Analysis of Correlation Effects of Double Mutations in Enzymes: A Revised Residual-Contact Network Clique Model
Abstract
1. Introduction
2. Results and Discussion
2.1. Additivity Analysis of Double Mutations in Staphylococcal Nuclease
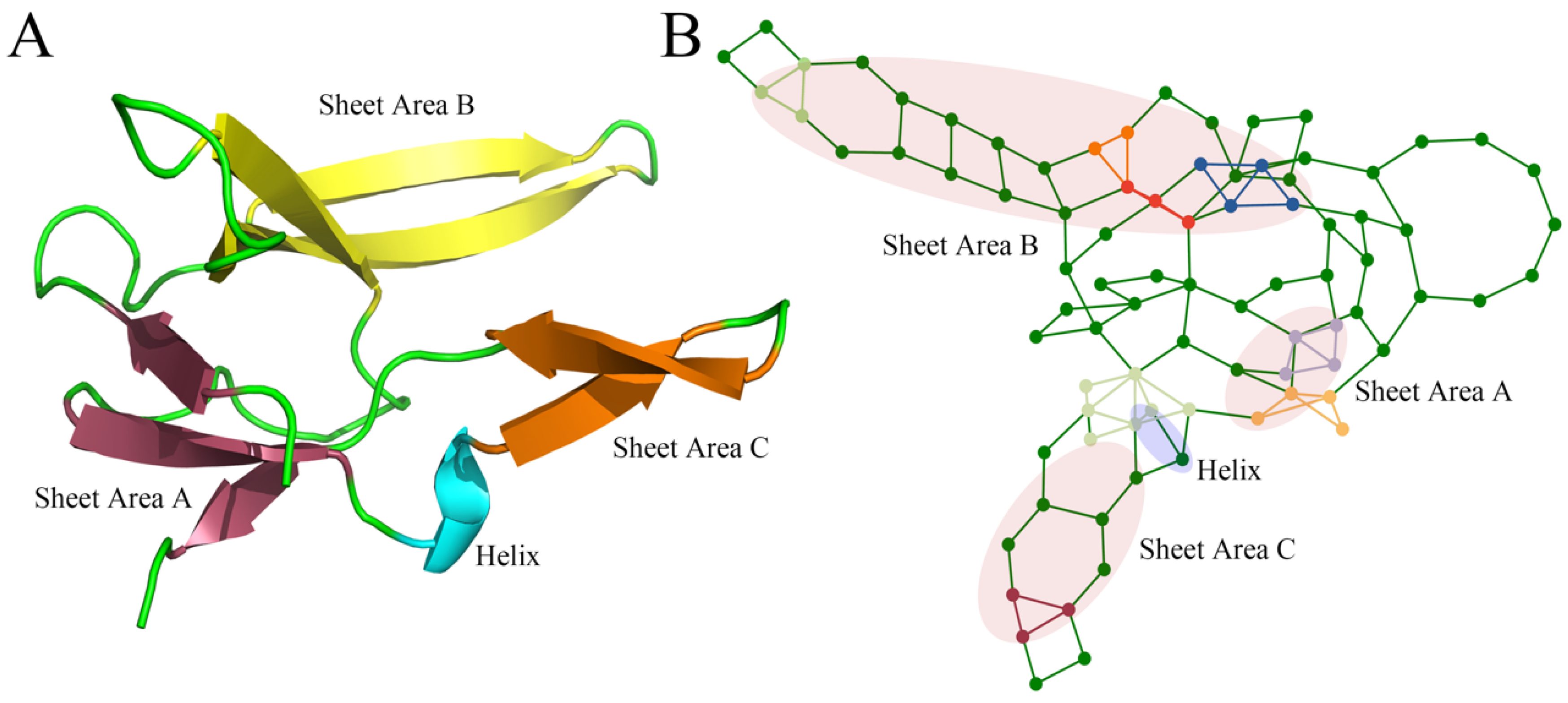
2.1.1. Clique Analysis of Double Mutation Correlation in Staphylococcal Nuclease
2.1.2. Perturbations in Clique Due to Local Structural Changes in Double Mutations
2.1.3. Hydrophobic Core and 2nd Structure Fragment in Additive Double Mutations in Clique Structures
2.1.4. Non-Additive Double Mutations in Clique Structures
2.1.5. Outside the Clique: Effect of the Third Amino Acid Site between Double Mutations
2.2. Double Mutations in Gene V
2.2.1. The 3-Clique Analysis of Gene V Protein
2.2.2. Clique Perturbation Due to Local Structural Change by Mutations
2.2.3. Prediction Bias Due to Different Bases for Judging Mutation Correlations
2.3. Improvement to the Model: Double Mutations in Phage T4 Lysozyme
3. Methods
3.1. Protein Mutation Datasets
3.2. Determining the Additivity and Non-Additivity of Double Mutations
3.3. AlphaFold Modeling of the Specific Structure of Double Mutations
3.4. Residual-Contact Network
3.5. The Clique Community and the Correlation Effects in Double-Site Mutations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dinmukhamed, T.; Huang, Z.; Liu, Y.; Lv, X.; Li, J.; Du, G.; Liu, L. Current advances in design and engineering strategies of industrial enzymes. Syst. Microbiol. Biomanuf. 2021, 1, 15–23. [Google Scholar] [CrossRef]
- Steinbrecher, T.; Zhu, C.; Wang, L.; Abel, R.; Negron, C.; Pearlman, D.; Feyfant, E.; Duan, J.; Sherman, W. Predicting the effect of amino acid single-point mutations on protein stability—Large-scale validation of MD-based relative free energy calculations. J. Mol. Biol. 2017, 429, 948–963. [Google Scholar] [CrossRef] [PubMed]
- Dehghanpoor, R.; Ricks, E.; Hursh, K.; Gunderson, S.; Farhoodi, R.; Haspel, N.; Hutchinson, B.; Jagodzinski, F. Predicting the effect of single and multiple mutations on protein structural stability. Molecules 2018, 23, 251. [Google Scholar] [CrossRef]
- Wells, J.A. Additivity of Mutational Effects in Proteins. Biochemistry 1990, 29, 8509–8517. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.A.; Clay, C.J.; Luce, K.S.; Edgell, M.H.; Lee, A.L. Detection of native-state nonadditivity in double mutant cycles via hydrogen exchange. J. Am. Chem. Soc. 2010, 132, 8010–8019. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sondek, J.; Shortle, D. Structural and Energetic Differences between Insertions and Substitutions in Staphylococcal Nuclease. Proteins-Struct. Funct. Genet. 1992, 13, 132–140. [Google Scholar] [CrossRef]
- Green, S.M.; Shortle, D. Patterns of Nonadditivity between Pairs of Stability Mutations in Staphylococcal Nuclease. Biochemistry 1993, 32, 10131–10139. [Google Scholar] [CrossRef]
- Chen, J.M.; Stites, W.E. Energetics of side chain packing in staphylococcal nuclease assessed by systematic double mutant cycles. Biochemistry 2001, 40, 14004–14011. [Google Scholar] [CrossRef]
- Sandberg, W.S.; Terwilliger, T.C. Engineering Multiple Properties of a Protein by Combinatorial Mutagenesis. Proc. Natl. Acad. Sci. USA 1993, 90, 8367–8371. [Google Scholar] [CrossRef]
- Skinner, M.M.; Terwilliger, T.C. Potential use of additivity of mutational effects in simplifying protein engineering. Proc. Natl. Acad. Sci. USA 1996, 93, 10753–10757. [Google Scholar] [CrossRef]
- Baase, W.A.; Liu, L.; Tronrud, D.E.; Matthews, B.W. Lessons from the lysozyme of phage T4. Protein Sci. 2010, 19, 631–641. [Google Scholar] [CrossRef]
- Jemimah, S.; Gromiha, M.M. Exploring additivity effects of double mutations on the binding affinity of protein-protein complexes. Proteins-Struct. Funct. Bioinform. 2018, 86, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Ming, D.M.; Chen, R.; Huang, H. Amino-Acid Network Clique Analysis of Protein Mutation Non-Additive Effects: A Case Study of Lysozyme. Int. J. Mol. Sci. 2018, 19, 1427. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, H.; Becktel, W.; Matthews, B. Enhanced protein thermostability from designed mutations that interact with α-helix dipoles. Nature 1988, 336, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-J.; Baase, W.A.; Shoichet, B.K.; Wilson, K.P.; Matthews, B.W. Enhancement of protein stability by the combination of point mutations in T4 lysozyme is additive. Protein Eng. Des. Sel. 1995, 8, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Daopin, S.; Alber, T.; Baase, W.; Wozniak, J.; Matthews, B. Structural and thermodynamic analysis of the packing of two α-helices in bacteriophage T4 lysozyme. J. Mol. Biol. 1991, 221, 647–667. [Google Scholar] [CrossRef]
- Reetz, M.T. The Importance of Additive and Non-Additive Mutational Effects in Protein Engineering. Angew. Chem.-Int. Ed. 2013, 52, 2658–2666. [Google Scholar] [CrossRef]
- Hollmann, F.; Martinez, J.S.; Reetz, M.T. Learning from Protein Engineering by Deconvolution of Multi-Mutational Variants. Angew. Chem. Int. Ed. Engl. 2024, e202404880. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Istomin, A.Y.; Gromiha, M.M.; Vorov, O.K.; Jacobs, D.J.; Livesay, D.R. New insight into long-range nonadditivity within protein double-mutant cycles. Proteins-Struct. Funct. Bioinform. 2008, 70, 915–924. [Google Scholar] [CrossRef]
- Nikam, R.; Kulandaisamy, A.; Harini, K.; Sharma, D.; Gromiha, M.M. ProThermDB: Thermodynamic database for proteins and mutants revisited after 15 years. Nucleic Acids Res. 2021, 49, D420–D424. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Del Conte, A.; Camagni, G.F.; Clementel, D.; Minervini, G.; Monzon, A.M.; Ferrari, C.; Piovesan, D.; E Tosatto, S.C. RING 4.0: Faster residue interaction networks with novel interaction types across over 35,000 different chemical structures. Nucleic Acids Res. 2024, 52, W306–W312. [Google Scholar] [CrossRef]
- Hagberg, A.; Swart, P.J.; Schult, D.A. Exploring Network Structure, Dynamics, and Function Using NetworkX; Los Alamos National Laboratory (LANL): Los Alamos, NM, USA, 2008. [Google Scholar]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Pan, Q.; Pires, D.E.V.; Rodrigues, C.H.M.; Ascher, D.B. DDMut: Predicting effects of mutations on protein stability using deep learning. Nucleic Acids Res. 2023, 51, W122–W128. [Google Scholar] [CrossRef] [PubMed]


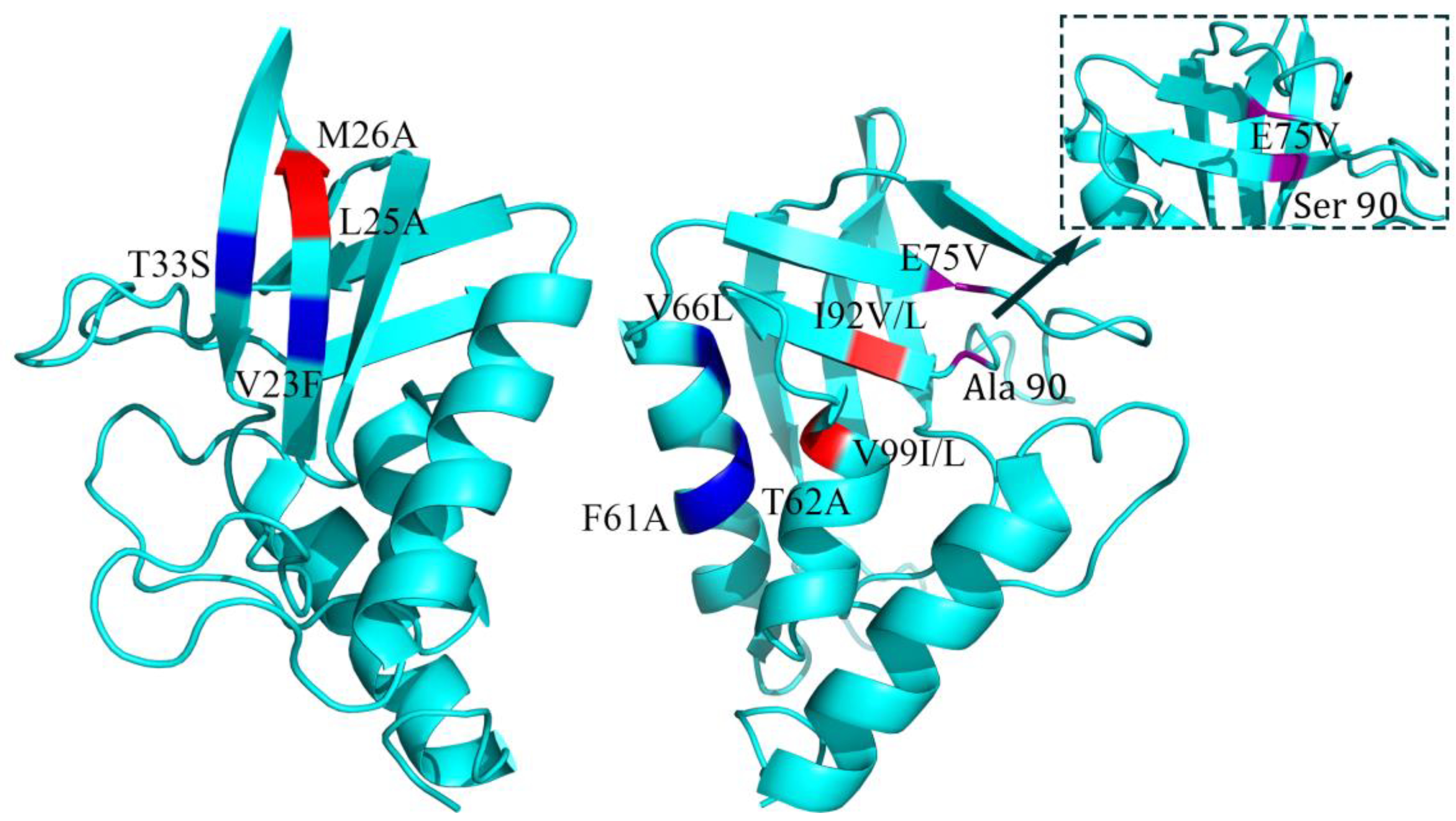
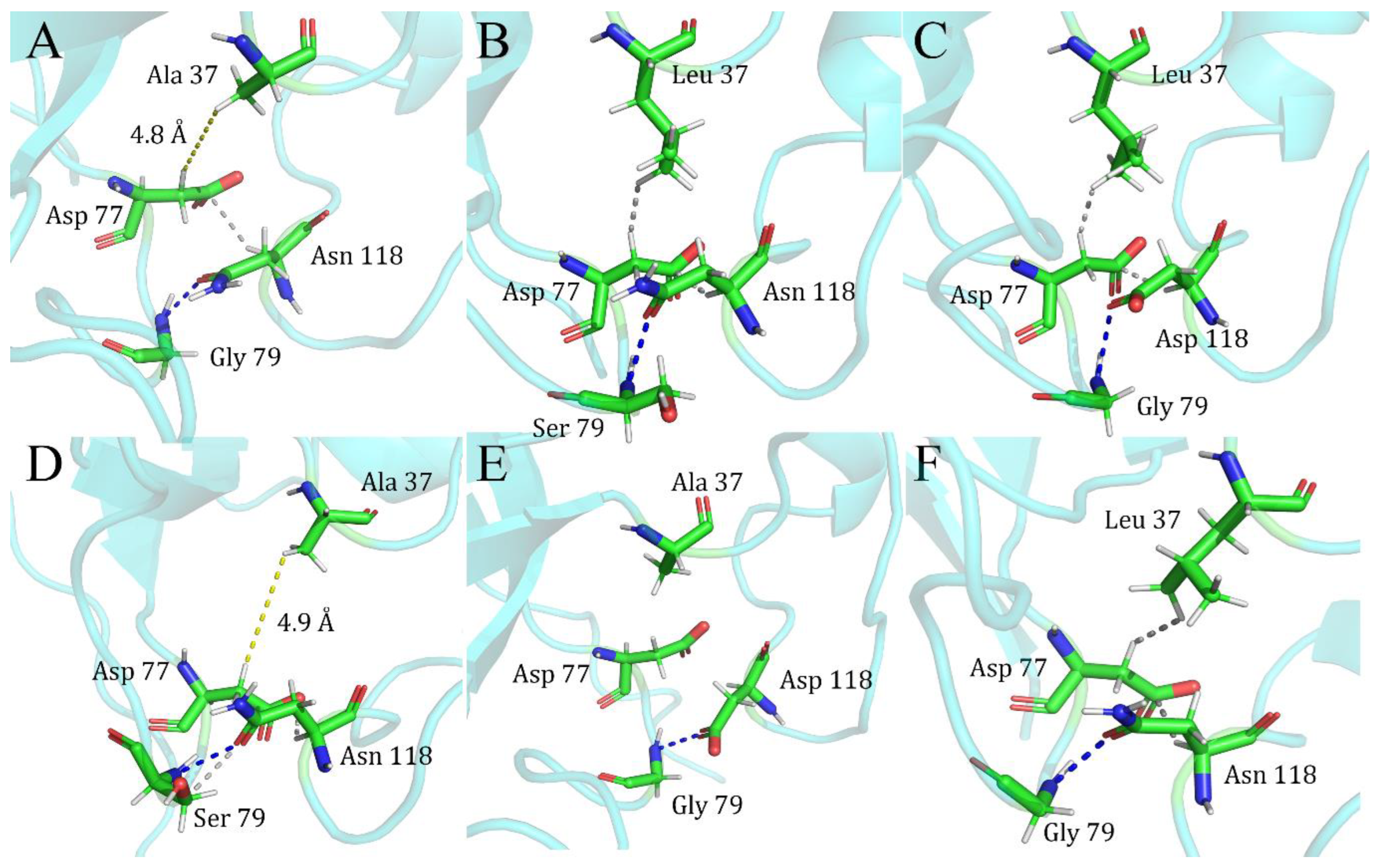
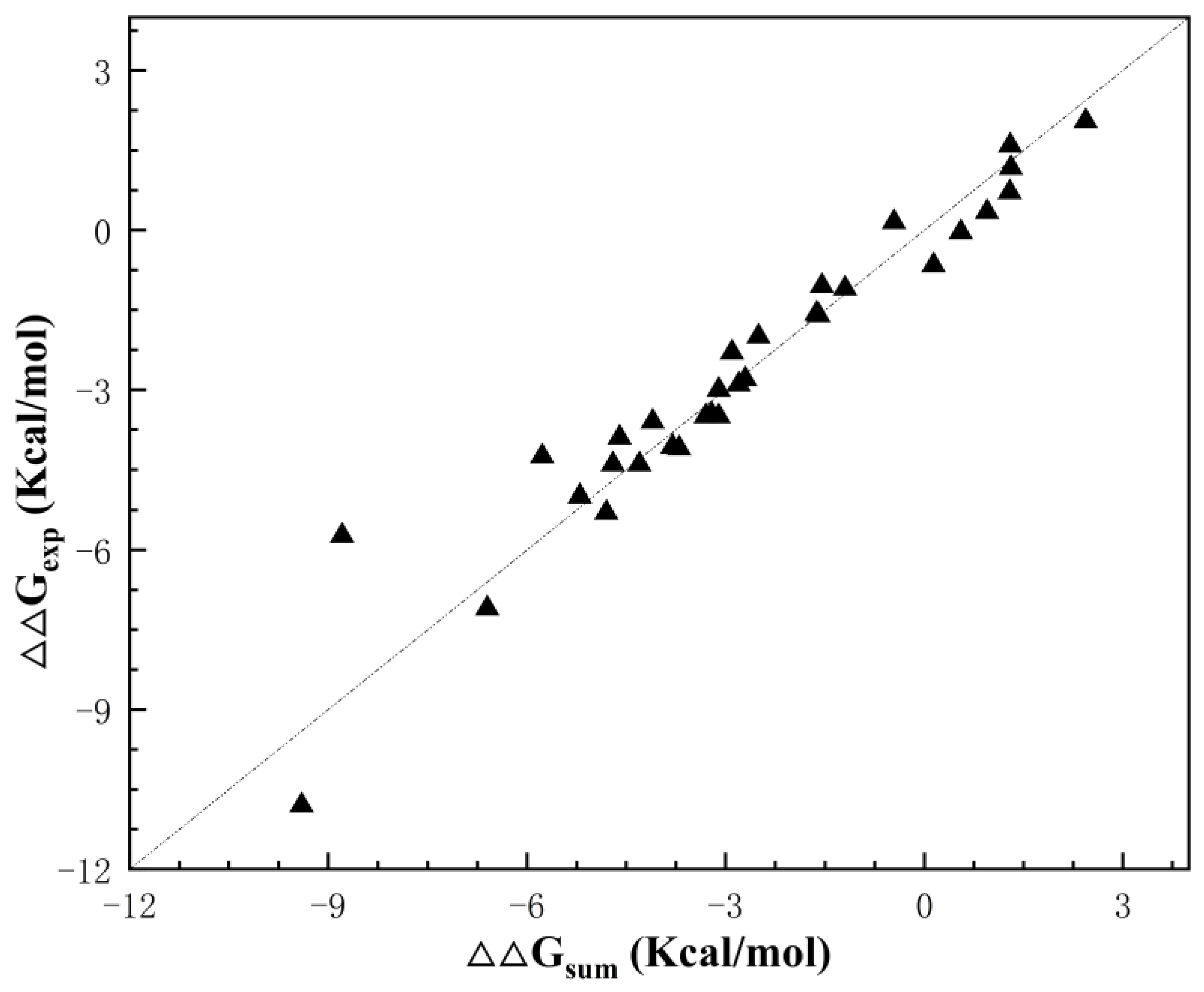
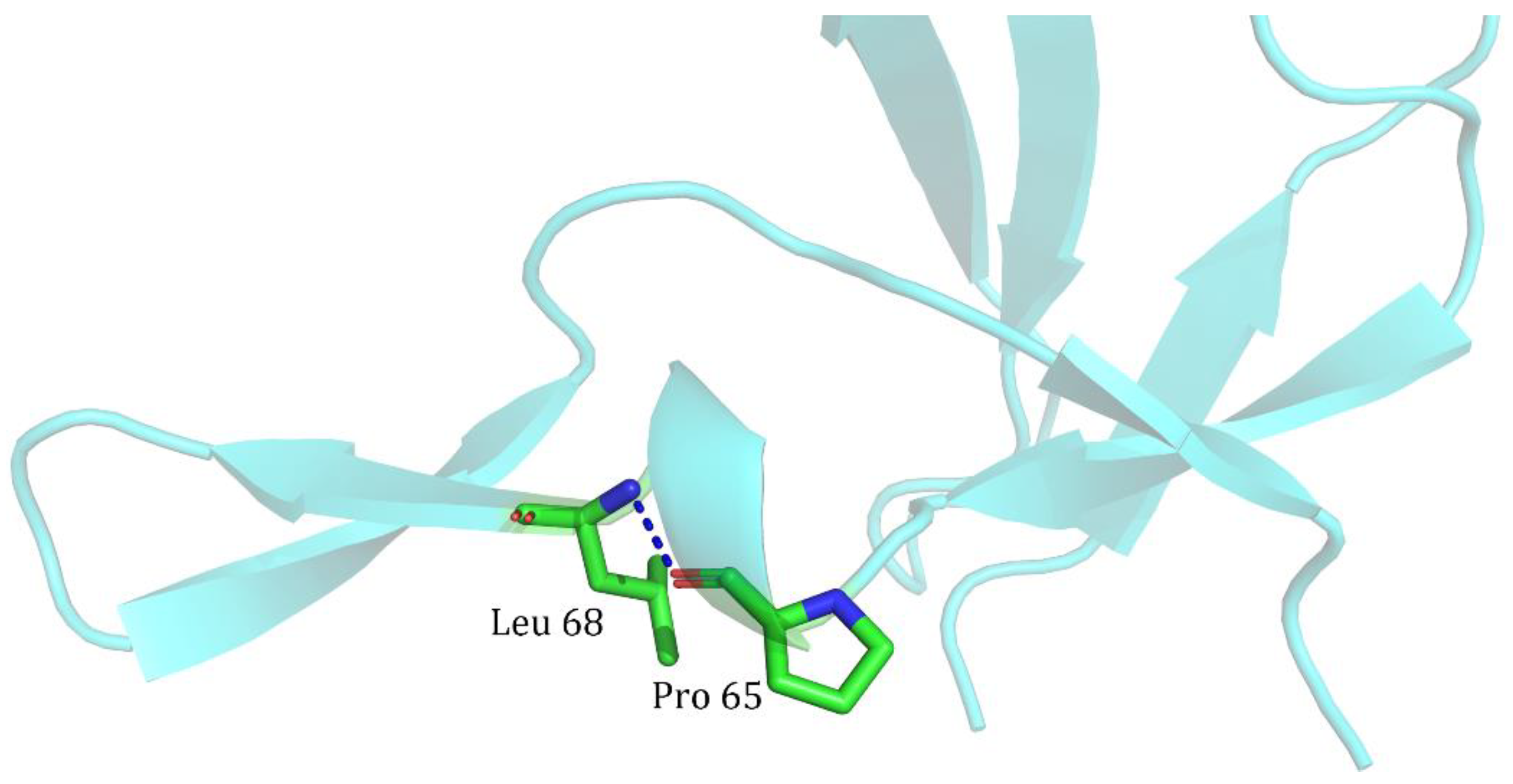
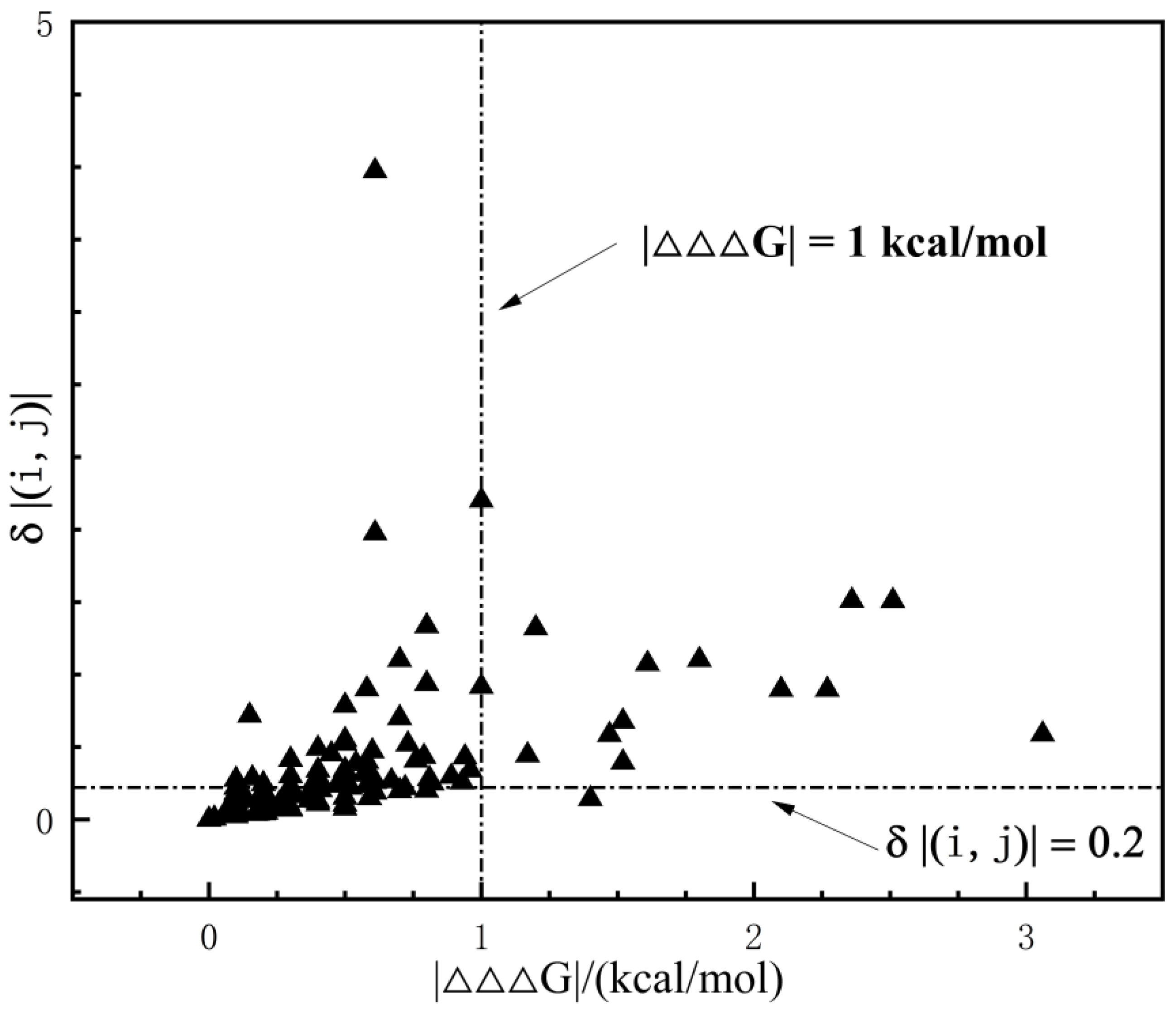
| Index | Mutation | ∆∆Gsum | ∆∆Gexp | ∆∆∆G | 3-Clique Community |
|---|---|---|---|---|---|
| 1 | L37A/A90S | −3.58 | −3.32 | 0.26 | 99, 100, 37, 36, 90, 92 |
| 2 | T62A/V66L | −2.5 | −2.4 | 0.1 | 64, 65, 66, 67, 68, 69, 102, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63 |
| 3 | I92V/V99I | −0.6 | −0.7 | −0.1 | 98, 99, 100, 101, 103, 104, 105, 92, 93 |
| 4 | I92L/V99I | −0.8 | −0.9 | −0.1 | 98, 99, 100, 101, 37, 103, 104, 36, 105, 102, 106, 92 |
| 5 | I92V/V99L | −0.7 | −0.9 | −0.2 | 98, 99, 100, 101, 102, 103, 104, 105, 106, 93, 92, 61 |
| 6 | I92L/V99L | −0.9 | −1.3 | −0.4 | 99, 100, 36, 37, 92 |
| 7 | F61A/T62A | −4.8 | −4.6 | 0.2 | 64, 65, 66, 67, 69, 60, 61, 62, 63 |
| 8 | L25A/M26A | −4.4 | −4.4 | 0 | 25, 26, 12, 13 |
| 9 | E75V/A90S | −4.21 | −3.6 | 0.61 | 91, 90, 75 |
| 10 | V23F/T33S | −3.3 | −2.72 | 0.58 | 33, 34, 23 |
| Index | Mutation | ΔΔG1 | ΔΔG2 | ΔΔGsum | ΔΔGexp | ΔΔΔG | 3-Clique Community |
|---|---|---|---|---|---|---|---|
| 1 | L37A/G79S | −1.68 | −2.66 | −4.34 | −1.83 | 2.51 | \ |
| 2 | L37A/N118D | −1.68 | −2.4 | −4.08 | −1.72 | 2.36 | \ |
| 3 | G79S/N118D | −2.66 | −2.4 | −5.06 | −2.79 | 2.27 | 79, 80, 118 |
| 4 | G79D/N118D | −2.28 | −2.4 | −4.68 | −2.58 | 2.1 | 79, 80, 118 |
| 5 | L7A/L37A | −1.58 | −1.68 | −3.26 | −1.65 | 1.61 | \ |
| 6 | L7A/G79S | −1.58 | −2.66 | −4.24 | −2.77 | 1.47 | \ |
| 7 | L37A/E75V | −1.68 | −2.31 | −3.99 | −2.47 | 1.52 | \ |
| 8 | P117L/N118D | 0.2 | −2.4 | −2.2 | −1 | 1.2 | 77, 78, 79, 117, 118, 119, 120 |
| 9 | V23F/I72V | −2.3 | −1.77 | −4.07 | −2.9 | 1.17 | \ |
| Index | Mutation | ΔΔGsum | ΔΔGexp | ΔΔΔG | 3-Clique Community |
|---|---|---|---|---|---|
| 1 | E45A/K48A | −0.55 | 0.01 | 0.56 | 33, 38–50 |
| 2 | N116A/M120A | −0.03 | 0.21 | 0.24 | 115–125 |
| 3 | R119A/Q123A | −0.4 | −0.17 | 0.23 | 84, 111, 114–125 |
| 4 | M120A/Q122A | −0.44 | −0.25 | 0.19 | 114–123 |
| 5 | T115A/R119A | −0.32 | −0.17 | 0.15 | 114–123 |
| 6 | E128A/V131A | 0.43 | 0.44 | 0.01 | 117, 127–129, 131–133 |
| 7 | N116D/R119M | 0.7 | 0.6 | −0.1 | 115–116,119–120 |
| 8 | T115A/S117A | 1.13 | 0.95 | −0.18 | 114–125 |
| 9 | A98V/T152S | −7.5 | −4.8 | 2.7 | 2–12, 88–106, 145, 148–155,161 |
| 10 | M102L/V111F | −2.51 | −2.11 | 0.4 | 102–103, 106–107, 108,111 |
| 11 | L99A/E108V | −3.3 | −3.1 | 0.2 | 92, 95–109, 111–112, 114–125, 137–138, 140–142, 145–146, 153 |
| 12 | L99G/E108V | −5.6 | −5.6 | 0 | 1–2, 5–7, 9–11, 92, 95–103, 105–109, 111–112, 114, 138, 145–146, 148–150, 153–154 |
| 13 | M6I/R96H | −4.2 | −5.08 | −0.88 | 1–3, 5–7, 9–11, 85–93, 95–106, 130, 144–145, 148–150, 152–154 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Xu, J.; Ming, D. Analysis of Correlation Effects of Double Mutations in Enzymes: A Revised Residual-Contact Network Clique Model. Int. J. Mol. Sci. 2024, 25, 9114. https://doi.org/10.3390/ijms25169114
Zhang X, Xu J, Ming D. Analysis of Correlation Effects of Double Mutations in Enzymes: A Revised Residual-Contact Network Clique Model. International Journal of Molecular Sciences. 2024; 25(16):9114. https://doi.org/10.3390/ijms25169114
Chicago/Turabian StyleZhang, Xianbo, Junpeng Xu, and Dengming Ming. 2024. "Analysis of Correlation Effects of Double Mutations in Enzymes: A Revised Residual-Contact Network Clique Model" International Journal of Molecular Sciences 25, no. 16: 9114. https://doi.org/10.3390/ijms25169114
APA StyleZhang, X., Xu, J., & Ming, D. (2024). Analysis of Correlation Effects of Double Mutations in Enzymes: A Revised Residual-Contact Network Clique Model. International Journal of Molecular Sciences, 25(16), 9114. https://doi.org/10.3390/ijms25169114






