activity by GABP/NRF2. International Journal of Molecular Sciences. 2024; 25(16):9095. https://doi.org/10.3390/ijms25169095
Chicago/Turabian StyleStanca, Eleonora, Francesco Spedicato, Anna Maria Giudetti, Laura Giannotti, Benedetta Di Chiara Stanca, Fabrizio Damiano, and Luisa Siculella. 2024. "EPA and DHA Enhance CACT Promoter Activity by GABP/NRF2" International Journal of Molecular Sciences 25, no. 16: 9095. https://doi.org/10.3390/ijms25169095





